Abstract
The Sup35 protein (Sup35p) of Saccharomyces cerevisiae is a translation termination factor of the eRF3 family. The proteins of this family possess a conservative C–terminal domain responsible for translation termination and N–terminal extensions of different structure. The N–terminal domain of Sup35p defines its ability to undergo a heritable prion-like conformational switch, which is manifested as the cytoplasmically inherited [PSI+] determinant. Here, we replaced the N–terminal domain of S.cerevisiae Sup35p with an analogous domain from Pichia methanolica. Overexpression of hybrid Sup35p induced the de novo appearance of cytoplasmically inherited suppressor determinants manifesting key genetic and biochemical traits of [PSI+]. In contrast to the conventional [PSI+], ‘hybrid’ [PSI+] showed lower mitotic stability and preserved their suppressor phenotype upon overexpression of the Hsp104 chaperone protein. The lack of Hsp104 eliminated both types of [PSI+]. No transfer of prion state between the two Sup35p variants was observed, which reveals a ‘species barrier’ for the [PSI+] prions. The data obtained show that prion properties are conserved within at least a part of this protein family.
Keywords: evolutionary conservation/Pichia methanolica/prion/release factor eRF3/Saccharomyces cerevisiae
Introduction
Prions are a unique class of infectious agents whose infectivity is related solely to protein. In mammals they cause fatal neurodegenerative diseases, such as Creutzfeldt–Jacob disease of humans and bovine spongiform encephalopathy (for a review, see Horwich and Weissman, 1997; Prusiner et al., 1998). According to the prion hypothesis, the infectious process relies on the presence of host-encoded prion protein (PrP), which is able to exist in two structurally and functionally different forms. The infectious form (PrPSc) can catalyze the conversion of the normal cellular form (PrPC) into PrPSc. A related phenomenon was described for the yeast Saccharomyces cerevisiae, where the prion states of some proteins manifest themselves as cytoplasmic genetic determinants with unusual properties (Wickner, 1994). The best studied of these determinants are [PSI+] and [URE3], which correspond to the prion states of the yeast proteins Sup35 (Sup35p) and Ure2 (Ure2p). These determinants may be eliminated upon growth at low concentrations of guanidine hydrochloride (GuHCl), which is a protein denaturant when used at ∼1000-fold higher concentrations (Cox et al., 1988; Wickner, 1994). The prion determinants can reappear with high frequency upon overexpression of Sup35p or Ure2p, respectively (Chernoff et al., 1993; Wickner, 1994). Study of the biochemical properties of Sup35p and Ure2p showed that they aggregate in the strains with [PSI+] or [URE3], respectively, while being soluble in strains lacking these determinants (Patino et al., 1996; Paushkin et al., 1996; Edskes et al., 1999). While prions represent disease in mammals, this may not be so in lower eukaryotes. For example, the Het-s prion-like determinant of the filamentous fungus Podospora anserina represents an important part of the system for vegetative incompatibility (Coustou et al., 1997). If the prion mechanisms in yeast are useful, the prion properties of the respective proteins are likely to be conserved in other species. To check this, we decided to test such properties of Sup35p of the distantly related yeast Pichia methanolica (formerly called Pichia pinus).
Sup35p is a yeast homolog of the eRF3 translation termination factor of higher eukaryotes (Stansfield et al., 1995; Zhouravleva et al., 1995). It represents a multidomain protein in which the C–terminal (C) domain of amino acids 254–685 is similar to translation elongation factor EF-1α and essential for translation termination and cell viability. The N–terminal part of Sup35p is inessential for viability and may be subdivided into the middle (M) domain of unknown function and the N–terminal (N) domain of 123 amino acids, involved in [PSI+] maintenance (Ter-Avanesyan et al., 1993, 1994). The N domain plays a key role in the [PSI+] phenomenon, being required for [PSI+] propagation in vivo and solely responsible for Sup35p prion conversion and oligomerization into amyloid-like fibrils in vitro (Glover et al., 1997; King et al., 1997; Paushkin et al., 1997a,b). The Sup35 protein of P.methanolica (Sup35Pp) shares the three-domain structure of S.cerevisiae Sup35p (Kushnirov et al., 1990). Although the similarity of N domains is fairly low, the N domain of P.methanolica Sup35p is rich in glutamine and asparagine residues (Figure 1), a trait that distinguishes the prion-forming domains of S.cerevisiae Sup35p and Ure2p, and thus could possess prion properties (Kushnirov et al., 1990; Coschigano and Magasanik, 1991).
Fig. 1. Comparison of amino acid sequences of the N domains of Sup35 proteins from S.cerevisiae and P.methanolica. Sequences were aligned by introducing gaps (–).
Here, we created and expressed in S.cerevisiae a fusion of the P.methanolica Sup35p N domain with the M and C domains from S.cerevisiae. This protein (Sup35PSp) was able to adopt the prion state, resulting in the [PSI+]-like nonsense suppressor determinants. Hybrid Sup35PSp showed the key biochemical properties of yeast prions. At the same time, ‘hybrid’ [PSI+] ([PSI+PS]) was distinguished from the prototype by lower stability and by preservation of the suppressor phenotype upon overexpression of the Hsp104 chaperone protein (Hsp104p).
Results
De novo generation of the hybrid [PSI+PS]
To test the prion properties of the Sup35p N domain of P.methanolica we used S.cerevisiae, since P.methanolica lacks a genetic system for detection of the [PSI+] phenotype. The protein encoded by the P.methanolica SUP35 gene (SUP35P) is not as efficient in translation termination in S.cerevisiae as native Sup35p, since replacement of the chromosomal SUP35 gene with SUP35P resulted in a weak nonsense suppressor phenotype (Zadorski and Inge–Vechtomov, 1998). This is probably related to both the SUP35P promoter strength and the properties of the C domain of Sup35Pp. This prompted us to create a hybrid gene, encoding a fusion of the N domain of P.methanolica Sup35p with the M and C domains of S.cerevisiae Sup35p under the control of the S.cerevisiae SUP35 promoter (SUP35-PS gene). The hybrid gene was used to replace the chromosomal SUP35 in strain 5V–H19. No suppression of the ade2-1 nonsense mutation was detected in the resulting PS-5V–H19 strain, indicating efficient termination of translation. Using an antibody specific to the C domain of Sup35p, which is common for the Sup35 and Sup35PS proteins, we observed that these proteins were expressed in 5V–H19 and PS-5V–H19 at approximately equal levels (not shown).
It is known that overexpression of Sup35p causes [PSI+] generation de novo (Chernoff et al., 1993). Here, we observed that transformants of PS-5V–H19 [psi–] with multicopy plasmids encoding Sup35PSp or Sup35Pp produced Ade+ clones with high frequency on adenine omission medium. Most of these clones were convertible to adenine dependence by GuHCl treatment and thus carried [PSI+PS]. Surprisingly, overexpression of Sup35Pp in strain 5V–H19 [psi–] resulted in the appearance of conventional [PSI+], albeit with low frequency (not shown). Since this strain also expresses Sup35p, it is not clear which of the two Sup35 proteins played a key role in [PSI+] generation.
To avoid such ambiguity, further studies of [PSI+] generation by Sup35PSp were performed in strains with the chromosomal SUP35-C allele encoding Sup35Cp lacking N and M domains (Kochneva-Pervukhova et al., 1998). In such strains, only the plasmid-encoded Sup35p variants can form prion seeds, but the [PSI+] phenotype is not expressed due to the presence of non-prion Sup35Cp. Strain 9V–H70 (SUP35-C) was transformed with multicopy plasmids carrying the SUP35P, SUP35-PS or SUP35 genes. To score the appearance of [PSI+], cytoplasm of the transformants was transferred to the tester strains PS-5V–H19 [psi–] and 5V–H19 [psi–] using a cytoduction procedure. The transformants were crossed to testers, which resulted in cytoplasmic mixing without nuclear fusion and diploid formation due to kar1-1 mutation of 9V–H70, which blocks karyogamy (see Materials and methods). The appearance of Ade+ GuHCl-sensitive cytoductants was observed, but only in the cases when the transforming plasmids and the tester strains encoded Sup35p N domain of the same origin.
To quantify the de novo [PSI+] generation, strain 1-5V–H19 (SUP35-C) was transformed with multicopy plasmids encoding Sup35PSp or Sup35p. The induced [PSI+] were recovered by cytoplasmic transfer to the 3B-H72 and c10B-H49 [psi–] kar1-1 tester strains, respectively. These testers carry cycloheximide resistance mutations, which allow easy identification of cytoductants. Among 10 580 cytoductants of 3B-H72 6.3% carried [PSI+PS], whereas of 12 850 tested cytoductants of c10B-H49 only 3.1% were [PSI+]. Thus, Sup35PSp was ∼2–fold more efficient in the induction of prion state.
It is noteworthy that [PSI+] generation by overexpression of complete S.cerevisiae Sup35p requires the cytoplasmic [PIN+] determinant (Derkatch et al., 1997). In contrast, overexpression of P.methanolica Sup35Pp or hybrid Sup35PSp caused the appearance of [PSI+PS] irrespective of the presence of [PIN+] (not shown).
Variability of hybrid [PSI+PS]
Similarly to the conventional [PSI+], the newly generated isolates of hybrid [PSI+PS] showed ‘strain variation’, differing by their suppressor efficiency and mitotic stability. Isolates #1 and #2 showed strong suppressor phenotype (‘strong’ [PSI+PS]), being white on yeast extract/peptone/dextrose (YPD) medium and able to grow on adenine omission medium after 2 days of incubation. Isolates #3, #4 and #6 had weak suppressor phenotype (‘weak’ [PSI+PS]), being pink on YPD and growing on the adenine-less medium on the third or fourth day of incubation. Among the ‘strains’ of conventional [PSI+], more efficient suppression correlated with higher mitotic stability (Derkatch et al., 1996). The strains of [PSI+PS] generally followed this rule (Table I) with the exception of isolate #4, which showed high stability but only intermediate suppression level. In general, [PSI+PS] had significantly lower mitotic stability than conventional [PSI+] with equivalent suppressor efficiency. The conventional [PSI+] with strong suppressor phenotype in strain 5V–H19 were lost at a rate of <0.05% during growth on YPD medium (our unpublished data). In contrast, in the isogenic PS-5V–H19 strain even the most stable [PSI+PS] were lost with a frequency >0.2%.
Table I. Mitotic stability of different [PSI+PS] strains.
| [PSI+PS] strain | Total number of colonies | Number of colonies |
% of red colonies | % of red and sectored colonies | ||
|---|---|---|---|---|---|---|
| white or pink | with red sectors | red | ||||
| 1 | 513 | 495 | 15 | 3 | 0.58 ± 0.01 | 3.5 ± 0.4 |
| 3 | 877 | 553 | 234 | 90 | 10.3 ± 1.6 | 36.9 ± 7.9 |
| 4 | 868 | 863 | 3 | 2 | 0.23 ± 0.08 | 0.58 ± 0.15 |
| 6 | 844 | 172 | 561 | 111 | 13.2 ± 4.0 | 79.6 ± 10.0 |
The [PSI+PS] stability data represent an average of three independent clones. The SD is indicated.
The lack of transfer of prion state between Sup35p and Sup35PSp and [PSI+PS] behavior in diploids
Transformants of strains 5V–H19 [PSI+] and PS-5V–H19 [PSI+PS] (isolates #1 and #2) with centromeric plasmids encoding Sup35PSp and Sup35p, respectively, had a uniform antisuppressor phenotype, which indicated the lack of transfer of prion state to the heterologous plasmid-encoded Sup35 protein. To detect possible inhibitory effects on [PSI+] propagation, this experiment was repeated using multicopy plasmids encoding C–terminally truncated Sup35PSΔSp and Sup35ΔSp proteins. Since these proteins are non-functional in termination, the phenotypic expression of [PSI+] determinants in transformants should not be altered, allowing direct monitoring of [PSI+] loss. No red or red-sectored clones were observed among transformants of strain 5V–H19 [PSI+], while transformation of strain PS-5V–H19 [PSI+PS] produced red clones with low frequency not exceeding the rate of spontaneous [PSI+PS] loss (not shown). Therefore, the propagation of both [PSI+] and [PSI+PS] was not inhibited in the presence of heterologous Sup35 protein.
The absence of cross-transfer of prion state between Sup35p and Sup35PSp was also shown by studying heterozygous SUP35-PS/SUP35 diploids, which carried either [PSI+PS] or [PSI+]. The cross of strain PS-5V–H19 carrying [PSI+PS] #1 with 1A-H19 [psi–] resulted in diploid clones with a uniform red and adenine-requiring phenotype. This indicates that Sup35p was not inactivated by conversion into prion form. The random spore analysis has shown that among 10 diploid clones tested only three produced white Ade+ haploid segregants, which carried [PSI+PS], as confirmed by the GuHCl test. Even these three diploid clones possessed [psi–] cells, since they yielded some proportion of 0Ade+:4Ade– tetrads, indicating the [PSI+PS] loss in pre-meiotic diploid cells (Table II). This suggests that the stability of [PSI+PS] #1 in hetero– zygous diploids was significantly lower than in haploids. A reciprocal cross (PS-5V–H19 [psi–] × 1A-H19 [PSI+]) also produced diploid clones with antisuppressor phenotype, indicating the resistance of Sup35PSp to inactivation by [PSI+]. However, all six tested diploid clones segregated spore cultures with the suppressor phenotype and did not yield 0Ade+:4Ade– tetrads (Table III), which reflects high mitotic stability of the conventional [PSI+].
Table II. Data from the cross of PS-5V–H19 [PSI+PS] with 1A-H19 [psi–].
| Diploid clone | Number of tetrads with Ade+:Ade– ratio |
% of Ade+ segregants | ||||
|---|---|---|---|---|---|---|
| 0:4 | 1:3 | 2:2 | 3:1 | 4:0 | ||
| 3 | 4 | 1 | 4 | 0 | 0 | 25 |
| 7 | 9 | 7 | 4 | 0 | 0 | 19 |
| 8 | 7 | 6 | 4 | 0 | 0 | 21 |
| total | 20 | 14 | 12 | 0 | 0 | 21 |
Ade+ segregants expressed Sup35PSp while Ade– segregants expressed Sup35p, as shown by Western blot analysis (segregants from four 2Ade+:2Ade– tetrads were tested).
Table III. Data from the cross of PS-5V–H19 [psi–] with 1A-H19 [PSI+].
| Diploid clone | Number of tetrads with Ade+:Ade– ratio |
% of Ade+ segregants | ||||
|---|---|---|---|---|---|---|
| 0:4 | 1:3 | 2:2 | 3:1 | 4:0 | ||
| 1 | 0 | 9 | 17 | 0 | 0 | 41 |
| 8 | 0 | 2 | 8 | 0 | 0 | 45 |
| total | 0 | 11 | 25 | 0 | 0 | 42 |
Ade+ segregants expressed Sup35p while Ade– segregants expressed Sup35PSp, as shown by Western blot analysis (segregants from five 2Ade+:2Ade– tetrads were tested).
These experiments also showed that in meiosis [PSI+PS] behaves like a cytoplasmic determinant. This was further confirmed by studying its segregation in the diploids homozygous for SUP35-PS (Table IV). [PSI+PS] stability in homozygous diploids was higher than in heterozygous diploids, as evident from the comparison of Tables II and IV. This could be related to lower Sup35PSp levels in heterozygous diploids, where it constitutes only half of total Sup35p.
Table IV. Segregation of [PSI+PS] in diploids homozygous for SUP35-PS.
| Strain | Number of tetrads with Ade+:Ade– ratio |
% of Ade+ segregants | ||||
|---|---|---|---|---|---|---|
| 0:4 | 1:3 | 2:2 | 3:1 | 4:0 | ||
| 4V–H73 [psi–] | 0 | 0 | 1 | 1 | 9 | 93 |
| 17G-H73 [psi–] | 1 | 4 | 6 | 10 | 17 | 75 |
The diploids were obtained from crosses of the indicated strains with the PS-5V–H19 [PSI+PS] strain.
Sup35PSpPSI+ forms aggregates, shows increased resistance to proteases and seeds the prion conversion in vitro
The aggregation of Sup35p, typical of the [PSI+] state, can be observed microscopically (Patino et al., 1996). For such observations, the C domain of Sup35PSp inessential for aggregation was replaced with green fluorescent protein (GFP) and expressed in [PSI+PS] and [psi–] cells. In the former, GFP coalesced into multiple bright spots, which is typical of the [PSI+] state. In the latter, GFP fluorescence was distributed evenly throughout the cells (Figure 2).
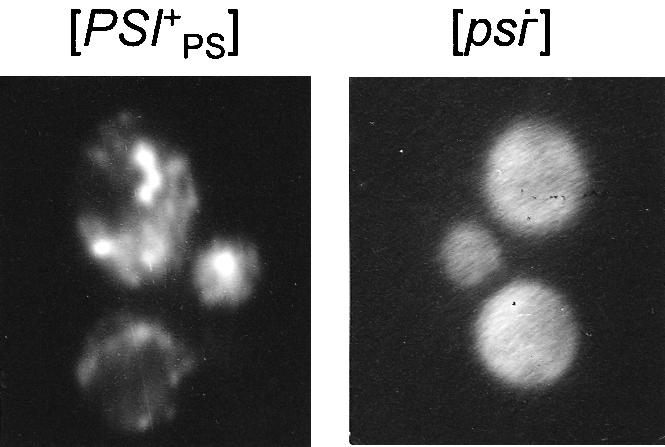
Fig. 2. The Sup35PSp–GFP fusion protein forms aggregates in [PSI+PS], but not in [psi–] strains. Fluorescent images of the [PSI+PS] and [psi–] cells of strain PS-5V–H19, transformed with centromeric plasmid encoding Sup35PSp–GFP fusion protein.
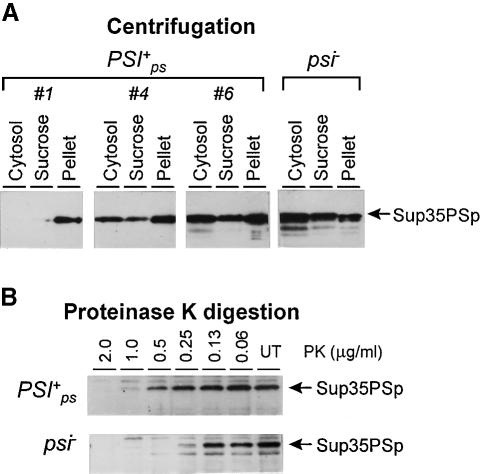
A centrifugation analysis of different [PSI+PS] isolates showed that in all cases most of the Sup35PSp was found in the pellet (Figure 3). The higher suppressor efficiency of the [PSI+PS] strain correlated with lower Sup35p levels in the soluble fraction. This indicates that the suppressor phenotype was caused by the depletion of soluble Sup35p and the variation of this phenotype was solely defined by the extent of Sup35PSp aggregation, which, in turn, is likely to result from different rates of Sup35p prion conversion, typical of different prion conformations of Sup35p. The antisuppressor phenotype of SUP35-PS/SUP35 diploids carrying either [PSI+] or [PSI+PS] indicated that Sup35pPSI+ did not seed conversion of Sup35PSppsi– into the prion state and vice versa. This was confirmed by centrifugation analysis: in both cases the protein found in aggregates corresponded to the type of [PSI+] present in the diploid. In the same diploid combining [PSI+] and [PSI+PS] both Sup35p and Sup35PSp were aggregated (not shown).
Fig. 3. Analysis of aggregation and proteinase K resistance of Sup35PSp in [PSI+PS] and [psi–] lysates. Immunoblotting with Sup35p antibody. (A) Lysates of the indicated [PSI+PS] derivatives of strain PS-5V–H19 were fractionated by centrifugation through a sucrose pad. Cytosol, sucrose and pellet, centrifugation fractions. (B) The lysates were treated with the indicated amounts of proteinase K (PK) for 30 min at 37°C. UT, untreated lysates.
An increased protease resistance is a characteristic feature of Sup35p from [PSI+] cells (Paushkin et al., 1996). A comparison of the proteinase K resistance of Sup35PSp from [PSI+PS] and [psi–] cells revealed ∼4-fold higher resistance in the former case (Figure 3).
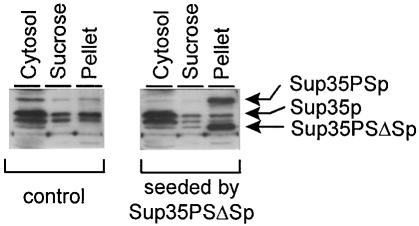
The prion properties of Sup35PSp were studied using the in vitro reactions of prion conversion. The addition of C–terminally truncated Sup35PSp from [PSI+PS] cells to the mixture of [psi–] lysates containing Sup35PSp and Sup35p caused the prion conversion to the aggregated form of Sup35PSp, but not of Sup35p (Figure 4).
Fig. 4. In vitro prion conversion of Sup35PSp. Equal amounts of lysates of the [psi–] strains PS-5V–H19 and 5V–H19 were mixed with the prion form of C–terminally truncated Sup35PSΔSp, obtained as described in Materials and methods. Control, the same mix of lysates without addition of prion seeds. The reactions were incubated for 2 h, fractionated by centrifugation and analyzed by immunoblotting. Cytosol, sucrose and pellet, centrifugation fractions.
Hsp104p effects on propagation and manifestation of [PSI+PS]
The Hsp104 protein is critical for [PSI+] propagation. The HSP104 disruption always eliminates [PSI+], while increased Hsp104p levels cause antisuppressor effect and gradual [PSI+] loss over successive generations (Chernoff et al., 1995; Paushkin et al., 1996). We tested these effects for hybrid [PSI+PS]. The HSP104 disruption invariably caused [PSI+PS] elimination in all strains carrying different isolates of hybrid [PSI+PS]. However, in contrast to conventional [PSI+], no antisuppressor phenotype was observed in [PSI+PS] strains with extra copies of HSP104. This phenotype is difficult to distinguish from the [psi–] one, which complicates the analysis of elimination of conventional [PSI+] because [PSI] status can be scored only after the loss of HSP104 plasmid. The transformation of different isolates of [PSI+PS] with multicopy HSP104 plasmid did not alter their original phenotype (white or pink, depending on [PSI+PS] ‘strength’). However, some colonies became red and some contained red sectors due to the [PSI+PS] loss. The rate of [PSI+PS] elimination in transformants depended on the strain of determinant, being significant only for ‘weak’ strains (Table V). Another distinction from conventional [PSI+] was that Hsp104p overproduction inhibited growth of ‘strong’ [PSI+PS]. The colonies of transformants of PS-5V–H19 [PSI+PS] #1 and #2 with multicopy HSP104 plasmid were 0.5 mm in diameter 4 days after transformation, while control transformants with pEMBLyex4 reached the same size in 2 days.
Table V. Influence of Hsp104p overexpression on the stability of different [PSI+PS] strains.
| [PSI+] strain | Plasmid | Number of transformants |
||
|---|---|---|---|---|
| white or pink | with red sectors | red | ||
| #1 | pFL44-HSP104 | 62 (96.9%) | 2 (3.1%) | 0 |
| pEMBLyex4 | 140 (97.9%) | 0 | 3 (2.1%) | |
| #2 | pFL44-HSP104 | 243 (97.9%) | 5 (2.1%) | 0 |
| pEMBLyex4 | 390 (100%) | 0 | 0 | |
| #3 | pFL44-HSP104 | 196 (78.7%) | 11 (4.4%) | 42 (16.9%) |
| pEMBLyex4 | 135 (88.2%) | 3 (2.0%) | 15 (9.8%) | |
| #4 | pFL44-HSP104 | 34 (49.3%) | 32 (46.3%) | 3 (4.4%) |
| pEMBLyex4 | 89 (98.9%) | 0 | 1 (1.1%) | |
The pEMBLyex4 vector lacking the HSP104 gene was used as a control. The data represent an average from three independent transformants. The percentage of transformants of each type is indicated in parentheses.
To study the effect of excess Hsp104p on the Sup35PSpPSI+ aggregates, we analyzed the size distribution of Sup35PSp in the lysates of ‘strong’ [PSI+PS] isolate #1. The presence of excess Hsp104p increased the levels of soluble Sup35PSp up to ∼20% of its total amount. A similar effect was observed for ‘strong’ conventional [PSI+] (Figure 5). The appearance of Sup35PSp in the soluble fraction was not due to the accumulation of [psi–] cells, since the latter constituted only a negligible fraction of the analyzed yeast culture (Table V). Thus, the excess of Hsp104p acts to dissolve Sup35PSpPSI+ aggregates, the same way as it acts on conventional Sup35pPSI+ (Patino et al., 1996; Paushkin et al., 1996). This may explain the accelerated loss of ‘weak’ [PSI+PS] upon overexpression of Hsp104p.
Fig. 5. Dependence of Sup35PSp and Sup35p prion aggregation on Hsp104p levels. The lysates of [PSI+] strains PS-5V–H19 or 5V–H19 transformed with multicopy HSP104 or control pEMBLyex4 plasmid were separated into five fractions by centrifugation and analyzed with Sup35p antibody.
Discussion
Sup35p is a yeast member of the eRF3 family of translation termination factors. The proteins of this family possess a highly conservative C domain responsible for the function in translation termination, and N–terminal extensions of varying structure for which no function was ascribed in most cases. The only exception is S.cerevisiae Sup35p, whose N domain is responsible for maintenance of the cytoplasmic [PSI+] determinant. This domain has properties similar to mammalian prions; in the prion state it aggregates, interfering with the Sup35p function in translation termination and causing the [PSI+] nonsense suppressor phenotype. This raises the question of whether the N–terminal extensions of eRF3 from other organisms have prion properties. The answer may help conclude whether the prion properties of Sup35p have biological significance.
To test the prion properties of the N domain of Sup35p of the methylotrophic yeast P.methanolica, this domain was used to replace the N domain of S.cerevisiae Sup35p, resulting in the hybrid Sup35PS protein. Similarly to Sup35p, overexpression of Sup35PSp caused the de novo generation of [PSI+]-like determinants. The Sup35PSp-based [PSI+PS] determinants obtained varied by the level of suppressor effect and showed the key genetic traits of yeast prions: they were cytoplasmically inherited and could be eliminated by GuHCl. Biochemical data revealed close similarity of the prion properties of Sup35PSp and Sup35p. Both proteins demonstrated a [PSI+]-dependent propensity to aggregate, increased protease resistance in [PSI+PS] strains and an ability to seed the prion self-conversion in vitro. However, some distinctions in the induction and propagation of Sup35PSp-based [PSI+PS] determinants were also noted. The induction of the prion state by overexpression of Sup35PSp occurred twice as frequently as by overexpression of Sup35p and independently of the [PIN+] factor. Another trait distinguishing [PSI+PS] was its decreased stability. These distinctions may reflect the individual properties of the Sup35Pp N domain. Alternatively, it is possible that the high stability of the prion state relies not only on the prion properties of a given protein, but also on the existence of the species-specific mechanisms, which influence the formation and stability of conventional [PSI+] but not of hybrid [PSI+PS]. For example, it was shown that the Sup35p N domain from S.cerevisiae, but not from P.methanolica, interacts with the cytoskeletal Sla1 protein, which increases the stability of conventional [PSI+] (Bailleul et al., 1999).
It is known that the prion diseases are poorly transferred between species. This phenomenon is known as a ‘species barrier’ and is related to small interspecies variations of PrP sequence (for a review, see Prusiner et al., 1998). The availability of Sup35p prion-forming domains from different yeast species allowed us to investigate whether there is a ‘species barrier’ for [PSI+] propagation. The sequence similarity between the N domains of Sup35p from S.cerevisiae and P.methanolica is significantly lower (∼40% identity) than within the mammalian PrP family, which should decrease the likelihood of interspecies prion conversion. Nevertheless, we observed that overexpression of Sup35Pp in strain 5V–H19 can induce Sup35p-based [PSI+], which could involve the transfer of prion state from overexpressed Sup35Pp to Sup35p. This prompted us to study the possibility of prion cross-conversion between the N domains of S.cerevisiae and P.methanolica Sup35p. Several lines of evidence presented in this study demonstrated the lack of transfer of prion state from Sup35p to Sup35PSp or in the opposite direction. The heterozygous SUP35-PS/SUP35 diploids possessing either [PSI+] or [PSI+PS] manifested antisuppressor phenotype, which indicates that the prion form of one protein can not seed conversion of another protein to the prion state. This conclusion was confirmed by centrifugation of the diploid lysates, by tetrad analysis of the diploids and by in vitro reactions of prion conversion. We also observed the lack of any inhibitory effects on propagation of the prion state of any one of these proteins from co-expression of the other protein. Thus, Sup35p and Sup35PSp are fully independent in propagation of their prion states.
This suggests that the induction of Sup35p-based [PSI+] by overexpression of Sup35Pp did not involve the transfer of prion state from Sup35Pp to Sup35p. It is possible that the excess of Sup35Pp alleviated [PSI+] formation by chromosomally encoded Sup35p by titrating some proteins that normally bind to S.cerevisiae Sup35p and prevent its spontaneous conversion into the prion form. One such protein could be Sup45p, whose ability to inhibit [PSI+] formation was shown by Derkatch et al. (1998).
The effects of Hsp104p on [PSI+PS] propagation were similar to those observed for conventional [PSI+]. The lack of Hsp104p caused elimination of [PSI+PS]; its overexpression could also promote [PSI+PS] loss, although the rate of loss was noticeable only for ‘weak’ [PSI+PS] strains. However, there was a significant difference in the effects of Hsp104p on phenotypic expression of conventional and hybrid [PSI+] determinants. Overexpression of Hsp104 caused antisuppressor ([psi–]-like) phenotype in conventional [PSI+] strains, but no such effect was observed in [PSI+PS] strains. Such an antisuppressor effect was considered an indication of a partial dissolution of Sup35pPSI+ aggregates by Hsp104p (Paushkin et al., 1996). It was proposed that Hsp104p breaks Sup35pPSI+ aggregates into smaller pieces, and this process produces some soluble Sup35p, especially when Hsp104p is overexpressed (Kushnirov and Ter-Avanesyan, 1998). Indeed, the fractionation of lysates of [PSI+PS] and [PSI+] cells overproducing Hsp104p revealed significant and comparable levels of soluble Sup35PSp and Sup35p proteins. However, since soluble Sup35PSp did not cause improved translation termination and antisuppression, it remains left to assume that it was mainly non-functional. Most likely, the inactivity of this Sup35PSp indicates that it originated from prion aggregates. The different properties of resolubilized Sup35p and Sup35PSp suggest the involvement of two different chaperone functions: resolubilization and refolding. It was shown recently that Hsp104p alone can not reactivate misfolded proteins; this requires the assistance of Hsp70 and Hsp40 chaperone proteins (Glover and Lindquist, 1998). The same probably applies to prion aggregates. We suggest that prion Sup35PSp is efficiently disaggregated by the Hsp104p excess, but is not reactivated by co-chaperones like Hsp70 and Hsp40. In agreement with this conclusion, we observed that extra copies of HSP104 caused growth inhibition of the yeast strains bearing strong [PSI+PS] (#1 and #2). This effect, not observed for conventional [PSI+], may be anticipated; by breaking Sup35PSpPSI+ aggregates, Hsp104p accelerates the prion conversion and depletes soluble Sup35PSp, which is not balanced by the appearance of functional refolded Sup35PSp molecules.
The results of this study demonstrate that the N domain of P.methanolica Sup35p manifests prion properties when expressed in a heterologous organism, S.cerevisiae, despite the fact that its amino acid sequence differs from that of the prionogenic domain of S.cerevisiae Sup35p. This suggests that P.methanolica Sup35p can exist in the prion state in its natural environment as well. The abundance of glutamine and the related amino acid asparagine is a characteristic feature of prion-forming domains of all known yeast prions: S.cerevisiae Ure2p and Sup35p, and Sup35p of P.methanolica. This feature is also exhibited by Sup35p of the yeast Candida albicans and the fungus Podospora anserina, but not by Sup35p homologs of higher eukaryotes including man, whose N–terminal regions resemble more the M domain of Sup35p by amino acid content (Entrez database; http://www.ncbi.nlm.nih.gov/Entrez/nucleotide.html). Thus, it is possible that prion properties of Sup35p are restricted by lower eukaryotes. The approach described in this paper may be used to study the prion properties of Sup35p from different species, as well as those of other potentially prionogenic proteins.
Materials and methods
Strains and genetic methods
The strains used are presented in Table VI. We used standard rich (YPD) and synthetic (SC) media for yeast (Sherman et al., 1986). Non-fermentable media contained glycerol (24 ml/l) as a sole carbon source. All solid media contained 2.5% (w/v) agar. Yeast cells were grown at 30°C. Standard methods of yeast genetics were used (Sherman et al., 1986). DNA transformation of lithium acetate-treated yeast cells was performed as described (Gietz et al., 1995). Yeast strains were cured, when required, of the [PSI+] and [PIN+] determinants by growth on YPD medium supplemented with 2 or 5 mM GuHCl, respectively (GuHCl test) (Tuite et al., 1981). The [psi–] colonies of ade2-1 SUQ5 strains were identified by their red color and adenine requirement, since the weak serine-inserting tRNA suppressor SUQ5 can suppress the ade2–1 ochre mutation only in the presence of [PSI+] determinant (Cox, 1965). The [pin–] colonies were identified by the inability of multicopy SUP35 plasmids to induce [PSI+] generation (Derkatch et al., 1997). The mitotic stability of different [PSI+] isolates in PS-5V–H19 was determined as the percentage of [PSI+] cells in their individual colonies grown on YPD medium.
Table VI. Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| 5V–H19a | MATa ade2-1 SUQ5 leu2-3,112 ura3-52 can-100 | Ter-Avanesyan et al. (1994) |
| 1-5V–H19 | MATa ade2-1 SUQ5 SUP35-C leu2-3,112 ura3-52 can-100 [psi–] [PIN+] | Ter-Avanesyan et al. (1994) |
| PS-5V–H19 | MATa ade2-1 SUQ5 SUP35-PS leu2-3,112 ura3-52 can-100 [psi–] [PIN+] | this study |
| 1A-H19a | MATα ade2-1 SUQ5 lys1-1 his3 leu2-3,112 | Ter-Avanesyan et al. (1994) |
| c10B-H49 | MATα ade2-1 SUQ5 kar1-1 his3 leu1 lys1-1 cyhR [psi–] [pin–] [rho–] | Kochneva-Pervukhova et al. (1998) |
| 9V-H70b | MATα ade2-1 SUQ5 SUP35-C kar1-1 leu2-3,112 ura3-52 his3 cyhR [psi–] | this study |
| 3B-H72 | MATα ade2-1 SUQ5 SUP35-PS kar1-1 leu1 leu2-3,112 lys1-1 cyhR [psi–] [PIN+] [rho–] | this study |
| 4V–H73 | MATα ade2-1 SUQ5 SUP35-PS his3 leu2-3,112 [psi–] | this study |
| 17G-H73 | MATα ade2-1 SUQ5 SUP35-PS his3 leu2-3,112 [psi–] | this study |
a[PSI+] and [psi–] variants of these strains were used.
b[PIN+] and [pin–] variants were used.
The ‘cytoduction’ experiments were performed as described (Ter–Avanesyan et al., 1994). Transformants of strain 9V–H70 were mated with the [psi–] [rho–] derivatives of strains 5V–H19 and PS-5V–H19. Strain 9V–H70 carried the kar1-1 mutation, which blocks karyogamy (nuclear fusion) and thus allows the transfer of only cytoplasmically located determinants between cells (Conde and Fink, 1976). The strains were mixed together on the surface of a YPD plate, incubated for 1 day, and [PSI+] [rho+] cytoductants were selected. To quantify the induction of [PSI+], transformants of the 1-5V–H19 [psi–] [PIN+] strain with multicopy SUP35 or SUP35-PS plasmids were crossed with the corresponding kar1-1 [psi–] [rho–] tester strains carrying the cyhR mutation. Cytoductants were selected on glycerol media containing 3 μg/ml cycloheximide and the frequency of [PSI+] induction was estimated as described by Kochneva-Pervukhova et al. (1998).
For the fluorescent microscopy, cells were grown overnight on medium selective for the SUP35-PS-GFP plasmid. Cells were photographed using an OptonIII microscope (Carl Zeiss, Germany) with a 63× objective.
Plasmid construction
A 3.6 kb genomic XhoI–XbaI fragment containing the SUP35 gene was cloned into the same sites of plasmids pRS315 (Sikorski and Hieter, 1989) and pEMBLyex4 (Cesareni and Murray, 1987) to yield pRS315-SUP35 and yex-SUP35 (Ter-Avanesyan et al., 1993). A part of yex-SUP35 plasmid encoding the Sup35p N domain was replaced with SmaI, BamHI and SacI cloning sites to yield yexSUP35-SBS plasmid. The sequence of the modified region is TCTGCCgatccccggggatccgagctcAAGGTATG, where the insertion is given in small letters and capitals denote original SUP35 sequence, with the first codon of the M domain (amino acid 124) italicized. A fragment of SUP35P starting from position –12 to the DraI site encoding amino acids 1–186 was amplified by PCR, and inserted into the SmaI site of yexSUP35SBS to yield yexSUP35PS plasmid. The plasmids yex-SUP35ΔS and yex-SUP35PSΔS were obtained from yex-SUP35 and yex-SUP35PS by deletion of the SalI fragment encoding amino acids 483–685 of Sup35p. The plasmid pRS315-SUP35PS was created by moving the XhoI–XbaI fragment containing SUP35PS from yex-SUP35PS to pRS315. The yex-SUP35P plasmid was obtained from yex-SUP35SBS by replacing the SmaI–HindIII fragment of SUP35 coding sequence with complete SUP35P coding sequence from position –12 to the HindIII site. pRS315-SUP35PS-eGFP was created by joining the XhoI–HpaI fragment encoding Sup35PSp N and M domains, the SmaI–XbaI fragment of pEGFP plasmid (Clontech) encoding GFP, and pRS315 plasmid cut by XhoI and XbaI. The plasmid pFL44L-HSP104 was a kind gift of M.Boguta.
Preparation, fractionation and analysis of yeast cell lysates
Yeast cultures were grown in liquid YPD medium or in a medium selective for plasmid marker to an OD600 of 1.5. The cells were harvested, washed in water, and lysed by vortexing with glass beads in 25 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol containing 1 mM phenylmethylsulfonyl fluoride (PMSF) to limit proteolysis. Cell debris was removed by centrifugation at 15 000 g for 10 min. To fractionate the lysates, they were loaded on 0.5 ml 30% sucrose pads made in buffer A and centrifuged in a Beckman SW50 rotor at 40 000 r.p.m. for 25 min at 4°C. The resulting supernatants, pellets and intermediate fractions were analyzed by Western blotting for Sup35p distribution. To obtain Sup35PSp seeds for the prion conversion reactions, strain 1-5V–H19 was transformed with multicopy yex-SUP35PSΔS plasmid encoding C–terminally truncated Sup35PSΔSp, which resulted in [PSI+PS] generation. Lysate of this strain was centrifuged in an SW50 rotor as above and the pellet was used to seed the prion conversion as described previously (Paushkin et al., 1997b). To study the protease resistance of Sup35p, lysates were prepared as described above, but without addition of PMSF. Each reaction contained 150 μg of total protein and 0.06–2.0 μg/ml of proteinase K (Boehringer Mannheim) in a volume of 50 μl. After 30 min incubation at 37°C, 4 μl aliquots were removed and analyzed by Western blotting.
Protein samples were separated on a 10–15% SDS–polyacrylamide gel according to Laemmli (1970), and electrophoretically transferred to nitrocellulose sheets (Towbin et al., 1979). Western blots were probed with polyclonal rabbit antibody against Sup35p. The expression levels of Sup35p and Sup35PSp were compared by probing serially diluted lysates with anti-Sup35p antibody affinity purified against the Sup35Cp fragment. Bound antibody was detected using the Amersham ECL system.
Acknowledgments
Acknowledgements
We thank A.I.Poznyakovski for constructing some plasmids and M.O.Agaphonov for helpful suggestions. The work was supported by grants from INTAS and the Russian Foundation for Basic Research (M.D.T.-A.) and the Wellcome Trust (V.V.K.).
References
- Bailleul P.A., Newnam, G.P., Steenbergen, J.N. and Chernoff, Y.O. (1999) Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae.Genetics, 153, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G. and Murray,A.H. (1987) Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In Setlow,J.K. (ed.), Genetic Engineering: Principles and Methods. Plenum Press, New York, NY, Vol. 4, pp. 135–154. [Google Scholar]
- Chernoff Y.O., Derkatch, I.L. and Inge-Vechtomov, S.G. (1993) Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist, S., Ono, B., Inge-Vechtomov, S.G. and Liebman, S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like ψ+. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Conde J. and Fink, G.R. (1976) A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl Acad. Sci. USA, 73, 3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano P.W. and Magasanik, B. (1991) The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol., 11, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V., Deleu, C., Saupe, S. and Begueret, J. (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci. USA, 94, 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B.S. (1965) psi, a cytoplasmic suppressor of super-suppressor in yeast. Heredity, 20, 505–521. [Google Scholar]
- Cox B.S., Tuite, M.F. and McLaughlin, C.S. (1988) The ψ factor of yeast: a problem in inheritance. Yeast, 4, 159–178. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff, Y.O., Kushnirov, V.V., Inge-Vechtomov, S.G. and Liebman, S.W. (1996) Genesis and variability of prion-like [PSI] factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley, M.E., Zhou, P., Chernoff, Y.O. and Liebman, S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics, 147, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley, M.E. and Liebman, S.W. (1998) Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl Acad. Sci. USA, 95, 2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K., Gray, V.T. and Wickner, R.B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA, 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl, R.H., Willems, A.R. and Woods, R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Glover J.R. and Lindquist, S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell, 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Kowal, A.S., Schirmer, E.C., Patino, M.M., Liu, J.-J. and Lindquist, S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of ψ+, a heritable prion-like factor of S.cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Horwich A.L. and Weissman, J.S. (1997) Deadly conformations—protein misfolding in prion diseases. Cell, 89, 499–510. [DOI] [PubMed] [Google Scholar]
- King C.-Y., Tittmann, P., Gross, H., Gebert, H., Aebi, M. and Wuthrich, K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA, 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Poznyakovski, A.I., Smirnov, V.N. and Ter–Avanesyan, M.D. (1998) C–terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae. Curr. Genet., 34, 146–151. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. and Ter-Avanesyan, M.D. (1998) Structure and replication of yeast prions. Cell, 94, 13–16. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. et al. (1990) Divergence and conservation of SUP2(SUP35) gene of yeast Pichia pinus and Saccharomyces cerevisiae. Yeast, 6, 461–472. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–689. [DOI] [PubMed] [Google Scholar]
- Patino M.M., Liu, J.J., Glover, J.R. and Lindquist, S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov, V.V., Smirnov, V.N. and Ter-Avanesyan, M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov, V.V., Smirnov, V.N. and Ter-Avanesyan, M.D. (1997a) Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol., 17, 2798–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov, V.V., Smirnov, V.N. and Ter-Avanesyan, M.D. (1997b) In vitro propagation of the prion-like state of the yeast Sup35 protein. Science, 277, 381–383. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Scott, M.R., De Armond, S.J. and Cohen, F.E. (1998) Prion protein biology. Cell, 93, 337–348. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sikorski R.S. and Hieter, P. (1989) A system of shuttle vectors and yeast host strains designated for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I. et al. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Kushnirov, V.V., Dagkesamanskaya, A.R., Didichenko, S.A., Chernoff, Y.O., Inge-Vechtomov, S.G. and Smirnov, V.V. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol., 7, 683–692. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Dagkesamanskaya, A.R., Kushnirov, V.V. and Smirnov, V.N. (1994) The SUP35 omnipotent suppressor is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics, 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin, T. and Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M.F., Mundy, C.R. and Cox, B.S. (1981) Agents that cause a high frequency of genetic change from [psi+] to [psi–] in Saccharomyces cerevisiae. Genetics, 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered Ure2 protein: evidence for prion analogue in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Zadorski S.P. and Inge-Vechtomov, S.G. (1998) The SUP35 gene of Pichia methanolica is the recessive suppressor in the yeast Saccharomyces cerevisiae. Dokl. Akad. Nauk, 361, 825–829. (in Russian) [PubMed] [Google Scholar]
- Zhouravleva G., Frolova, L., Le Goff, X., Le Guellec, R., Inge-Vechtomov, S.G., Kisselev, L. and Philippe, M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]