Abstract
Signaling by GDNF through the Ret receptor tyrosine kinase is required for the normal growth and morphogenesis of the ureteric bud (UB) during kidney development. Recent studies have sought to understand the precise role of Ret signaling in this process, and the specific responses of UB cells to GDNF. Surprisingly, the requirement for Gdnf and Ret was largely relieved by removing the negative regulator Spry1, revealing unexpected functional overlap between GDNF and FGF10. However, the kidneys that developed without Gdnf/Ret and Spry1 displayed significant branching abnormalities, suggesting a unique role for GDNF in fine-tuning UB branching. GDNF/Ret signaling alters patterns of gene expression in UB tip cells, and one critical event is upregulation of the ETS transcription factors Etv4 and Etv5. Mice lacking Etv4 and Etv5 fail to develop kidneys. Thus, these genes represent key components of a regulatory network downstream of Ret. Studies of chimeric embryos in which a subset of cells lack either Ret, Etv4/5 or Spry1 have revealed an important role for this pathway in cell movement. Ret signaling, via Etv4 and Etv5, promotes competitive cell rearrangements in the nephric duct, in which the cells with the highest level of Ret signaling preferentially migrate to form the first ureteric bud tip.
Introduction
The kidney is a complex organ, whose basic components include blood vessels, thousands of nephrons (the excretory units, each consisting of a proximal tubule, loop of Henle, distal tubule and connecting tubule) and a tree-like collecting system (including cortical and medullary collecting ducts, calyces, papilla and ureter) that conveys the urine from each nephron to the bladder. The renal collecting system in some ways resembles the highly branched epithelia of other organs, such as the lung, but also has many distinct features, such as the functions and properties of the differentiated epithelial cells, specific branching patterns and some of the signals and genes that regulate its development. Development of the collecting system has been investigated extensively because of its attractive features as a basic experimental system to study morphogenesis, the many types of birth defects that can affect it in humans and the possibility of eventual repair or regeneration to treat human kidney disease. This review will describe some of the molecular and cellular mechanisms underlying the formation of the collecting duct system, focusing on recent experiments in our laboratory that have addressed these issues.
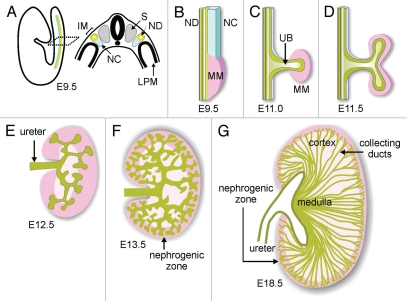
The nephrons and the collecting system both originate in the intermediate mesoderm (IM) (Fig. 1A). The dorsal portion of the IM forms a tube, called the nephric duct (ND) or Wolffian duct, whose tip elongates caudally between about E8.0 and E9.5, until it contacts and joins the cloaca. The ventral IM remains as a mesenchymal cell population called the nephrogenic cord (NC). The rostral ND and NC together generate the pronephros and mesonephros, primitive renal tubules that are transient organs in higher vertebrates and will not be further discussed here. The metanephric (or adult) kidney forms at the level of the hindlimb, through inductive interactions between the ND and a specialized region of the NC, called the metanephric mesenchyme (MM) (Fig. 1B and C). The MM contains progenitor cells that will later form the epithelia of the metanephric nephrons,1–3 and it also produces the inductive signals that promote and position the outgrowth of the ureteric bud (UB) from the ND and its subsequent branching (Fig. 1D–F).
Figure 1.
Schematic diagram of development of the renal collecting duct system. (A) Diagram illustrating location of intermediate mesoderm (IM), including the nephric duct (ND) and nephrogenic cord (NC) in the mouse embryo. S, somite; LPM, lateral plate mesoderm. (B) Formation of metanephric mesenchyme (MM). (C) Outgrowth of ureteric bud (UB). (D) First branching of the UB. (E and F) Continued UB branching and formation of the nephrogenic zone (dark pink). (G) Elongation of collecting ducts to form the medulla. Modified from reference 13.
Outgrowth of the UB is a crucial initiating event of kidney development, which depends on the prior differentiation of the MM from the IM. The UB gives rise to the epithelium of the collecting ducts, calyces, pelvis and ureter. Failure to make a UB invariably leads to renal agenesis. The correct rostral-caudal positioning of the UB is very important, as buds that form in the wrong position results in a ureter that fails to connect correctly to the bladder.4 After the UB penetrates the metanephric mesenchyme (E11.0), it begins to branch repeatedly, mostly by bifurcation at the tips.5 Other modes of branching (lateral budding and terminal trifurcation) also occur less frequently, at least in organ culture;5 the patterns of branching that occur in vivo have not been well documented in the mouse kidney, as they have been in the lung.6 Consistent with a specific role in growth and branching, the cells at the UB tip express many genes that are not expressed by cells in the trunks (i.e., the tubular portions) and vice versa.7,8
The UB branches for about 10 generations over the next 3–4 days as the kidney grows, then it enters a phase in which the UB trunks elongate extensively, followed by 1–2 more rounds of terminal branching9 (Fig. 1G). Since the UB tips continually induce new nephrons as they branch, mutations that even subtly reduce UB branching can lead to renal hypoplasia with reduced nephron number. As low nephron number is thought be a risk factor for hypertension and kidney failure,10–12 the control of UB branching morphogenesis is not only a fascinating biological problem, but a medically significant one.
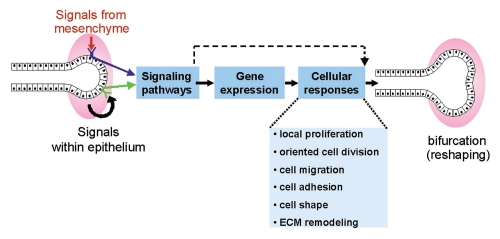
Ureteric bud branching morphogenesis is known to be largely controlled by signals from the surrounding mesenchyme, and also probably by signaling within the epithelium. But how do these signals result in changes in the size and shape of the UB epithelium? Presumably, they lead to changes in gene expression patterns in UB cells (as well as to more direct effects on the cell, e.g., via the cytoskeleton), and these in turn cause alterations in cellular behavior. Ultimately, in order for morphogenesis to occur, the properties of UB cells (e.g., cell size, shape, adhesion to other cells) or their behaviors (e.g., survival, division, migration) must be altered (Fig. 2). I will next describe some recent experiments that have sought to elucidate each of these steps (extracellular signals, gene expression changes and cellular behaviors) in the control of UB morphogenesis.
Figure 2.
Working model for some of the events underlying branching morphogenesis. Ureteric bud branching morphogenesis is controlled by signals (both promoting and inhibiting branching) from the metanephric mesenchyme and probably other signals within the epithelium. The receptors trigger intracellular signaling pathways, which lead to changes in gene expression, as well as transcription-independent changes in the cell. Some of the cellular responses that may underlie branching (i.e., changes in the shape of the epithelium) are listed.
Signals that Control Ureteric Bud Morphogenesis
Several signals from the MM, which together control UB outgrowth and branching, have been described (reviewed in ref. 13–15). GDNF is apparently the most important inducer of UB outgrowth, as the UB fails to form in most Gdnf−/− mutant embryos, as in those for Ret, the receptor tyrosine kinase (RTK) for GDNF or Gfra1, the GDNF co-receptor (reviewed in ref. 16 and 17). Similarly, mutations in many other upstream genes that reduce or eliminate Gdnf expression (e.g., Npnt, α8β1 integrin, Gdf11, Six2, Eya1, Pax2 or Hox11 paralogs) also cause UB outgrowth to fail. Conversely, most of the mutations in mice that cause multiple UBs (leading to duplex ureters and kidneys) do so by expanding rostrally the domain of Gdnf expression (Slit2, Robo2, FoxC2) or failing to negatively regulate the response to GDNF (Spry1, Bmp4).
Several other growth factors that signal through receptor tyrosine kinases also play a role in UB branching, but their importance for UB outgrowth from the ND remained unclear. Studies of the ND in organ culture show that several members of the FGF family are competent to induce ureteric budding.18 Lack of Fgf7 or Fgf10 in vivo causes reduced UB branching leading to renal hypoplasia, as does deletion of Fgfr2 in the UB epithelium.19 Similarly, VEGF-A,20 HGF and EGF signaling promote UB branching.21,22 However, none of these mutations in mice interfered with UB outgrowth from the ND, and their affects on renal branching were relatively minor. Thus, the prevailing view was that GDNF has a unique role in UB outgrowth in vivo, and that it then plays a continuing role, together with several other factors, to control the more complex growth and branching of the UB during renal development.
However, genetic studies in our lab have revealed an unexpected degree of overlap in function between Gdnf and Fgf10 (and perhaps other factors) in UB outgrowth in vivo. These experiments were designed to better understand the specific role GDNF/Ret signaling in UB branching. It had been postulated that GDNF acts as a chemoattractant, analogous to the role of Fgf10 in lung bud branching,23 to attract the two new tips of a branching UB.24 We first tried to test this model by generating mice that expressed GDNF ectopically in the UB epithelium, and not in its normal site, the MM.25 Our reasoning was that this was likely to eliminate any spatial gradients of GDNF that might normally provide positional information to the UB. These mice developed grossly normal kidneys, with typical T-shaped branching UB tips,25 showing that the mesenchymal expression of GDNF is not critical for UB branching. However, several caveats remained. First, for technical reasons we were unable to analyze the branching pattern in detail, to see if it was entirely normal. Second, while GDNF was not expressed in the MM, it remained conceivable that GDNF expressed by UB cells might bind to extracellular matrix in a way that regenerated the same (hypothetical) extracellular gradients as in wild-type kidneys.26
More recently, we have avoided these caveats by seeking conditions in which kidneys can develop in the complete absence of GDNF, and also by using better imaging techniques to visualize branching patterns. Sprouty1 (Spry1) is a negative regulator of RTK signaling whose expression is induced in the UB tips by GDNF/Ret signaling, and which acts in a negative feedback loop to modulate GDNF/Ret signaling.27 It was previously shown that heterozygosity for Spry1 could rescue the renal hypoplasia that occurs in Gdnf+/− heterozygotes,28 but double homozygotes had not been examined. While Gdnf−/− mice rarely make a ureter or a kidney (and the kidneys that sometimes develop are extremely rudimentary), surprisingly, we found that the absence of the negative regulator Spry1 restored ureter and kidney development in >90% of Gdnf−/−;Spry1−/− mice (Fig. 3A–C).29 The kidneys were somewhat reduced in size and nephron number (∼50–70% of normal), but quite normal in shape and histology. To exclude the possibility that another Ret ligand, such as neurturin,30 was able to substitute for GDNF, we also examined Ret−/−;Spry1−/− mice and these had kidneys similar to those of Gdnf−/−;Spry1−/− double mutants. These results suggested that a major role of GDNF/Ret signaling is to “balance” the negative regulation provided by Spry1 and when both are absent, other endogenous factors can replace GDNF as the inducer of UB outgrowth from the ND and continued branching.
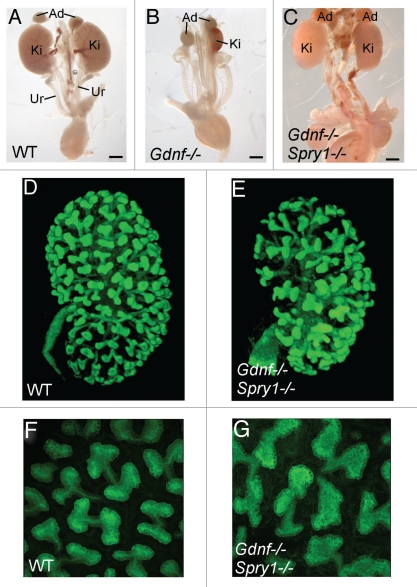
Figure 3.
Loss of Spry1 rescues kidney development in Gdnf−/− mice, but with an abnormal UB branching pattern. (A–C) excretory systems dissected from newborn mice of the indicated genotypes. Ad, adrenal; Ki, kidney; Ur, ureter. (D–G) 3D reconstruction of the branched UB in E15.5 wild-type and double-mutant kidneys (also carrying a Hoxb7/myrVenus transgene to allow the UB to be imaged). While the wild-type UB (D and F) shows a reiterative pattern of orthogonal bifurcation (F), with tips growing at right angles to their predecessors, the UB tips in the double mutant (G) instead display a variety of abnormal tip shapes and branching patterns. The 3D images were generated from confocal Z-stacks, using Volocity (D and E) or ImageJ (F and G). Modified from reference 29.
These findings left several questions: First, what other signals can replace GDNF (but only when Spry1 is absent)? Fgf10 was a strong candidate, as it is expressed in the MM, similarly to GDNF and is able to induce ectopic buds from cultured NDs, in the absence of any GDNF.18,29 When Fgf10 was eliminated in Gdnf−/−;Spry1−/− mice, UB outgrowth failed completely in the triple mutants. Thus, Fgf10 must be responsible, at least in part, for promoting and correctly positioning, UB outgrowth in Gdnf−/−;Spry1−/− mice. It is not yet clear if other factors (e.g., HGF, EGFs, VEGF-A) are also required under these conditions (to a greater extent than when Gdnf and Spry1 are present).
A second question was whether the specific pattern of branching was affected in Gdnf−/−;Spry1−/− or Ret−/−;Spry1−/− mice. Examining the UB at various stages of kidney development, with the aid of fluorescent protein-encoding transgenes31,32 and 3-D image-rendering, revealed that the UB branched extensively, but in a distinctly abnormal pattern. In a normal kidney at E15.5, the UB tips are evenly spaced, uniform in size and mostly branching in the “orthogonal bifurcation” pattern,6 in which each new pair of branches forms at a ∼90 degree angle from the parental branch (Fig. 3D and F). In contrast, the double mutant UB tips were irregular in size, shape and branch angle (Fig. 3E and G). This was not due to simply the absence of Spry1 (in Spry1−/−, the tips were swollen but otherwise appeared normally patterned; not shown) and instead seems to reflect a specific role of Gdnf in branch patterning.
The mechanism of this effect remains unclear. It is possible that gradients of GDNF normally pattern the UB tips, but if so, the GDNF protein distribution must be regulated by some mechanism other than gene expression,26 as GDNF mRNA is very diffusely expressed in the MM.16 Alternatively, GDNF signaling to the UB tips may stimulate a unique program of gene expression and cellular behavior that leads to the normal pattern of UB morphogenesis. A key question now is whether GDNF provides positional information in this context. Answering it will require better methods to visualize the normal spatial distribution of GDNF and its interaction with the ECM, as well as methods to subtly alter its spatial expression pattern in vivo.
Signaling Pathways and Gene Expression Changes Downstream of Ret
How does GDNF/Ret signaling induce UB growth and branching? Ret activates a number of intracellular signaling pathways, including Ras-Erk MAP kinase, PLCγ/Ca+ and PI3K-Akt pathways, as well as others.33 The importance of these three pathways has been studied using specific chemical inhibitors in organ cultures, or by making knock-in mice with mutations in Ret tyrosine residues whose phosphorylation upon GDNF signaling activates different downstream pathways. Inhibiting PI3K entirely blocks UB growth and branching,34 while, conversely, deleting PTEN (the PI3K phosphatase) causes irregular UB branching.35 Inhibiting MEK (Erk Map kinase-kinase) slows UB branching with a smaller effect on overall elongation.5,36 A substitution for Ret tyrosine 1062, which is linked to both the PI3K and Erk pathways (as well as several others), results in renal agenesis.37,38 Blocking the activation of PLCγ by mutating Ret tyrosine 1015 has complex effects, leading to multiple ureters but also smaller kidneys,38 apparently because this pathway is required for normal UB branching, but also for normal Spry1 expression. For the most part, it is not clear how these signaling pathways are coupled to the alterations in cellular properties and behaviors that underlie morphogenesis.
Ret signaling also leads to changes in gene expression in the UB,27,39,40 but compared to the large number of upstream genes known to regulate Gdnf or Ret expression,41,42 very few downstream genes were known. To address this question, we performed a microarray screen to identify genes whose expression in isolated ureteric buds (iUBs) was altered by culture in the presence/absence of GDNF.43 In addition to the three genes previously known to be upregulated by GDNF (Wnt11, Spry1, Ret), the screen identified many novel genes upregulated by GDNF, including the chemokine receptor Cxcr4, the cytokine Crlf1, the signaling inhibitors Dusp6 and Spred2 and the transcription factors Myb, Etv4 and Etv5. Crlf1 is believed to have a role in the induction of nephrogenesis by the UB tips.8 Cxcr4, the receptor for Cxcl12/Sdf-1, is important for cell migration in several developing systems and in the immune system.44,45 In the kidney, it is important for development of the glomerular tuft and renal vasculature,46 and inhibitor studies suggest a role in UB branching and nephrogenesis,47 but its role in UB morphogenesis in vivo is currently being investigated. The possible roles of Myb, Spred2 and Dusp6 in UB branching remain to be determined. The most important finding resulting from this screen was the crucial role of Etv4 and Etv5 in kidney development, downstream of Ret (and probably other RTKs).43
Etv4 and Etv5 encode two very closely related transcription factors of the ETS family, which are involved in neuronal development, spermatogenesis and limb development, where their expression is induced by GDNF, FGFs or HGF.48–51 In developing kidneys, Etv4 and Etv5 are both expressed strongly in the UB tips and more weakly in the MM and nascent nephrons; their expression in the UB requires normal levels of Ret signaling, via the PI3K pathway, as it is abolished in mice with a hypomorphic Ret mutation, or in wild-type kidneys cultured with a PI3K-inhibitor.43 Mice lacking either Etv4 or Etv5 had only rare defects in kidney development, but Etv4/Etv5 double mutants were severely affected, establishing a redundant role for these two genes. Etv4−/−;Etv5+/− mutants had either renal agenesis or hypodysplasia, while Etv4−/−;Etv5−/− newborn mice lacked kidneys entirely, although they often had ureters. The hypoplastic kidneys in Etv4−/−;Etv5+/− mutants had severe branching defects and UBs isolated from these kidneys branched very poorly in Matrigel cultures compared to controls,43 suggesting a cell-autonomous role in the UB. As Etv4 and Etv5 expression requires normal Ret signaling levels, their lack of expression likely contributes significantly to the failure of kidney development in Ret, Gdnf and Gfrα1 mutant mice.
Unlike a lack of Gdnf or Ret, the requirement for Etv4 and/or Etv5 in kidney development could not be overcome by removing Spry1.43 This suggests that Etv4 and Etv5 mediate the effects of other growth factors besides GDNF and while these individual growth factors are to some degree redundant, they all signal through Etv4 and Etv5. Thus, the two ETS transcription factors are key components of a gene network downstream of receptor tyrosine kinases that promotes and controls renal branching morphogenesis (Fig. 4).
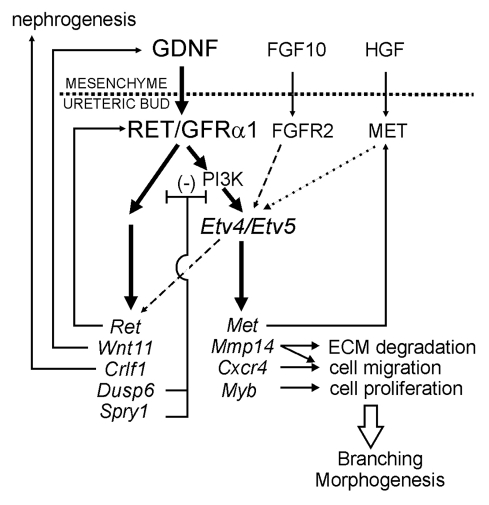
Figure 4.
Model for role of Etv4 and Etv5 in a gene network controlling ureteric bud branching morphogenesis. GDNF from the metanephric mesenchyme signals to the UB via GFRα1 and RET, thus upregulating a set of genes (bold arrows), including Etv4 and Etv5. Etv4 and Etv5 expression requires PI3-kinase activity, while the signaling pathways regulating other downstream genes are unknown. FGF10 signaling normally has a lesser role, if any, in regulating Etv4/Etv5 expression in the UB (dashed arrow). Several of the genes downstream of Ret (e.g., Met, Mmp14, Cxcr4, Myb) are apparently regulated via Etv4 and Etv5, as their expression is extinguished in Etv4−/−;Etv5+/− kidneys. Other GDNF-induced genes (e.g., Ret, Wnt11, Crlf1, Dusp6, Spry1) are relatively insensitive to Etv4/Etv5 levels (dashed arrow) and are likely regulated via other transcription factors. Collectively, these downstream genes compose part of a regulatory network that promotes UB branching morphogenesis and other aspects of kidney development. In three positive feedback loops, Ret signaling positively regulates its own expression, Wnt11 upregulates Gdnf,39 and Met encodes the receptor for HGF, which may also upregulate Etv4/Etv5 (dotted arrow). Spry1 and Dusp6 are inhibitors of RTK signaling that participate in negative feedback loops; Dusp6 is a MAP kinase phosphatase, while the signaling pathway(s) inhibited by Sprouty1 is unclear. Crlf1 may participate in the induction of nephrogenesis. Mmp14, Cxcr4 and Myb are likely to contribute to cellular processes important for UB morphogenesis. Reproduced from reference 43.
To identify some of the target genes of Etv4 and Etv5 in the kidney, we first tested several of the genes that are regulated by GDNF/Ret signaling. Expression of Cxcr4 and Myb was greatly reduced in Etv4−/−;Etv5+/− mutant kidneys (as in Ret-hypomorphic kidneys), suggesting that these genes are normally upregulated by GDNF/Ret signaling via Etv4 and Etv5. However, Ret, Crlf1, Dusp6 and Wnt11 were expressed at normal levels in Etv4−/−; Etv5+/− kidneys, indicating that their expression is relatively insensitive to Etv4/Etv5 (or at least that a single Etv5 allele is sufficient for their normal expression).
We also examined Met and Mmp14, two genes known to regulated by Etv4 in other cell types and found that their expression requires Etv4/5 in the UB. Upregulation of Met (encoding the receptor tyrosine kinase for HGF) by Ret signaling is probably important to render the UB responsive to HGF, and thus to promote further growth and branching.52 MMP14 (MT1-MMP) is a membrane-type matrix metalloproteinase, which has been implicated in UB branching in culture,53,54 as well as by analysis of Mmp14−/− mice (Zent R and Riggins K, personal communication). MMPs may promote branching morphogenesis in several ways: through their enzymatic activity, they may remodel the extracellular matrix (ECM) to allow UB growth and also release bound growth factors.55 MMP14 may also function via an autocrine signaling mechanism that promotes cell migration in vitro.56
While these candidate gene studies yielded some insight into the possible effector genes downstream of Etv4/Etv5, the full set of target genes remains to be defined by more comprehensive approaches. It is also important to address the phenotypic effects of Etv4/5 expression at the cellular level, a issue that has been investigated by some of the experiments described below.
The identification of many genes whose expression is altered by Ret signaling in the UB and of Etv4 and Etv5 as key transcription factors in this regulatory network, provides a large set of candidate genes that may play some role in UB morphogenesis. The most straightforward genetic approach to examine their functions (i.e., examining the phenotypes of individual knockout mice) is expensive and time consuming and better methods are needed to screen large numbers of genes for effects on kidney development. Hopefully, a combination of organ culture systems (whole kidneys or isolated UBs) and improved methods to manipulate gene expression in these systems (such as viral vectors and RNA interference) can provide a higher throughput assay for gene function in the developing kidney.
Cellular Events in UB Morphogenesis
Signals to the UB, and the resulting changes in intracellular signaling and gene expression patterns, must ultimately be translated into changes in the behavior of UB epithelial cells in order for the UB to grow and branch in specific ways. Some of the cellular events likely to be relevant include cell proliferation, oriented cell division, changes in cell shape, changes in adhesive properties, cell migration and remodeling of the ECM (Fig. 2). While several cell culture models for UB branching are available, they do not appear to accurately reflect developmental events in vivo. Explanted kidneys or isolated UBs developing in organ culture57 provide a more authentic view of the cellular events that occur in vivo. Several additional methods and experimental “tools” have been developed that are useful for this purpose.
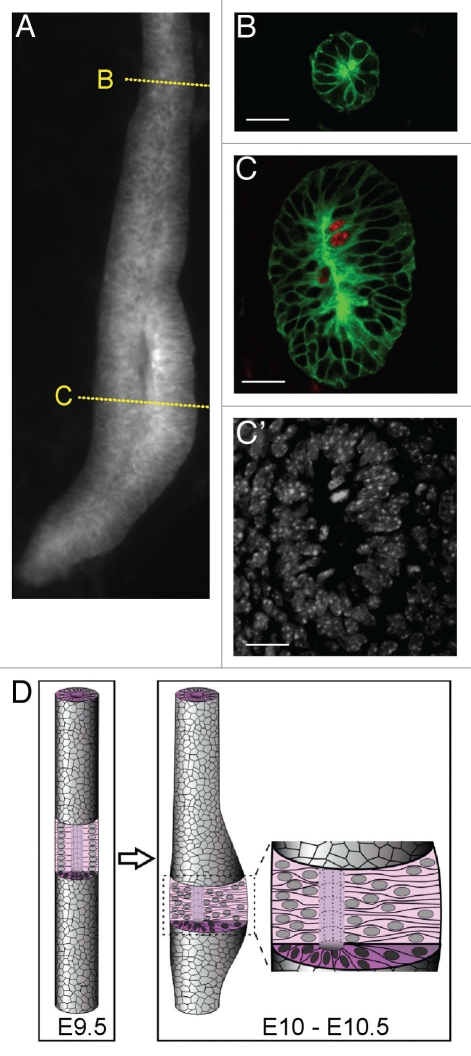
One such tool we have recently described is a mouse strain that expresses a membrane-bound form of yellow fluorescent protein myrVenus in the ND and UB under the Hoxb7 promoter, allowing individual cell outlines to be seen clearly in live specimens with a confocal microscope.32 This allowed us to observe a change in cell shape in the caudal ND preceding ureteric budding, which to our knowledge had not been previously reported. At E10.0, the caudal portion of the ND thickens and forms a pseudostratified epithelium58 (Fig. 5), i.e., one in which the nuclei lie at different levels along the apical-basal axis, but every cell contacts the basement membrane. Locally pseudostratified regions were known to occur in several types of epithelia before they produce outgrowths, including the mammary line,59 otic placodes,60 and the liver and thyroid buds.61,62 The significance of the pseudostratified domain of the ND remains unclear, as do the signals that induce its formation. We speculate that the high density of nuclei generated in the pre-budding ND epithelium during this process might permit rapid expansion of a localized region of the ND to form the UB. An examination of the literature revealed that the ND apparently fails to become pseudostratified in Osr1 mutant mice (see Fig. 7 in ref. 63). Since Osr1 is expressed in the IM and MM but not the ND, this suggests the possibility that a signal from the MM causes the caudal ND to undergo this alteration. However, GDNF/Ret signaling is not required, as Ret−/− NDs become pseudostratified similarly to wild-type.58 By E14.5, after the bud has grown out and branched a few times, the UB reverts to a simple cuboidal epithelium,54,58 while continuing to branch. Therefore both pseudostratified and simple epithelia can undergo similar branching events.
Figure 5.
The nephric duct forms a (transiently) pseudostratified epithelium before ureteric bud formation. (A) Whole mount image of wild-type Nephric duct carrying Hoxb7/myr-Venus transgene at E10.0. Yellow lines indicate approximate planes of the sections at right, which show rostral ND in (B) and pseudostratified caudal ND in (C). (B and C) were stained with anti-GFP to detect myr-Venus (green) and anti-pH3 (red). C′, Hoechst nuclear stain of the section in (C). (D) Schematic diagram of the formation of pseudostratified epithelium preceding ureteric budding. The pseudostratified epithelium persists during early UB branching, but reverts to a simple epithelium before E14.5.58 Scale bars 20 µM. Modified from reference 58.
A valuable method we have developed to study cell behaviors during kidney development is the generation of chimeras using embryonic stem (ES) cells carrying transgenes driven by the Hoxb7 promoter, which express fluorescent reporter proteins (e.g., GFP) specifically in the ND/UB lineage.58,64 When injected into blastocysts of a different strain, the ES cells contribute to all tissues in the resulting chimeric embryos, but because the GFP is only expressed in ND and UB cells, it is possible to clearly visualize the ND/UB cells without interference by the overlying mesenchyme. We first examined the development of chimeric kidneys made with Hoxb7/GFP cells that were wild-type (except for the transgene), which were injected into wild-type (unmarked, GFP-negative) embryos.64 The behaviors of the GFP+ and GFP− cells showed that there was extensive cell movement in the UB epithelium, although whether it was random or directed was not apparent. They also showed that some GFP+ UB tip cells divided to yield both GFP+ tip cells (self-renewal) and GFP+ trunk cells (differentiation). This suggested that tip cells are the bipotential progenitors of new tips and trunks,64 a model supported by recent inducible genetic fate mapping studies (Paul Riccio and F. Costantini, unpublished data).
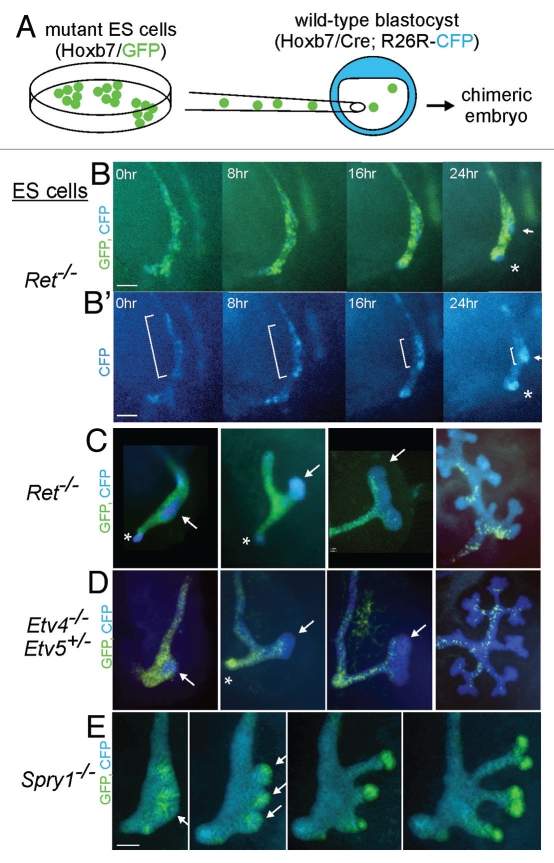
We next generated chimeras using ES cells that were Ret−/− and carried Hoxb7/GFP, allowing us to examine the specific cellular defects caused by lack of Ret. In the early UB (E11.0), the tip was devoid of mutant (GFP+) cells, while the trunk contained both wild-type and mutant cells; as the UB branched, the mutant cells failed to contribute to any of the new tips, and were soon “diluted out” by the wild-type cells.58,64 To examine the earlier events that lead to the exclusion of Ret−/− cells from the UB tips, we analyzed additional chimeras in which the host embryo expressed cyan fluorescent protein (CFP) in the ND and UB, while the ES-derived (Ret−/−) cells expressed GFP (Fig. 6A).58 This allowed us to perform time-lapse imaging of bud formation and outgrowth. We found that at E10.0 the mutant and wild type cells were randomly arranged in the ND, but before budding, wild type cells moved together into a cluster in the dorsal ND (E10.5, Fig. 6B and B'), which next emerged as the tip of the UB (E11.0, Fig. 6C), while mutant cells were largely excluded from this region. We called this region the “primary UB tip domain” because it gives rise to the first UB tip, as well as to later tips.58 Thus, the reason that Ret−/− cells fail to contribute to the primary UB tip domain is apparently a defect in cell movement in the ND. There was no difference in proliferation or survival between the Ret−/− and wild-type cells in the ND.58
Figure 6.
Time-lapse chimera analysis of the behavior of cells with mutations affecting Ret signaling. (A) ES cells were derived from mouse embryos with mutations in Ret, Etv4 and Etv5 or Spry1 and carrying a Hoxb7/GFP transgene expressed in the ND and UB lineage. The ES cells were injected into blastocysts carrying Hoxb7/Cre68 and R26R-CFP, a Cre-reporter strain that expresses CFP in any cells expressing Cre and their descendants.69 Thus, in the ND and UB of the chimeric embryos, mutant cells (derived from ES cells) appear “green” and wild-type cells (derived from host embryo) appear “blue.” Images from left to right show successive stages of ND development (B and B′) and UB outgrowth and branching (C–E). (B and B′) time-lapse imaging of development of a Ret−/−↔wild-type chimera starting at ∼E10.0. (B) shows CFP and GFP images merged, revealing interspersion of wild-type (CFP+) and Ret−/− (GFP+) cells in the nephric duct at 0 hr, but enrichment of wild-type cells at the primary UB tip domain (arrow) and the CND (*) by 24 hr. (B′) shows only the CFP channel, revealing that wild-type cells undergo rearrangements to form the UB tip domain (brackets) and the CND (*). (C) additional Ret−/−↔wild-type chimeras at successive stages (starting at E10.5) of UB formation, outgrowth and branching. When the UB grows out and branches, the Ret−/− cells contribute to the trunks, but not to the UB tips. (D) Etv4−/−, Etv5+/− cells behave virtually identically to Ret−/− cells in the UB, but they can contribute to the CND (*). (E) Spry1−/− cells behave oppositely to Ret−/− or Etv4−/−, Etv5+/− cells, preferentially contributing to the primary UB tip domain and then to the UB tips. Note that formation of multiple UBs is a common property of Spry1−/− embryos.27 Modified from references 58 and 67.
Further studies with chimeric embryos made using ES cells and host embryos with different levels of Ret signaling revealed that ND cells can compete, based on their level of signaling, to contribute to the UB tip domain. For example, cells lacking Spry1 (which have elevated signaling), prevailed over wild-type cells to form the UB tip domain (Fig. 6E). This competition is reminiscent of that observed in the developing fly respiratory epithelium, where cells compete based on FGF-receptor signaling levels to form the tip of the air sac or the tracheal branch.65,66 In normal, non-chimeric mouse embryos, all ND cells express the Ret gene at similar levels, but the level of Ret signaling (as reflected by di-phosphorylated Erk MAP kinase, a downstream indicator of Ret signaling) was very heterogeneous. Furthermore, time-lapse imaging showed that normal ND cells can move independently of their neighbors during bud formation. Together, the data suggest a model in which a subset of ND cells have elevated Ret signaling and these cells migrate past their neighbors to form the first UB tip.58 Why might similar competitive cell behaviors occur in branching epithelia in both flies and mammals? Perhaps competition limits the number of cells involved in branch formation or budding, thus forming a small, discrete bud rather than a massive swelling.
As several of the genes we had identified downstream of Ret and Etv4/5 are implicated in cell migration (in particular, Cxcr4, Met and Mmp14), we asked whether Etv4/Etv5 mutant cells would also show a defect in the chimeric assay.67 Indeed, Etv4−/−;Etv5+/− cells behaved identically to Ret−/− cells, failing to contribute to the UB tip domain in chimeras with wild-type cells (Fig. 6D). Etv4−/−;Etv5−/− cells were even more defective, contributing mainly to the rostral ND, less often to the caudal ND and almost never to the tip or trunk of the UB.67 Thus, the effects of GDNF/Ret signaling on ND cell movement appear to be mediated by Etv4 and Etv5; and the more severe defect in Etv4−/−; Etv5−/− cells than Ret−/− cells supports the model that Etv4/5 also mediate additional signals, most likely from FGFRs or other RTKs.
While this work illustrates the power of chimera analysis to reveal the roles of specific genes in cell behaviors, it raises as many questions as it answers. Why are signaling levels heterogeneous among ND cells? What guides the movement of some ND cells towards the site where the UB tip domain forms—chemoattraction by GDNF or some other guidance mechanism that is activated downstream of Ret signaling? Does this process involve cell sorting due to differences in cell adhesion? Do similar cell rearrangements play a role in continued UB branching, or is this a phenomenon specific to the initial bud formation from the ND? A variety of experimental approaches will be required to address these issues.
Questions and Answers
Dr. David Ornitz, Alumni Endowed Professor of Developmental Biology, Washington University School of Medicine: What do you make of the GDNF expression pattern?
Dr. Frank Costantini, Professor of Genetics and Development, Columbia University Medical Center: The distribution of GDNF mRNA appears very diffuse throughout the metanephric mesenchyme. So if there is any truth to the idea that GDNF is localized and can pattern the UB by chemoattraction, as FGF10 is thought to do in lung development, it has to be occurring at the protein level, perhaps by its ability to bind to extracellular matrix molecules such as heparan sulfate proteoglycans.
Dr. Ornitz: Do you think the regulation of epithelial branching patterning is intrinsic to the mesenchyme?
Dr. Costantini: Well, we know that if you culture the ureteric bud in Matrigel, without mesenchyme (Sanjay Nigam's work, primarily), it will branch with a certain pattern, but the pattern doesn't really look like that in a normal kidney.
Dr. Ornitz: Does the mesenchyme pattern epithelial branching or is the branching intrinsic to the epithelium.
Dr. Costantini: The epithelium has an intrinsic ability to branch as shown by its ability to branch in Matrigel in the presence of GDNF and other soluble factors, but the pattern seems to be different, so I think that the mesenchyme fine-tunes the pattern of branching. While this depends on GDNF signaling through Ret, whether it does it through chemoattraction by GDNF or by another mechanism, is not yet clear.
Dr. Jianghui Hou, Assistant Professor of Medicine, Washington University School of Medicine: From a cell biology point of view, tubular cell proliferation or tubular elongation requires the mitotic spindles of proliferating cells to align in the same direction as the elongation angle. Defects in aligning the mitotic spindle may affect tubular branching. Have you found any gene related to this process downstream of the Ret signaling pathway that might cause tubular branching defects?
Dr. Costantini: That is a very good question. In later kidney development it is established that mitotic orientation is involved in the elongation of the collecting duct, so I think it could have a role in branching, but we don't know.
Dr. Hila Barak, Postdoctoral Fellow in Developmental Biology, Washington University School of Medicine: Is it known whether cells in the stroma play a role in ureteric bud branching?
Dr. Costantini: We know the stoma is important for normal ureteric bud branching, based mainly on studies with Foxd1 (Bf2) mutant mice, but to my knowledge the specific factors that are made by the stroma are not really known.
Dr. Barak: In your presentation you showed us a list of genes that have the most elevated expression in the ureteric buds which were cultured with GDNF. I saw that this list includes genes that have a role in proliferation. Have you examined the role of those genes in the ureteric bud?
Dr. Costantini: If you go further down on the list of genes upregulated by GDNF (than what I showed in my slide) there are a lot of genes that are proliferation-associated. This is consistent with other evidence that one of the effects of GDNF is to promote ureteric bud cell proliferation.
Dr. Robert Heuckeroth, Associate Professor of Pediatrics and Developmental Biology, Washington University School of Medicine: I was struck by these beautiful positive and negative feedback loops. What is known about what turns this process off? Why does the kidney not keep growing forever and what happens in postnatal life? As a child gets bigger, does the ureteric bud continue to branch or do ducts just elongate?
Dr. Costantini: One paper that addresses this question was from Larry Patterson in Cincinnati.70 They looked at markers of nephrogenesis and ureteric bud branching and found that kidney development continues for three days after birth in the mouse. Then the nephrogenic mesenchyme, which produces GDNF, disappears and markers of Ret signaling in the UB, like Wnt11, are also lost.
So it appears that branching and nephron induction continue a little bit after birth and then they stop and you mainly get duct elongation and nephron maturation thereafter.
Dr. Heuckeroth: What is the underlying mechanism that stops branching? Dr. Costantini: I don't think that is known, but the result just mentioned suggests that it may be the loss of GDNF-producing mesenchyme.
Dr. Raphael Kopan, Professor of Medicine and Developmental Biology Washington University School of Medicine: Mouse cells heterozygous for Ret are behaving like wild type. Can I then assume that human conditions are due to homozygous recessive mutations?
Dr. Costantini: In humans, heterozygous RET loss of function mutations cause Hirschsprung's disease, a developmental defect of the enteric nervous system and sometimes kidney defects. In contrast, Ret heterozygous mice are essentially normal and only homozygotes have both severe kidney and enteric nervous system defects.
Dr. Kopan: So in humans, heterozygotes have a phenotype and in mice, they do not. Do you think you could expose this if you were to lower Etv4 by one copy, then also eliminate one copy of Ret.
Dr. Costantini: We did that experiment and didn't see any genetic interaction. But when Cristina Cebrian in the lab combined a Gdnf heterozygote (which has an incompletely penetrant kidney phenotype) with either Etv4 homozygosity or Etv4/Etv5 double heterozygosity (neither of which had much of an effect alone), we did see a big effect.
Dr. Kopan: The Fgf10 rescue was incomplete. Do you think that if you were to delete Spry1 only in the ureteric bud you would repair the effect. How do you feel about it? Have you tried recombination of normal mesenchyme?
Dr. Costantini: The problem with that is when you do the recombination, in the best of circumstances, wild type-wild type, you don't really get a very nice branching pattern, so that would not be a good way to look at it. Combining the Spry1 ureteric bud specific knockout and a Gdnf knockout could be done, but we haven't. So you are suggesting that the Spry1 might have an effect in mesenchyme? Albert Basson and Jonathan Licht did a UB-specific Spry1 knockout as well as the complete Spry1 knockout and the phenotypes were the same. So based on that, there is no evidence as yet for a role of Spry1 in mesenchyme.
Dr. Susan Kiefer, Assistant Professor of Medicine, St. Louis University School of Medicine: I was struck by the behavior of ETV4 and 5 cells in chimeric experiments being identical to Ret in the ureteric bud region, but it didn't look like it was identical in the common nephric duct.
Dr. Costantini: I didn't go into that, but that is exactly right. In the Ret chimera, the Ret−/− cells don't contribute to the common nephric duct. But the Etv4−/−, Etv5+/− cells do. So although that process is Ret dependent, it is less dependent on Etv4 and Etv5 (“less” because the cells still have one Etv5 allele).
Dr. Kiefer: Do you think there is going to be a totally different set of Ret dependent genes in the common nephric duct region?
Dr. Costantini: Even in the kidney, there are some genes that are Ret dependent but expressed normally in Etv4−/−, Etv5+/− mutant kidneys. So the genes regulated by Ret in the common nephric duct might be some of these genes or could be different genes than those regulated by Ret in the kidney.
Dr. Sanjay Jain, Assistant Professor of Medicine and Immunology and Pathology, Washington University School of Medicine: Regarding Spry1 and Ret, it seems that in the double knock out there is rescue of the individual phenotypes, at least morphologically. Have you seen any differentiation problems in the branching tips.
Dr. Costantini: As I showed, we looked at several ureteric bud tip markers and all we looked at (Ret, Wnt11, Etv4) were normal. We looked at only one trunk marker, Wnt7b and it was also normal.
Dr. Jain: What I mean is in differentiation markers of the collecting duct.
Dr. Costantini: We did not look at markers of the differentiated collecting ducts.
Dr. Marc Hammerman, Chromalloy Professor of Medicine, Washington University School of Medicine: Do you know what renal function is like in those animals?
Dr. Costantini: Well they don't live, because the removal of Spry1 doesn't rescue all of the other problems caused by loss of Ret or Gdnf, so they die within a day of birth.
Acknowledgements
I would like to thank the past and present members of my lab who have carried out the experiments described here: Naoya Asai, Cristina Cebrian, Xuan Chi, Richard Kuo, Satu Kuure, Benson Lu, Odyssé Michos, Paul Riccio, Reena Shakya, Shankar Srinivas, Tomoko Watanabe, Linda Williams and Zaiqi Wu. I also thank our many collaborators (too numerous to list here) who provided mice, expert advice and experimental contributions and Ellen Freed for helpful comments on the manuscript. This work has been supported by grants from the NIH (DK055388, DK075578, DK082715, DK083289) and fellowships from the National Kidney Foundation, American Heart Association, Sigrid Juselius Foundation and Finnish Culture Foundation.
Edited transcripts of research conferences sponsored by Organogenesis and the Washington University George M. O'Brien Center for Kidney Disease Research (P30 DK079333) are published in Organogenesis. These conferences cover organogenesis in all multicellular organisms including research into tissue engineering, artificial organs and organ substitutes and are participated in by faculty at Washington University School of Medicine, St. Louis, MO.
Footnotes
Previously published online: www.landesbioscience.com/journals/organogenesis/article/12680
References
- 1.Saxen L. Organogenesis of the Kidney. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- 2.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, et al. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetani N, Bouchard M. Plumbing in the embryo: developmental defects of the urinary tracts. Clin Genet. 2009;75:307–317. doi: 10.1111/j.1399-0004.2009.01175.x. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Costantini F. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol. 2004;271:98–108. doi: 10.1016/j.ydbio.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caruana G, Cullen-McEwen L, Nelson AL, Kostoulias X, Woods K, Gardiner B, et al. Spatial gene expression in the T-stage mouse metanephros. Gene Expr Patterns. 2006;6:807–825. doi: 10.1016/j.modgep.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, et al. Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol. 2005;16:1993–2002. doi: 10.1681/ASN.2004121127. [DOI] [PubMed] [Google Scholar]
- 9.Cebrian C, Borodo K, Charles N, Herzlinger DA. Morphometric index of the developing murine kidney. Dev Dyn. 2004;231:601–608. doi: 10.1002/dvdy.20143. [DOI] [PubMed] [Google Scholar]
- 10.Poladia DP, Kish K, Kutay B, Bauer J, Baum M, Bates CM. Link between reduced nephron number and hypertension: studies in a mutant mouse model. Pediatr Res. 2006;59:489–493. doi: 10.1203/01.pdr.0000202764.02295.45. [DOI] [PubMed] [Google Scholar]
- 11.Hoy WE, Bertram JF, Denton RD, Zimanyi M, Samuel T, Hughson MD. Nephron number, glomerular volume, renal disease and hypertension. Curr Opin Nephrol Hypertens. 2008;17:258–265. doi: 10.1097/MNH.0b013e3282f9b1a5. [DOI] [PubMed] [Google Scholar]
- 12.Kett MM, Bertram JF. Nephron endowment and blood pressure: what do we really know? Curr Hypertens Rep. 2004;6:133–139. doi: 10.1007/s11906-004-0089-2. [DOI] [PubMed] [Google Scholar]
- 13.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costantini F. Renal branching morphogenesis: concepts, questions and recent advances. Differentiation. 2006;74:402–421. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- 15.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 16.Costantini F, Shakya R. Gdnf/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–127. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- 17.Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ. Renal aplasia in humans is associated with RET mutations. Am J Hum Genet. 2008;82:344–351. doi: 10.1016/j.ajhg.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeshima A, Sakurai H, Choi Y, Kitamura S, Vaughn DA, Tee JB, et al. Glial cell-derived neurotrophic factor independent ureteric bud outgrowth from the Wolffian duct. J Am Soc Nephrol. 2007;18:3147–3155. doi: 10.1681/ASN.2007060642. [DOI] [PubMed] [Google Scholar]
- 19.Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Pediatr Nephrol. 2007;22:343–349. doi: 10.1007/s00467-006-0239-7. [DOI] [PubMed] [Google Scholar]
- 20.Tufro A, Teichman J, Banu N, Villegas G. Crosstalk between VEGF-A/VEGFR2 and GDNF/RET signaling pathways. Biochem Biophys Res Commun. 2007;358:410–416. doi: 10.1016/j.bbrc.2007.04.146. [DOI] [PubMed] [Google Scholar]
- 21.Davies JA, Fisher CE. Genes and proteins in renal development. Exp Nephrol. 2002;10:102–113. doi: 10.1159/000049905. [DOI] [PubMed] [Google Scholar]
- 22.Ishibe S, Karihaloo A, Ma H, Zhang J, Marlier A, Mitobe M, et al. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development. 2009;136:337–345. doi: 10.1242/dev.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 24.Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- 25.Shakya R, Jho EH, Kotka P, Wu Z, Kholodilov N, Burke R, et al. The role of Gdnf in patterning the excretory system. Dev Biol. 2005;283:70–84. doi: 10.1016/j.ydbio.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Basson MA, Watson-Johnson J, Shakya R, Akbulut S, Hyink D, Costantini FD, et al. Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol. 2006;299:466–477. doi: 10.1016/j.ydbio.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 29.Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, et al. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies JA, Millar CB, Johnson EM, Jr, Milbrandt J. Neurturin: an autocrine regulator of renal collecting duct development. Dev Genet. 1999;24:284–292. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<284::AID-DVG11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas S, Goldberg MR, Watanabe T, D'Agati V, al-Awqati Q, Costantini F. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–251. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 32.Chi X, Hadjantonakis AK, Wu Z, Hyink D, Costantini F. A transgenic mouse that reveals cell shape and arrangement during ureteric bud branching. Genesis. 2009;47:61–66. doi: 10.1002/dvg.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- 34.Tang MJ, Cai Y, Tsai SJ, Wang YK, Dressler GR. Ureteric bud outgrowth in response to RET activation is mediated by phosphatidylinositol 3-kinase. Dev Biol. 2002;243:128–136. doi: 10.1006/dbio.2001.0557. [DOI] [PubMed] [Google Scholar]
- 35.Kim D, Dressler GR. PTEN modulates GDNF/RET mediated chemotaxis and branching morphogenesis in the developing kidney. Dev Biol. 2007;307:290–299. doi: 10.1016/j.ydbio.2007.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher CE, Michael L, Barnett MW, Davies JA. Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development. 2001;128:4329–4338. doi: 10.1242/dev.128.21.4329. [DOI] [PubMed] [Google Scholar]
- 37.Wong A, Bogni S, Kotka P, de Graaff E, D'Agati V, Costantini F, Pachnis V. Phosphotyrosine 1062 is critical for the in vivo activity of the Ret9 receptor tyrosine kinase isoform. Mol Cell Biol. 2005;25:9661–9673. doi: 10.1128/MCB.25.21.9661-9673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain S, Encinas M, Johnson EM, Jr, Milbrandt J. Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev. 2006;20:321–333. doi: 10.1101/gad.1387206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepicelli CV, Kispert A, Rowitch DH, McMahon AP. GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol. 1997;192:193–198. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- 40.Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- 41.Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 42.Bouchard M. Transcriptional control of kidney development. Differentiation. 2004;72:295–306. doi: 10.1111/j.1432-0436.2004.07207001.x. [DOI] [PubMed] [Google Scholar]
- 43.Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murdoch C. CXCR4: chemokine receptor extraordinaire. Immunol Rev. 2000;177:175–184. doi: 10.1034/j.1600-065x.2000.17715.x. [DOI] [PubMed] [Google Scholar]
- 45.Schier AF. Chemokine signaling: rules of attraction. Curr Biol. 2003;13:192–194. doi: 10.1016/s0960-9822(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 46.Takabatake Y, Sugiyama T, Kohara H, Matsusaka T, Kurihara H, Koni PA, et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J Am Soc Nephrol. 2009;20:1714–1723. doi: 10.1681/ASN.2008060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueland J, Yuan A, Marlier A, Gallagher AR, Karihaloo A. A novel role for the chemokine receptor Cxcr4 in kidney morphogenesis: an in vitro study. Dev Dyn. 2009;238:1083–1091. doi: 10.1002/dvdy.21943. [DOI] [PubMed] [Google Scholar]
- 48.Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 49.Brent AE, Tabin CJ. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131:3885–3896. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- 50.Helmbacher F, Dessaud E, Arber S, deLapeyriere O, Henderson CE, Klein R, et al. Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron. 2003;39:767–777. doi: 10.1016/s0896-6273(03)00493-8. [DOI] [PubMed] [Google Scholar]
- 51.Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Chattopadhyay N, Qin S, Szekeres C, Vasylyeva T, Mahoney ZX, et al. Coordinate integrin and c-Met signaling regulate Wnt gene expression during epithelial morphogenesis. Development. 2009;136:843–853. doi: 10.1242/dev.027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanwar YS, Ota K, Yang Q, Wada J, Kashihara N, Tian Y, et al. Role of membrane-type matrix metalloproteinase 1 (MT-1-MMP), MMP-2 and its inhibitor in nephrogenesis. Am J Physiol. 1999;277:934–947. doi: 10.1152/ajprenal.1999.277.6.F934. [DOI] [PubMed] [Google Scholar]
- 54.Meyer TN, Schwesinger C, Bush KT, Stuart RO, Rose DW, Shah MM, et al. Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: toward a model of branching through budding in the developing kidney. Dev Biol. 2004;275:44–67. doi: 10.1016/j.ydbio.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 55.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 56.Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci USA. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiao J, Sakurai H, Nigam SK. Branching morphogenesis independent of mesenchymal-epithelial contact in the developing kidney. Proc Natl Acad Sci USA. 1999;96:7330–7335. doi: 10.1073/pnas.96.13.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn. 2004;229:349–356. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- 60.Meier S. Development of the embryonic chick otic placode. I. Light microscopic analysis. Anat Rec. 1978;191:447–458. doi: 10.1002/ar.1091910405. [DOI] [PubMed] [Google Scholar]
- 61.Bort R, Signore M, Tremblay K, Martinez Barbera JP, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev Biol. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Fagman H, Andersson L, Nilsson M. The developing mouse thyroid: embryonic vessel contacts and parenchymal growth pattern during specification, budding, migration and lobulation. Dev Dyn. 2006;235:444–455. doi: 10.1002/dvdy.20653. [DOI] [PubMed] [Google Scholar]
- 63.Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 65.Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- 66.Cabernard C, Affolter M. Distinct roles for two receptor tyrosine kinases in epithelial branching morphogenesis in Drosophila. Dev Cell. 2005;9:831–842. doi: 10.1016/j.devcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Kuure S, Chi X, Lu B, Costantini F. The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development. 2010;137:1975–1979. doi: 10.1242/dev.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–5312. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
- 69.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartman HA, Lai HL, Patterson LT. Cessation of renal morphogenesis in mice. Dev Biol. 2007;310:379–387. doi: 10.1016/j.ydbio.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]