Abstract
The LQT1 locus (KCNQ1) has been correlated with the most common form of inherited long QT (LQT) syndrome. LQT patients suffer from syncopal episodes and high risk of sudden death. The KCNQ1 gene encodes KvLQT1 α–subunits, which together with auxiliary IsK (KCNE1, minK) subunits form IKs K+ channels. Mutant KvLQT1 subunits may be associated either with an autosomal dominant form of inherited LQT, Romano–Ward syndrome, or an autosomal recessive form, Jervell and Lange-Nielsen syndrome (JLNS). We have identified a small domain between residues 589 and 620 in the KvLQT1 C–terminus, which may function as an assembly domain for KvLQT1 subunits. KvLQT1 C–termini do not assemble and KvLQT1 subunits do not express functional K+ channels without this domain. We showed that a JLN deletion–insertion mutation at KvLQT1 residue 544 eliminates important parts of the C–terminal assembly domain. Therefore, JLN mutants may be defective in KvLQT1 subunit assembly. The results provide a molecular basis for the clinical observation that heterozygous JLN carriers show slight cardiac dysfunctions and that the severe JLNS phenotype is characterized by the absence of KvLQT1 channel.
Keywords: KvLQT1 channels/long QT syndrome/potassium channels
Introduction
Long QT (LQT) syndrome is a rare disease, which is characterized by a prolongation of the QT interval on the electrocardiogram. Patients suffer from syncopal episodes due to ventricular arrhythmias like torsade de pointes and a high risk of sudden death (reviewed in Wang et al., 1998). Inherited LQTs are the autosomal dominant Romano–Ward syndrome (RWS) (Romano, 1963; Ward, 1964) and the autosomal recessive Jervell and Lange-Nielsen syndrome (JLNS) (Jervell and Lange-Nielsen, 1957). In addition to the cardiac phenotype, JLN patients have severe bilateral congenital deafness.
It has been shown that the inherited LQTs are associated with mutations in genes that encode cardiac ion channels. So far, four LQT genes have been identified, an additional one has been mapped to chromosome 4q25-27 (Wang et al., 1998). The LQT1 locus is responsible for the most common form of this inherited cardiac arrhythmia. It encodes the KvLQT1 (KCNQ1) potassium channel, which displays the typical topology of Shaker-type Kv α–subunits with cytoplasmic N- and C–termini flanking a membrane-inserted core region. It comprises six putative transmembrane segments and a P loop domain bearing the K+ signature sequence (Wang et al., 1996). When expressed alone, KvLQT1 elicits a rapidly activating K+ current (Barhanin et al., 1996; Sanguinetti et al., 1996). The KvLQT1 protein interacts with the auxiliary K+ channel subunit IsK (KCNE1, minK), which has a single transmembrane region and cannot form K+ channels on its own (Takumi et al., 1988; Attali et al., 1993; Lesage et al., 1993; Attali, 1996; Busch and Suessbrich, 1997; Kacmarek and Blumenthal, 1997). KvLQT1 and IsK form the functional IKs channel complex, which is characterized by a slow activation after depolarization. It does not inactivate and displays a slow deactivation (Barhanin et al., 1996; Sanguinetti et al., 1996). An accumulation of open IKs channels at high stimulation frequencies due to the slow deactivation might be of (patho–)physiological relevance. Defects in the corresponding genes lead to decreased potassium outward currents within the plateau phase of the cardiac action potential. The mechanisms of interaction between KvLQT1 and IsK, and the stoichiometry of the IKs channel complex have not yet been elucidated.
Mutations in KCNQ1 also cause the recessive form of the LQT syndrome, JLNS (Neyroud et al., 1997). Heterozygous mutations in KCNQ1 display less cardiac dysfunctions, whereas in the homozygous trait a severe cardiac phenotype and bilateral deafness can be detected. Therefore, KvLQT1 is supposed to be involved not only in ventricular repolarization, but also in the control of endolymph homeostasis of the inner ear (Vetter et al., 1996). It has been shown that mutations in the KCNE1 gene also result in JLNS (Schulze-Bahr et al., 1997; Splawski et al., 1997; Tyson et al., 1997; Duggal et al., 1998). Thus, the genetic data demonstrated that mutations may affect KvLQT1/IsK1 channel activity in a dominant or recessive manner.
Mutations in either of the two genes associated with a dominant RWS phenotype have been intensively investigated by expressing the corresponding mutant channel subunit in heterologous expression systems. The results showed that the mutant subunits appear to suppress dominantly the activity of wild-type subunits. Most likely, assembly of mutant and wild-type subunits rendered KvLQT1/IsK channels inactive (Chouabe et al., 1997; Splawski et al., 1997; Wollnik et al., 1997). In contrast, mutant subunits mimicking a recessive JLNS mutation did not dominantly suppress the activity in an in vitro expression system.
Co-expression of this C–terminal JLNS mutation with wild-type (wt) subunits yielded currents ∼50% of the wild-type KvLQT1 (Wollnik et al., 1997). The data suggested that KvLQT1 subunits with a JLNS-type mutation cannot form functional KvLQT1/IsK channels by themselves.
Most likely, KvLQT1 α–subunits assemble to functional Kv channels in the form of tetramers, as is known for Shaker (Sh) channels. Therefore, we reasoned that the recessive nature of some JLNS mutations might be due to a dysfunctional assembly domain in the KvLQT1 α–subunit. To test this hypothesis, we identified biochemically and functionally a cytoplasmic KvLQT1 α–subunit domain, which has properties similar to the previously identified tetramerization domain (T1–domain) in the cytoplasmic N–terminus of Sh channels (Li et al., 1992), but is located within the C–terminus of the protein (amino acids 589–620). The KvLQT1 assembly domain sequence is similar, but not identical, among KCNQ family members. Our results show that the recessive nature of the C–terminal JLN mutations is due to a mutation of the assembly domain resulting in the failure of the mutant α–subunits to associate with wt α–subunits.
Results
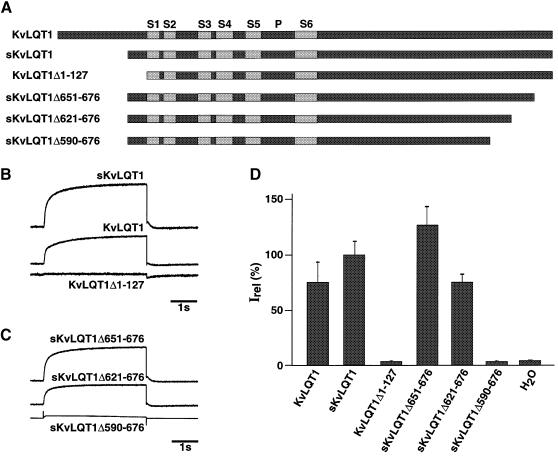
The majority of LQT syndrome mutations associated with defective KvLQT1 channels have been found in the membrane-integrated core region (Li et al., 1998). However, several RWS and JLNS mutations have also been described, which alter the cytoplasmic C–terminal sequence of KvLQT1 subunits (Chouabe et al., 1997; Wollnik et al., 1997; Neyroud et al., 1999). This suggests that cytoplasmic domains are critical for functional KvLQT1 channel expression. In a first screen for critical cytoplasmic KvLQT1 domains, we truncated both the cytoplasmic N- and C–termini of KvLQT1 subunits. The deletion constructs were expressed in the heterologous Xenopus oocyte expression system and also in Chinese hamster ovary (CHO) tissue culture cells by transient transfection. We chose to express the KvLQT1 constructs in the absence of auxiliary minK subunits in order not to complicate the interpretation of our expression studies. Two different KvLQT1 open reading frames (ORFs) have been reported. The derived protein sequences differ in their N–terminal part up to amino acid residue 107 and then are identical up to the C–terminal end (Sanguinetti et al., 1996; Chouabe et al., 1997). In agreement with previously published data (Sanguinetti et al., 1996; Chouabe et al., 1997; Lee et al., 1997; Demolombe et al., 1998), both KvLQT1 ORFs produced KvLQT1 channels, which apparently did not differ in their gating properties. This indicated that the first 95 N–terminal amino acid residues in KvLQT1 protein (KvLQT1) were not important for functional KvLQT1 channel expression in Xenopus oocytes (Figure 1A, B and D). The shorter KvLQT1 (sKvLQT1) displayed the same characteristics as full-length KvLQT1. In contrast, a complete deletion of the cytoplasmic N–terminus and the first seven amino acid residues of membrane-spanning segment S1 (KvLQT1Δ1–127) produced non-functional subunits (Figure 1A, B and D), similar to the data reported for iso2 KvLQT1 (Demolombe et al., 1998).
Fig. 1. Effect of N- and C–terminal deletions on expression of KvLQT1 currents in Xenopus oocytes. (A) Diagram illustrating KvLQT1 constructs used for heterologous expression of cRNA in Xenopus oocytes. Two KvLQT1 cDNA versions encoding full-length KvLQT1 and a shorter (sKvLQT1) ORF have been reported (Sanguinetti et al., 1996; Yang et al., 1997). Grey boxes indicate putative transmembrane regions S1–S6; P refers to the P–domain being involved in pore formation. Numbers in deletion constructs refer to amino acid residues of full-length KvLQT1 protein. (B) Currents measured after injection of cRNA encoding KvLQT1, sKvLQT1 and KvLQT1Δ1–127, respectively. (C) Currents were measured after injection of cRNA encoding, sKvLQT1Δ651–676, sKvLQT1Δ621–676 and sKvLQT1Δ590–676. (D) Data pooled from a number of oocytes (n = 6–28) including the constructs illustrated in (A) and an H2O control. The 100% Irel value corresponds to the steady-state current amplitude of sKvLQT1 after a 4 s test pulse to +40 mV.
When we expressed sKvLQT1 constructs with a truncated C–terminus (Figure 1A), we could only delete a small part of the C–terminal protein sequence without losing expression. Injection of sKvLQT1Δ651–676 and sKvLQT1Δ621–676 mRNA into Xenopus oocytes led to the expression of functional KvLQT1 channels with characteristics and mean currents that were similar to wild type (Figure 1A, C and D). Similar data were obtained in the CHO cell expression system (not shown). Further deletion of the KvLQT1 C–terminus (sKvLQT1Δ590–676) produced KvLQT1 subunits that were unable to express functional KvLQT1 channels (Figure 1A, C and D). The residual current amplitudes that were observed in sKvLQT1Δ590–676 mRNA-injected oocytes (4 ± 0.81% at +40 mV, n = 6) were not significantly different from H2O-injected control oocytes (Figure 1D). This indicated that the 56 C–terminal amino acid residues of KvLQT1 protein could be deleted and were not essential for functional KvLQT1 channel expression. The results further indicated that in the vicinity of amino acid residue 590, KvLQT1 subunits may contain domain(s) critical for the expression of functional KvLQT1 channels in heterologous expression systems. The observation fitted well with the genetic data that missense mutations at KvLQT1 amino acid residues 587 and 591 are associated with LQT syndrome (Itoh et al., 1998; Neyroud et al., 1999).
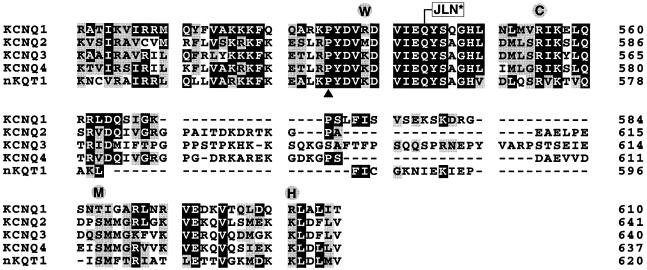
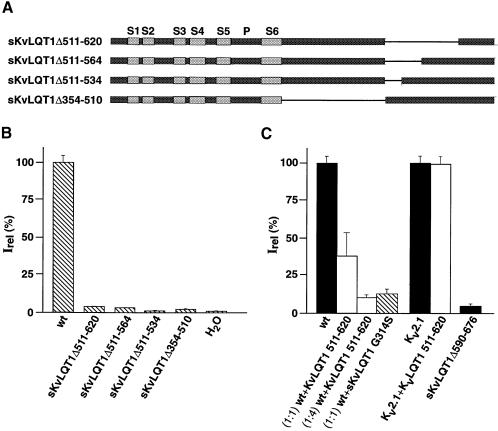
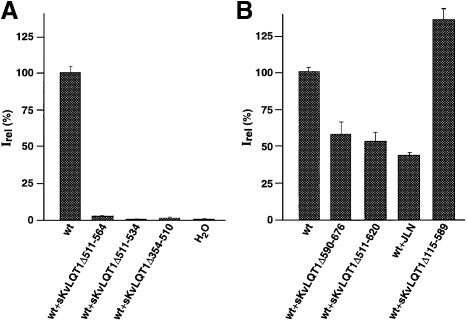
Amino acid residue 590 of KvLQT1 is flanked on both sides by domains with a significant degree of sequence conservation among the KCNQ family members (see Figure 2). We constructed internal KvLQT1 deletion mutants where this conserved sequence had been completely (sKvLQT1Δ511–620) or partially deleted (sKvLQT1Δ511–564; sKvLQT1Δ511–534) (Figure 3A). None of the internal deletion constructs produced functional KvLQT1 channels after injection of the corresponding cRNA into Xenopus oocytes (Figure 3B). Even a relatively small deletion of the first 23 amino acid residues of the conserved C–terminal sequence (sKvLQT1Δ511–534) produced non-functional KvLQT1 subunits (Figure 3B). These results suggested that the KvLQT1 C–terminus contained a domain(s) between residues 511 and 620 that was critical for the expression of functional KvLQT1 channels.
Fig. 2. Alignment of C–terminal sequences conserved in the KCNQ family. KCNQ1 encoding the KvLQT1 protein (Chouabe et al., 1997; Yang et al., 1997), KCNQ2 (Biervert et al., 1998; Singh et al., 1998), KCNQ3 (Charlier et al., 1998), KCNQ4 (Kubisch et al., 1999) and nKQT1 (Wei et al., 1996) sequences were aligned using the clustalw algorithm of the Meglign (DNAstar Inc.) program. Numbers on the right side refer to the last amino acid residue in each lane. Residues that are identical in at least three out of the five sequences are boxed in black, structurally conserved residues are boxed in grey. KCNQ1 point mutations associated with the LQT syndrome (Chouabe et al., 1997; Itoh et al., 1998; Neyroud et al., 1999) are circled. The deletion–insertion mutation at the amino acid residue linked to JLN syndrome (Neyroud et al., 1997) is indicated by a boxed JLN*. An insertion mutation (▴) in the homologous region of KCNQ2 has been linked to the occurrence of benign familial neonatal convulsions (Biervert et al., 1998).
Fig. 3. Effect of internal deletions on expression of KvLQT1 currents. (A) Diagram illustrating sKvLQT1 constructs used for heterologous expression in Xenopus oocytes (B) and in CHO cells (C). Numbers in deletion constructs refer to amino acid residues of full-length KvLQT1 protein (see Figure 1). (B) Currents were measured after injection of cRNA encoding sKvLQT1, sKvLQT1Δ511–564, sKvLQT1Δ511–534 and sKvLQT1Δ354–510, respectively, and for control H2O. In each case, currents were evoked with voltage jumps to +40 mV from a holding potential of –80 mV. Data pooled from 4–20 oocytes. The 100% Irel value corresponds to the steady-state current amplitude of sKvLQT1, which was measured at +40 mV 4 s after voltage jump. (C) Currents were measured in transfected CHO cells in the whole-cell configuration of the patch–clamp technique. Cells were transfected with KvLQT1 DNA, sKvLQT1Δ590–676 DNA, Kv2.1 DNA (black bars) or mixtures of KvLQT1 DNA with KvLQT1 511–620 (white bars) or KvLQT1 G314S (hatched bar) as indicated under each column bar. For the control, CHO cells were transfected with a 1:1 mixture of Kv2.1 and KvLQT1 511–620 DNAs. Current amplitudes were measured after voltage jumps from –80 to +40 mV. Data were pooled from 4–16 cells. Current amplitudes were related to each other by setting wt KvLQT1 current amplitude (82 pA/pF) to 100% and Kv2.1 current amplitude (711 pA/pF) to 100%.
The expression results obtained with the C–terminal KvLQT1 deletion constructs resembled those that we had previously obtained with ether-à-go-go (eag) Kv channels (Ludwig et al., 1997). We showed that eag subunits with C–terminal deletions were non-functional because a C–terminal assembly domain (cad) had been deleted. Also, co-expression of cad with wt eag subunits suppressed functional eag channel expression. Similar observations were reported for co-expression experiments of Sh subunits with the cytoplasmic tetramerization (assembly) domain of Sh channels (Li et al., 1992). Most likely, the cytoplasmic assembly domains suppressed functional Kv channel expression by inhibiting the assembly of wt subunits to functional tetrameric channels, in a dominant-negative fashion. Because of the resemblance of our KvLQT1 data to those of eag, we hypothesized that the conserved C–terminal KvLQT1 domain might represent a cytoplasmic KvLQT1 assembly domain. Accordingly, we tested whether co-expression of this domain with wt KvLQT1 subunits would suppress the expression of functional KvLQT1 channels. Co-injection of KvLQT1 511–620 mRNA with sKvLQT1 RNA in a 1:5 molar ratio resulted in a suppression of KvLQT1 current. Compared with controls, the residual current amplitude at +40 mV was 35 ± 10% (n = 6) (data not shown). Similar data were obtained when we co-transfected CHO cells with wt KvLQT1 and KvLQT1 511–620 DNA constructs (Figure 3C). Transient transfections with a 1:1 ratio yielded KvLQT1 current amplitudes at +40 mV, which were 46.9 ± 15.7% (n= 5) of wt control current amplitudes (82.2 ± 13.1 pA/pF; n = 16). Transient transfections with a 1:4 ratio of wt KvLQT1 DNA versus KvLQT1 511–620 DNA virtually suppressed the expression of wt current (Figure 3C). The residual current amplitudes were 8.8 ± 2.3 pA/pF (n = 9) at +40 mV. For comparison, we co-transfected CHO cells with wt KvLQT1 DNA and a mutant KvLQT1 DNA carrying a missense mutation (G314S) in the KvLQT1 P–domain. Previously, it had been shown that the KvLQT1 G314S mutation was correlated with an RWS phenotype as well as with a dominant-negative suppression of functional KvLQT1 channel expression (Chouabe et al., 1997; Wollnik et al., 1997). Co-transfection of CHO cells with wt KvLQT1 and KvLQT1 G314S DNAs at a 1:1 ratio suppressed KvLQT1 current expression by 83.5 ± 4.3% (n= 7) (Figure 3C). The residual current amplitude of 13.6 ± 3.5 pA/pF (n = 7) at +40 mV was comparable to that obtained with CHO cells co-transfected with wt KvLQT1/KvLQT1 511–620 DNA at a 1:4 ratio.
For a control, we compared the current amplitudes in CHO cells transfected with Kv2.1 DNA with or without KvLQT1 511–620 DNA. The results showed that KvLQT1 511–620 DNA expression did not affect the amplitude of currents mediated by an unrelated Kv channel (Figure 3C). At +40 mV, the current amplitudes were 711.1 ± 142.7 pA/pF (n = 10) and 763.8 ± 196.7 pA/pF (n = 11) for Kv2.1 and Kv2.1 plus KvLQT1 511–620, respectively. The collective results indicated that a conserved C–terminal KvLQT1 domain was required for functional KvLQT1 channel expression. Co-expression of this domain with wt KvLQT1 subunits in Xenopus oocytes or in CHO cells suppressed the formation of functional KvLQT1 channels, possibly by interfering with the assembly of wt KvLQT1 subunits.
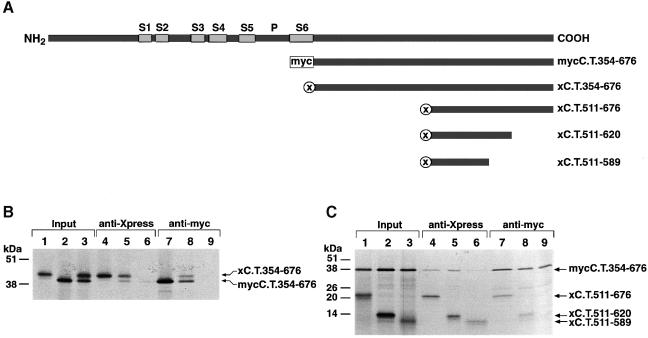
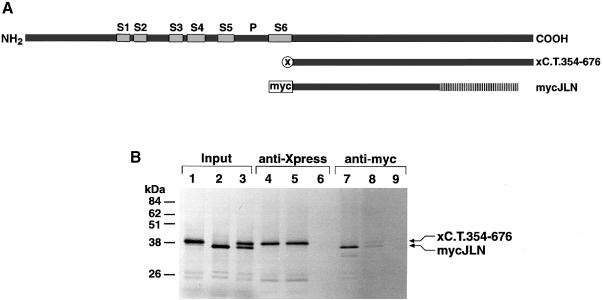
Cytoplasmic assembly domains of Shaker (Sh) and eag channels have the important property of self-interacting (Li et al., 1992; Ludwig et al., 1997). If the C–terminal KvLQT1 domain were an assembly domain, the KvLQT1 C–termini should also bind to each other in vitro. In a first attempt we tried to study the interaction of the putative KvLQT1 assembly domain in Escherichia coli expression systems similar to previous studies, which successfully expressed tetrameric complexes of the Sh assembly domain in E.coli (Shen and Pfaffinger, 1995). However, several attempts to express the KvLQT1 C–terminal domain(s) stably in E.coli expression systems failed. Therefore, we turned to a different approach and studied the self-interaction of KvLQT1 C–termini in co-immunoprecipitation experiments. For this purpose, we cloned the KvLQT1 C–terminus (amino acid residues 354–676) into myc–pcDNA3 and Xpress–pcDNA3HisC in order to obtain KvLQT1 C–termini tagged either with a myc tag (mycC.T.354–676) or with an Xpress tag (xC.T.354–676) (Figure 4A). In a coupled reaction, the corresponding cDNAs of mycC.T.354–676 and xC.T.354–676 were transcribed in vitro and translated in vitro using a reticulocyte lysate supplemented with [35S]methionine. The 35S–labelled protein translation products (Figure 4B) were co-immunoprecipitated either with anti-myc or anti-Xpress antibodies. Immunoprecipitates were analysed by SDS–PAGE followed by autoradiography. In controls, we showed that anti-myc antibodies precipitated only the myc-tagged and anti-Xpress antibodies only the Xpress-tagged KvLQT1 C–terminus (Figure 4B). When the myc- and Xpress-tagged C–termini of KvLQT1 were co-translated, the results showed that they were co-immunoprecipitated using either anti-myc or anti-Xpress antibodies (Figure 4B). This demonstrated that in vitro translated KvLQT1 C–termini interact with each other, consistent with the idea that the conserved C–terminal KvLQT1 domain constitutes an assembly domain.
Fig. 4. Co-assembly of tagged 35S–labelled C–terminal KvLQT1 fragments. (A) Diagram illustrating protein constructs tagged with myc (mycC.T.354–676) or Xpress (xC.T.354–676). Full-length KvLQT1 protein is shown on top as in Figure 1A. (B) 35S–labelled myc- and Xpress-tagged C–termini were obtained by in vitro translation (Input). They were precipitated either with anti-Xpress antibodies (anti-Xpress) or with anti-myc antibodies (anti-myc). Proteins were separated by SDS–PAGE followed by autoradiography. Molecular weight markers are on the left. Lanes 1, 4 and 9 contained only xC.T.354–676, lanes 2, 6 and 7 only mycC.T.354–676, and lanes 3, 5 and 8 xC.T.354–676 and mycC.T.354–676 as starting material. (C) 35S–labelled mycC.T.354–676 was co-translated with xC.T.511–676, xC.T.511–620 or xC.T.511–589. Lanes 1–3 show the mycC.T.354–676/xC.T.354–676 proteins (Input), which were precipitated either with anti-Xpress antibodies (lanes 4–6) or with anti-myc antibodies (lanes 7–9). SDS–PAGE and autoradiography were as in (B).
The self-interaction properties were tested further with constructs xC.T.511–676, xC.T.511–620 and xC.T.511–589 (Figure 4A). The constructs corresponded to either half the C–terminus, the conserved ∼100-amino-acid-long domain or a truncated version of the conserved domain, respectively. The corresponding cDNAs were transcribed and translated in vitro together with mycC.T.354–676 cDNA as above. The 35S–labelled protein translation products (Figure 4C) were immunoprecipitated with anti-myc or, alternatively, with anti-Xpress antibodies. Analysis of the immunoprecipitated protein materials showed that C–terminal sequence(s) between amino acid residues 511 and 620 was sufficient to interact with the myc-tagged C–terminus of KvLQT1 (Figure 4C). In contrast, the truncated C–terminus derived from the xC.T.511–589 construct did not bind detectably to the KvLQT1 C–terminus (Figure 4C). In agreement with the expression data showing that the vicinity of amino acid residue 590 is important for functional KvLQT1 channel expression, the co-immunoprecipitation experiments showed that the same region of KvLQT1 protein, and more specifically the domain between amino acids 590 and 620, is important in vitro for C–terminal complex formation. Collectively, the results suggested that KvLQT1 subunits with a C–terminal deletion are dysfunctional because of a defect in KvLQT1 subunit assembly.
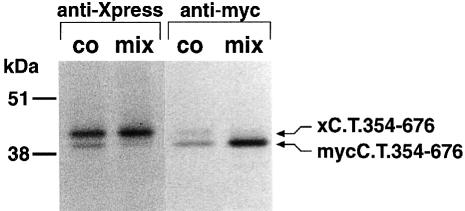
Co-assembly of differently tagged assembly domains of Sh channels took place only when they were translated together, not when the assembly domains had been synthesized separately and then mixed afterwards (Shen and Pfaffinger, 1995). A similar experiment is shown in Figure 5. 35S–labelled myc- and Xpress-tagged KvLQT1 C–termini were translated together in the same reaction tube and then immunoprecipitated. Alternatively, the two tagged KvLQT1 C–termini were synthesized in separate reaction tubes. Subsequently, they were mixed and immunoprecipitated. The results showed that the myc- and Xpress-tagged KvLQT1 C–termini were co-immunoprecipitated only when they had been co-translated. Both peptides were not co-immunoprecipitated when they had been synthesized separately and then mixed afterwards. Presumably, the C–terminal self-interaction is so strong that the C–terminal binding domains cannot be freely exchanged in mixing experiments.
Fig. 5. Interaction of 35S–labelled KvLQT1 C–termini with each other. cRNAs encoding Xpress-labelled (xC.T.354–676) or myc-labelled (mycC.T.354–676) KvLQT1 C–termini (see Figure 4A) were co-translated in vitro (co) or were translated in separate experiments and mixed afterwards (mix). Then they were precipitated either with anti-Xpress antibodies (anti-Xpress) or with anti-myc antibodies (anti-myc). Precipitates were analysed by SDS–PAGE followed by autoradiography. Molecular weight markers are on the left.
Mutations in the KvLQT1 gene have been found to be responsible for both inherited RWS and JLNS (Neyroud et al., 1997). This observation was striking, because patients with RWS suffer from an inherited autosomal dominant disorder, whereas JLNS constitutes a recessively transmitted disease. One JLNS mutation has been described that corresponds to a deletion–insertion at amino acid residue 544 in the conserved C–terminal KvLQT1 domain, leading to an alteration of the following ORF and a premature stop codon (Chouabe et al., 1997; Neyroud et al., 1997; Wollnik et al., 1997). We introduced this JLNS mutation into our mycC.T.354–676 construct (mycJLN) (Figure 6A) and investigated the capacity of the myc-tagged JLN C–terminus to assemble with Xpress-tagged wt C–terminus. 35S–labelled JLN and wt C–termini could not be co-immunoprecipitated (Figure 6B). The data showed that the JLNS-linked alteration of the C–terminal reading frame eliminated the binding activity of the C–terminal KvLQT1 assembly domain. In agreement with a previous report (Chouabe et al., 1997), introduction of the JLNS deletion–insertion mutation into KvLQT1 (KvLQT1–JLN) produced non-functional KvLQT1 subunits in CHO cell and Xenopus oocyte expression systems (data not shown), corroborating our observations that the conserved C–terminal KvLQT1 domain between residues 511 and 620 plays an important role in functional KvLQT1 expression.
Fig. 6. Effect of C–terminal JLN mutation on assembly of KvLQT1 C–terminus. (A) Diagram illustrating protein constructs tagged with Xpress (xC.T.354–676) or with myc (mycJLN). Full-length KvLQT1 protein is shown on top as in Figure 4A. The deletion–insertion mutation at KvLQT1 residue 544 (see Figure 2) is indicated by hatching. (B) 35S–labelled Xpress-tagged KvLQT1 and myc-tagged JLN C–termini were obtained by in vitro translation (Input). They were precipitated either with anti-Xpress antibodies (anti-Xpress) or with anti-myc antibodies (anti-myc). Proteins were separated by SDS–PAGE followed by autoradiography. Molecular weight markers are on the left. Lanes 1, 4 and 9 contained only xC.T.354–676, lanes 2, 6 and 7 only mycJLN, and lanes 3, 5 and 8 xC.T.354–676 and mycJLN as starting material.
We tested various non-functional KvLQT1 deletion constructs in co-expression experiments to assay their capacity to interfere with functional wt KvLQT1 channel expression. The results showed that co-expression of sKvLQT1Δ511–564, sKvLQT1Δ511–534 or sKvLQT1Δ354–510 subunits with wt KvLQT1 led to a complete suppression of wt KvLQT1 current expression (Figure 7A). In contrast, co–expression of wt KvLQT1 subunits with sKvLQT1Δ590–676 or sKvLQT1Δ511–620 led only to a partial reduction of KvLQT1 current amplitudes (∼50 ± 10% of control amplitudes; n = 5) (Figure 7B) and co-expression of wt KvLQT1 and sKvLQT1Δ115–589 produced normal KvLQT1 current amplitudes (Figure 7C). Also, co-expression of wt KvLQT1 and sKvLQT1 JLN at a ratio of 1:1 led to 64 ± 10% of control current amplitude (n = 5), while at a respective ratio of 2:1 it produced 109 ± 13% of control current amplitude (n = 6) (Figure 7B). We concluded from these results that the deletion mutants sKvLQT1Δ511–564, sKvLQT1Δ511–534 and sKvLQT1Δ354–510, though not functional, still bear the assembly domain and, thus, effectively interfered with KvLQT1 assembly. In contrast, the KvLQT1 mutants lacking residues between amino acids 590 and 620 did not effectively interfere with wt KvLQT1 channel expression, most likely because of defective assembly domain(s).
Fig. 7. Co-expression of sKvLQT1 with C–terminal KvLQT1 deletion constructs suppressed functional expression of KvLQT1 currents. (A) Data pooled from a number of experiments in which cRNAs encoding the deletion constructs sKvLQT1Δ511–564, sKvLQT1Δ511–534 and sKvLQT1Δ354–510 were co-expressed with sKvLQT1 (wt) cRNA in Xenopus oocytes. Peak amplitudes were measured following a 4 s test pulse to +40 mV from a holding potential of –80 mV. The bars represent the mean of measurements from 4–20 different oocytes. The 100% Irel value is as in Figure 1D. (B) Experiments were carried out as in (A) except that wt sKvLQT1 cRNA was co-injected with sKvLQT1Δ590–676, sKvLQT1Δ511–620, sKvLQT1Δ115–589 and sKvLQT1 JLN. Bars are as for (A). Means were derived from between 7 and 36 cells.
Discussion
In the present study we have investigated subunit assembly requirements of KvLQT1, a member of the KCNQ family of Kv channels, using the complementary methods of protein binding (co-immunoprecipitation assays) and functional expression of wt and mutant KvLQT1 subunits in heterologous expression systems (Xenopus oocytes and CHO cells). Previous expression studies with wt and an N–terminally truncated KvLQT1 isoform suggested that the cytoplasmic N–terminus of KvLQT1 did not contain a tetramerization domain like Sh channels (Demolombe et al., 1998). Our results show that assembly of KvLQT1 channels involves a cad that resides in a sequence region highly conserved among KCNQ family members, most likely between amino acids 590 and 620. The principal evidence for this conclusion is drawn from a number of observations: (i) differently tagged, 35S–labelled KvLQT1 C–termini bind to each other as shown by co-immunoprecipitation assays; in contrast, C–termini with a mutated or truncated cad do not bind; (ii) injections of subunits truncated from the C–terminus beyond residue 620 do not result in the expression of functional KvLQT1 channels; (iii) the cad domain exerts a dominant-negative effect on KvLQT1 channel expression; and (iv) cad mutations or deletions affect functional KvLQT1 channel expression.
Alternative explanations for the failure of KvLQT1 mutant constructs to produce functional KvLQT1 channels in heterologous expression systems might be: (i) inability of the cell to transport the protein to the plasma membrane; (ii) a default in the folding of the KvLQT1 polypeptide; and (iii) lack of post-translational modifications necessary for channel function. However, these reasons are unlikely to explain the dominant-negative effect of the C–terminal KvLQT1 region on KvLQT1 current expression since the N–terminus, which is present in the KvLQT1Δ115–589 construct, does not have this effect on KvLQT1. In addition, the KvLQT1 511–620 peptide has no dominant-negative effect on Kv2.1 channels expressed in the same heterologous expression systems. Furthermore, the co-immunoprecipitation experiments give a direct indication of binding to the KvLQT1 C–terminus and are therefore not subject to the above considerations.
In both the co-immunoprecipitation and the functional expression assays, it was apparent that deletion of residues 590–620 eliminated the cad–C–terminus interaction, abolished functional KvLQT1 channel expression and attenuated the dominant-negative effect on KvLQT1 channel expression in co-expression experiments. Thus, we have defined a minimal cad region for C–terminal KvLQT1 subunit interaction between residues 590 and 620. Conversely, co-expression of the cad region with wt KvLQT1 subunits exerted a dominant-negative effect on KvLQT1 channel expression, suggesting that important elements for the assembly of KvLQT1 subunits are contained within this region. Consistent with this idea are the observations that KvLQT1 subunits lacking a functional cad (e.g. sKvLQT1Δ590–676 and sKvLQT JLN) did not express KvLQT1 channels. Comparable cad mutations abolished binding to the wt KvLQT1 C–terminus in the co-immunoprecipitation assays and also dramatically affected the dominant-negative effect seen with mutant KvLQT1 subunits in the co-expression assay.
Previous studies with mutant Sh subunits suggest that Kv channel subunits may assemble in a random fashion to tetramers (Lichtinghagen et al., 1990; MacKinnon, 1991). Then, a dominant-negative effect on Kv channel expression implies that the presence of one non-functional subunit in a tetramer is sufficient to produce an inactive channel. In such a situation, the expression of wt and mutant subunits drastically suppresses 1:1 functional Kv channel expression. It is likely that this provides a molecular basis for understanding KvLQT1 subunit mutations associated with dominant forms of LQT syndrome. Our results showed that non-functional KvLQT1 subunits carrying deletions of C–terminal sequences between residues 354 and 564 (e.g. KvLQT1Δ354–510 and KvLQT1Δ511–564) behaved as subunits, which exert a dominant-negative effect on KvLQT1 channel expression. Indeed, these mutant subunits, though not functional, still possess the assembly domain and, thus, effectively interfered with KvLQT1 assembly. The results are in agreement with reports that missense mutations in the C–terminal 366–539 region of KvLQT1 protein are associated with dominant-negative forms of LQT syndrome (Donger et al., 1997; Tanaka et al., 1997). We did not find indications in the co-immunoprecipitation assays that the 366–539 region is involved in KvLQT1 subunit assembly. Other defaults, e.g. in gating, transport or surface expression, may be responsible for the observations that KvLQT1 subunits with deletions or missense mutations in the 366–539 region are non-functional. The exact mechanisms by which mutations in the 366–539 C–terminal KvLQT1 region cause a suppression of KvLQT1 channel function have yet to be determined.
Our results may provide a molecular explanation for the recessiveness of JLN mutations that were discovered in the KvLQT1 cad region (Chouabe et al., 1997; Neyroud et al., 1997). In particular, we have studied a JLN deletion–insertion mutation at residue 544, which leads to a modification of the following 107 amino acid residues and a premature stop codon (Neyroud et al., 1997). The mutation eliminates important parts of KvLQT1 cad, producing non-functional KvLQT1 subunits. Our co-immunoprecipitation data show that the cad of KvLQT1 JLN does not bind to the KvLQT1 C–terminus and that KvLQT1 JLN subunits do not suppress KvLQT1 channel expression in a dominant-negative manner. The results are comparable to a previous report that JLN mutations do not produce functional KvLQT1 channels, regardless of whether minK subunits are absent or present (Chouabe et al., 1997). Also, JLN subunits do not markedly affect the expression of wt KvLQT1 subunits (Chouabe et al., 1997; Wollnik et al., 1997; Mohammad-Panah et al., 1999). The notion that JLN mutants may be defective in subunit assembly is also consistent with the clinical observation that heterozygous JLN carriers only show slight cardiac dysfunctions (Fraser et al., 1964; Neyroud et al., 1997) and that the severe JLNS phenotype is characterized by the complete or almost complete absence of KvLQT1 channel.
Materials and methods
Molecular biology and cloning
The deletion and truncation mutants of human KvLQT1 were constructed using standard PCR techniques. N- and C–terminal primers carried a ribosomal binding site (Kozak, 1986) followed by a translation initiation codon and a termination codon, respectively. KvLQT1 deletion constructs are designated KvLQT1Δx–y, where x and y indicate the first and last deleted KvLQT1 residue according to the complete human KvLQT1 sequence (Chouabe et al., 1997; Yang et al., 1997). All constructs were sequenced using an ABI 377 DNA sequencer.
For expression in Xenopus oocytes, the PCR fragments were cloned with blunt ends into the SmaI site of pGEMHE. The mRNAs were prepared from NotI-linearized KvLQT1 wild-type and mutant constructs in pGemHE using an Ambion T7 m–Message Machine kit according to the manufacturer's instructions. RNA concentration was determined using the RiboGreen RNA quantitation kit (Molecular Probes) and inspected for purity by gel electrophoresis.
For immunoprecipitation experiments, deletion/truncation mutants lacking the N–terminus and the core region were fused to a myc tag (EQKLISEEDLN) and cloned into the pcDNA3.1 plasmid or fused to an Xpress tag (DLYDDDDK) via cloning into pcDNA3.1HisA,B,C plasmids (Invitrogen).
In vitro translation and immunoprecipitation experiments
35S–labelled proteins were synthesized from the respective cDNAs by in vitro translation in the TNT® Coupled Reticulocyte Lysate System (Promega). Differently tagged entire C–termini and mutants were either co-translated or translated separately. Aliquots of the translation reaction were diluted 40–fold into 1% Triton buffer (10 mM Tris pH 7.9, 140 mM NaCl, 1% Triton X-100, 0.1% bovine hemoglobin) in pre-coated reaction tubes (Triton buffer with 1% bovine hemoglobin). The proteins were immunoprecipitated by the addition of anti-myc (9E10; Boehringer Mannheim) or anti-Xpress (Invitrogen) antibody, respectively, for 90 min, followed by protein A–Sepharose (Pharmacia) for 90 min at room temperature. Immunoprecipitates were washed twice in high-salt Triton buffer (2 M NaCl), followed by Triton buffer, low-salt buffer (10 mM Tris pH 7.9, 140 mM NaCl) and 50 mM Tris pH 6.8. The samples were boiled in SDS sample buffer, and analysed on 12 or 15% SDS–polyacrylamide gels. Labelled proteins were visualized by autoradiography.
Cell culture and transfection
CHO cells were seeded on poly-d-lysine-coated glass coverslips in a 24-multiwell plate and grown in Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, 10% fetal calf serum (FCS) and antibiotics. Transfection was performed using 2 μl of lipofectamine (Gibco–BRL) according to the manufacturer's protocol and with 0.5 μg of the respective channel cDNA plasmids together with 0.5 μg of pIRES–CD8 (kindly provided by Dr A.Patel) as a marker for transfection. Transfected cells were visualized 48 h following transfection, using the anti-CD8 antibody-coated beads method (Jurman et al., 1994).
RNA injection into Xenopus oocytes
Frogs were anaesthetized with 0.12% tricaine (Sigma). Pieces of ovary were surgically removed and digested with 1.3 mg/ml collagenase A (Boehringer Mannheim) in Ca2+-free OR2 solution (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES; pH 7.5 NaOH) for 2 h to remove follicular cells. Stage IV and V oocytes were injected with 50 nl of cRNA or cRNA mixtures. Concentrations in experiments were 0.025–0.1 μg/μl (1–5 ng/oocyte). In co-expression experiments the wt cRNA concentration was kept constant and equal molar amounts of mutant cRNA were added. Oocytes were maintained at 18°C in Modified Barth's Solution [88 mM NaCl, 1 mM KCl, 0.4 mM CaCl2, 1 mM MgSO4, 0.33 mM Ca(NO3)2, 2.4 mM NaHCO3, 10 mM HEPES, pH 7.5 NaOH] supplemented with 50 μg/ml gentamycin (Sigma).
Electrophysiological recordings
Current measurements in Xenopus oocytes were performed 3 days after cRNA injection. Oocytes were bathed in an ND96 (96 mM NaCl, 2 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 5 mM HEPES, pH 7.5 NaOH) solution. Standard two-electrode voltage experiments were carried out at room temperature using an OC–725C amplifier (Warner Instruments) connected to a PowerMac computer via an ITC–16 interface (Instrutech Corp.) Microelectrodes were filled with 2 M KCl and had resistances of 0.5–1.0 MΩ. Voltage protocols were applied using PULSE 8.30 software (HEKA). Four-second voltage pulses to +40 mV were applied and the maximum amplitude was measured using PULSEFIT 8.30 software (HEKA) and normalized with respect to the wild-type control. The holding potential was held at –80 mV. Data are expressed as means ± SEM (n is the number of oocytes).
Current measurements in CHO cells were performed 48 h following transfection, using the whole-cell configuration of the patch–clamp technique (Hamill and Sakman, 1981). Signals were amplified using an Axopatch 200B patch–clamp amplifier (Axon Instruments), sampled at 1–5 kHz and filtered at 0.4–2 kHz via a 4–pole Bessel low pass filter, according to the experimental protocol. Data were acquired using pClamp 6.0.2 software (Axon Instruments) and an IBM-compatible 486 computer in conjunction with a DigiData 1200 interface (Axon Instruments). The patch pipettes were pulled from borosilicate glass (fibre filled) with a resistance of 4–8 MΩ and were filled with (in mM): 110 potassium gluconate, 2 KCl, 1 MgCl2, 5 KATP, 5 EGTA and 10 HEPES at pH 7.4. The external solution contained (in mM): 140 NaCl, 2 KCl, 1.8 CaCl2, 1.2 MgCl2, 11 glucose and 5.5 HEPES at pH 7.4. All data were expressed as the mean ± SEM.
Acknowledgments
Acknowledgements
We thank Susan Hoffmann for cloning KvLQT1 and KvLQT1Δ1–127, and Dörte Clausen for producing the figures. This work was supported by the German–Israeli Foundation for Scientific Research and Development (to O.P. and B.A.), the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie (O.P.), and the Minerva Foundation and the Israeli Ministry of Health (B.A.).
References
- Attali B. (1996) A new wave for heart rhythms. Nature, 384, 24–25. [DOI] [PubMed] [Google Scholar]
- Attali B., Guillemare, E., Lesage, F., Honore, E., Romey, G., Lazdunski, M. and Barhanin, J. (1993) The protein IsK is a dual activator of K+ and Cl– channels. Nature, 365, 850–852. [DOI] [PubMed] [Google Scholar]
- Barhanin J., Lesage, F., Guillemare, E., Fink, M., Lazdunski, M. and Romey, G. (1996) K(V)LQT1 and IsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature, 384, 78–80. [DOI] [PubMed] [Google Scholar]
- Biervert C., Schroeder, B.C., Kubisch, C., Berkovic, S.F., Propping, P., Jentsch, T.J. and Steinlein, O.K. (1998) A potassium channel mutation in neonatal human epilepsy. Science, 279, 403–406. [DOI] [PubMed] [Google Scholar]
- Busch A.E. and Suessbrich, H. (1997) Role of the ISK protein in the IminK channel complex. Trends Pharmacol. Sci., 18, 26–29. [DOI] [PubMed] [Google Scholar]
- Charlier C., Singh, N.A., Ryan, S.G., Lewis, T.B., Reus, B.E., Leach, R.J. and Leppert, M. (1998) A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nature Genet., 18, 53–55. [DOI] [PubMed] [Google Scholar]
- Chouabe C., Neyroud, N., Guicheney, P., Lazdunski, M., Romey, G. and Barhanin, J. (1997) Properties of KvLQT1 K+ channel mutations in Romano–Ward and Jervell and Lange-Nielsen inherited cardiac arrhythmias. EMBO J., 16, 5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demolombe S. et al. (1998) A dominant negative isoform of the long QT syndrome 1 gene product. J. Biol. Chem., 273, 6837–6843. [DOI] [PubMed] [Google Scholar]
- Donger C. et al. (1997) KvLQT1 C–terminal missense mutation causes a forme fruste long-QT syndrome. Circulation, 96, 2778–2781. [DOI] [PubMed] [Google Scholar]
- Duggal P., Vesely, M.R., Wattanasirichaigoon, D., Villafane, J., Kaushik, V. and Beggs, A.H. (1998) Mutation of the gene for IsK associated with both Jervell and Lange-Nielsen and Romano–Ward forms of Long-QT syndrome. Circulation, 97, 142–146. [DOI] [PubMed] [Google Scholar]
- Fraser G.R., Frogatt, P. and Murphy, T. (1964) Genetical aspects of the cardioauditory syndrome of Jervell and Lange-Nielsen (congenital deafness and electrocardiographic abnormalities). Ann. Hum. Genet., 28, 133–157. [DOI] [PubMed] [Google Scholar]
- Hamill O.P. and Sakmann, B. (1981) Multiple conductance states of single acetylcholine receptor channels in embryonic muscle cells. Nature, 294, 462–464. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tanaka, T., Nagai, R., Kikuchi, K., Ogawa, S., Okada, S., Yamagata, S., Yano, Y. and Nakamura, Y. (1998) Genomic organization and mutational analysis of KvLQT1, a gene responsible for familial long QT syndrome. Hum. Genet., 103, 290–294. [DOI] [PubMed] [Google Scholar]
- Jervell A. and Lange-Nielsen, F. (1957) Congenital deaf mutism, functional heart disease with prolongation of the QT interval and sudden death. Am. Heart J., 54, 59–78. [DOI] [PubMed] [Google Scholar]
- Jurman M.E., Boland, L.M., Liu, Y. and Yellen, G. (1994) Visual identification of individual transfected cells for electrophysiology using antibody-coated beads. Biotechniques, 17, 876–881. [PubMed] [Google Scholar]
- Kacmarek L.K. and Blumenthal, E.M. (1997) Properties and regulation of the minK potassium channel protein. Physiol. Rev., 77, 627–641. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kubisch C., Schroeder, B.C., Friedrich, T., Lutjohann, B., El–Amraoui, A., Marlin, S., Petit, C. and Jentsch, T.J. (1999) KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell, 96, 437–446. [DOI] [PubMed] [Google Scholar]
- Lee M.P., Hu, R.J., Johnson, L.A. and Feinberg, A.P. (1997) Human KvLQT1 gene shows tissue-specific imprinting and encompasses Beckwith–Wiedemann syndrome chromosomal rearrangements. Nature Genet., 15, 181–185. [DOI] [PubMed] [Google Scholar]
- Lesage F., Attali, B., Lakey, J., Honore, E., Romey, G., Faurobert, E., Lazdunski, M. and Barhanin, J. (1993) Are Xenopus oocytes unique in displaying functional IsK channel heterologous expression? Receptors Channels, 1, 143–152. [PubMed] [Google Scholar]
- Li H. et al. (1998) New mutations in KvLQT1 potassium channel that cause long-QT syndrome. Circulation, 97, 1264–1269. [DOI] [PubMed] [Google Scholar]
- Li M., Jan, Y.N. and Jan, L.Y. (1992) Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science, 257, 1225–1230. [DOI] [PubMed] [Google Scholar]
- Lichtinghagen R., Stocker, M., Wittka, R., Boheim, G., Stühmer, W., Ferrus, A. and Pongs, O. (1990) Molecular basis of altered excitability in Shaker mutants of Drosophila melanogaster.EMBO J., 9, 4399–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig J., Owen, D. and Pongs, O. (1997) Carboxy-terminal domain mediates assembly of the voltage-gated rat ether-à-go-go potassium channel. EMBO J., 16, 6337–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon R. (1991) New insights into the structure and function of potassium channels. Curr. Opin. Neurobiol., 1, 14–19. [DOI] [PubMed] [Google Scholar]
- Mohammad-Panah R., Demolombe, S., Neyroud, N., Guicheney, P., Kyndt, F., van den Hoff, M., Baró, I. and Escande, D. (1999) Mutations in a dominant-negative isoform correlate with phenotype in inherited cardiac arrhythmias. Am. J. Hum. Genet., 64, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyroud N. et al. (1997) A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nature Genet., 15, 186–189. [DOI] [PubMed] [Google Scholar]
- Neyroud N. et al. (1999) Genomic organization of the KCNQ1 K+ channel gene and identification of C–terminal mutations in the long-QT syndrome. Circ. Res., 84, 290–297. [DOI] [PubMed] [Google Scholar]
- Romano C. (1963) Congenital cardiac arrhythmias. Lancet, 1, 658. [DOI] [PubMed] [Google Scholar]
- Sanguinetti M.C., Curran, M.E., Zou, A., Shen, J., Spector, P.S., Atkinson, D.L. and Keating, M.T. (1996) Coassembly of KvLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature, 384, 80–83. [DOI] [PubMed] [Google Scholar]
- Schulze-Bahr E., Haverkamp, W., Wedeking, H., Rubie, C., Hordt, M., Borggrefe, M., Assman, G., Breithardt, G. and Funke, H. (1997) Autosomal recessive long-QT syndrome (Jervell Lange-Nielsen syndrome) is genetically heterogeneous. Hum. Genet., 100, 573–576. [DOI] [PubMed] [Google Scholar]
- Shen N.V. and Pfaffinger, P.J. (1995) Molecular recognition and assembly sequences involved in the subfamily-specific assembly of voltage-gated K+ channel subunit proteins. Neuron, 14, 625–633. [DOI] [PubMed] [Google Scholar]
- Singh N.A. et al. (1998) A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nature Genet., 18, 25–29. [DOI] [PubMed] [Google Scholar]
- Splawski I., Tristani-Firouzi, M., Lehmann, M.H., Sanguinetti, M.C. and Keating, M.T. (1997) Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nature Genet., 17, 338–340. [DOI] [PubMed] [Google Scholar]
- Takumi T., Ohkubo, H. and Nakanishi, S. (1988) Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science, 242, 1042–1045. [DOI] [PubMed] [Google Scholar]
- Tanaka T. et al. (1997) Four novel KVLQT1 and four novel HERG mutations in familial long-QT syndrome. Circulation, 95, 565–567. [DOI] [PubMed] [Google Scholar]
- Tyson J. et al. (1997) IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause Jervell and Lange-Nielsen syndrome. Hum. Mol. Genet., 6, 2179–2185. [DOI] [PubMed] [Google Scholar]
- Vetter D.E. et al. (1996) Inner ear defects induced by null mutation of the isk gene. Neuron, 17, 1251–1264. [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. (1996) Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nature Genet., 12, 17–23. [DOI] [PubMed] [Google Scholar]
- Wang Q., Chen, Q. and Towbin, J.A. (1998) Genetics, molecular mechanisms and management of long QT syndrome. Ann. Med., 30, 58–65. [DOI] [PubMed] [Google Scholar]
- Ward O.C. (1964) A new familial cardiac syndrome in children. J. Ir. Med. Assoc., 54, 103–106. [PubMed] [Google Scholar]
- Wei A., Jegla, T. and Salkoff, L. (1996) Eight potassium channel families revealed by the C.elegans genome project. Neuropharmacology, 35, 805–829. [DOI] [PubMed] [Google Scholar]
- Wollnik B., Schroeder, B.C., Kubisch, C., Esperer, H.D., Wieacker, P. and Jentsch, T.J. (1997) Pathophysiological mechanisms of dominant and recessive KvLQT1 K+ channel mutations found in inherited cardiac arrhythmias. Hum. Mol. Genet., 6, 1943–1949. [DOI] [PubMed] [Google Scholar]
- Yang W.P., Levesque, P.C., Little, W.A., Conder, M.L., Shalaby, F.Y. and Blanar, M.A. (1997) KvLQT1, a voltage-gated potassium channel responsible for human cardiac arrhythmias. Proc. Natl Acad. Sci. USA, 94, 4017–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]