Abstract
Peptides are preferred for designing inhibitors because of their high activity and specificity. Seven cyclopentapeptide inhibitors were designed in this study against dengue virus type 2 (DEN-2) NS3-NS2B protease: CKRRC, CGRRC, CRGRC, CRTRC, CTRRC, CKRKC and CRRKC. Docking analysis was performed to study the enzyme-inhibitor binding interactions. The free energy binding and estimated Ki values for all the inhibitors were found to be small (within micromolar range), indicating that the inhibitors bind considerably well to the binding site. The results showed that the cyclopentapeptide CKRKC was the best peptide inhibitor candidate with estimated free binding energy of -8.39 kcal/mol and Ki of 0.707 µM when compared to the standard inhibitor Bz-Nle-Lys-Arg-Arg-H that has been experimentally tested and shown to exhibit Ki value of 5.8 µM. Several modes of weak interactions were observed between the cyclopentapeptide CKRKC and the active site of DEN-2 NS3-NS2B protease. Thus, the cyclopentapeptide is proposed as a potential inhibitor to the NS3-NS2B protease activities of DEN-2. While these preliminary results are promising, further experimental investigation is necessary to validate the results.
Keywords: Dengue virus, DEN-2 NS3-NS2B protease, docking, inhibitor, peptide
Background
Dengue disease, caused by dengue virus infection, has emerged as an endemic in over 100 countries, especially in the tropical and subtropical regions, with more than 2.5 billion people or 40% of the world population at risk. Transmitted mainly by the Aedes aegypti mosquito, dengue virus is maintained in a cycle that involves human and the mosquito [1]. The infection produces clinical illness ranging from harmless typical fever (DF) to a fatal hemorrhagic fever (DHF) characterized by capillary leakage and thrombocytopenia. Moreover, it may manifest into failure of the circulation system and shock, termed as dengue shock syndrome (DSS) that may lead to death [2]. It is estimated that about 100 million dengue fever (DF) cases occur per year and about 250,000 ‐ 500,000 cases are of the severe form dengue hemorrhagic fever (DHF) [3].
Dengue virus is a member of the genus Flavivirus and family Flaviviridae that consists of four antigenically distinct viral serotypes, DEN-1, DEN-2, DEN-3 and DEN-4 [4]. Lifelong immunity to one serotype will prevent infection by the same serotype but does not provide protection from a secondary infection of a different serotype. The antibody-dependent enhancement (ADE) pathogenesis hypothesis proposes that secondary infection with a different serotype increases the risk of developing DHF and DSS [5].
The dengue virus genome is a single-stranded positive-sense RNA that consists of 10,723 nucleotides and encodes a single polyprotein precursor of 3,391 amino acids. The polyprotein is arranged in the order of C-prM-ENS1-NS2A-NS2B-NS3-NS4A- NS4B-NS5 consisting of three structural proteins (C, prM and E) and seven non-structural proteins (NS) that are cleaved co- and post-translationally [6]. The correct cleavage of this polyprotein requires both host cell and virus protease. The NS3 protease is essential for the polyprotein processing which needs the presence of NS2B cofactor for its optimal catalytic activity. It has been indicated that the catalytic triad of this protease is His51, Asp75 and Ser135, which is typical of a serine protease family [7].
The conjugate NS3-NS2B has been shown to cleave the precursor polyprotein at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B, and NS4B/NS5 junctions as well as at internal sites within C, NS2A, NS3, and NS4A [8]. These sites have two basic amino acid residues (Arg-Arg, Arg-Lys, Lys-Arg, or occasionally Gln-Arg) at the P2 and P1 positions, respectively [9]. Other studies have indicated that the P1 position has a strong preference for basic amino acid residues (Arg/Lys), whereas the preference for P2 is Arg > Thr > Gln/Asn/Lys, while for P3 is Lys > Arg > Asn [10] (see Schecter and Berger for the abbreviation of P1, P2, and P3). Until today, there is no vaccine or anti-dengue viral drug currently available in the market against dengue infection. Therefore, there is a need for the design of antiviral agents to fight this disease. One of the antiviral agent strategies involves blocking and inhibiting viral enzyme activity, thus preventing virus replication [2, 11].
Several groups have synthesized various types of compound to inhibit the NS3-NS2B protease activities. One of them is a peptidic α-keto-amide compound Ac-FAAGRR-α-keto-SL-CONH2, with Ki value of 47 μM [12], and some others include aldehyde compounds resulting in low micromolar Ki value of up to 1.5 μM [13]. Some natural compounds, such as 'chalcones, isolated from Boesenbergia rotunda, have shown noncompetitive inhibitory activity against the NS3-NS2B protease. Its cyclohexenyl chalcone derivates, 4-hydroxypanduratin A and panduratin A, showed good competitive inhibitory activities against DEN-2 virus NS3-NS2B protease, exhibiting Ki values of 21 and 25 μM, respectively [14].
In this in silico study, we designed peptide-based cyclic inhibitors which showed inhibitory activities toward DEN-2 NS3-NS2B protease. The binding interactions between these inhibitors and NS3-NS2B protease were studied by docking methods using AutoDock 4.0 software. The aim of this study was to get a better peptide-based drug ligand that could inhibit polyprotein processing of DEN-2, and also to better understand the interactions between the inhibitor and the enzyme's binding sites via computational docking methods. We hope that the results from this study will provide insight about the design of antiviral drugs for dengue virus.
Methodology
Designing peptide-based ligands
The peptide ligands were designed based on substrate specificity for NS3- NS2B protease and the analysis of binding pocket of NS3-NS2B protease catalytic site. The specific binding site on the enzyme results in selectivity for positively charged amino acid residues of the substrates. Jun Li, et al., [10] reported that the P1 position of the enzyme's binding site has a strong preference for basic amino acid residues (Arg/Lys), whereas the preference for the P2 is Arg > Thr > Gln/Asn/Lys and for P3 is Lys > Arg > Asn. We, therefore, used this combination to determine our peptide ligands. Our ligands consist of five amino acid residues (pentapeptide) cyclized by an SS disulfide bridge. As a positive control, we used the standard ligand BzNle-Lys-Arg-Arg-H that has been experimentally tested and shown to exhibit Ki value of 5.8 µM [13].
Building ligand structures
The peptide ligand structures were built using Hyperchem Pro 8.0 software. Amino acid residues were chosen from the dialog box in the database menu to draw the two dimensional structure of the ligands and thereafter the ligands were modeled into three dimensional structures.
Preparing enzyme structure
The NS2B cofactor is essential for the protease catalytic activity of NS3. The three dimensional structure of this protease associated with NS2B cofactor available in the Protein Data Bank (http://www.rcsb.org/pdb) (accession code: 2FOM) was used for this study. Water, chlorine atoms and glycerol molecules were removed from the crystal structure to reduce the complexities in the binding studies.
Ligand structures optimization
The ligand structures were minimized with Hyperchem Pro 8.0 software using conjugate gradient Polak-Ribiere method to obtain the most stable structure geometry.
Enzyme structure optimization
All hydrogen atoms were added to the enzyme structure to define the correct ionization and tautomeric states of amino acid residues such as Asp, Ser, Glu, Arg and His using HyperChem Pro 8.0 software [15]. Enzyme structure was minimized with the same software using the steepest descent and conjugate gradient methods. This two-step procedure was set up to bring the energy level of the system to minimum and structurally stable.
Protein-ligand docking
The docking of ligands to the catalytic triad of NS3-NS2B protease was performed using AutoDock 4.0 software (http://autodock.scripps.edu/). AutoDock is reported to be the most popular docking program [16] and is reliable [17]. Using the software, polar hydrogen atoms were added to the enzyme and its nonpolar hydrogen atoms were merged, whereas for the ligand, nonpolar hydrogen atoms were merged and Gasteiger charges were added. All rotatable bonds of ligands were set to be rotatable. All calculation for protein-fixed ligand-flexible docking was done using the Lamarckian Genetic Algorithm (LGA) method. A population size of 150 and 10 millions energy evaluations were used for 100 search runs. The grid box with a dimension of 60 × 60 × 60 points and 0.375 Å grid spacing was used around the catalytic triad to cover the entire enzyme binding site and accommodate ligands to move freely. After the docking searches were completed, clustering histogram analysis was performed based on an RMSD (root mean square deviation) of not more than 1.5 Å. The best conformation was chosen from the most populated cluster with the lowest docked energy. The interactions of complex enzyme-ligand conformations, including hydrogen bond and other interactions, were analysed using Viewerlite (www.accelrys.com) and UCSF Chimera software (www.cgl.ucsf.edu/chimera/).
Discussion
Designing peptide-based ligands
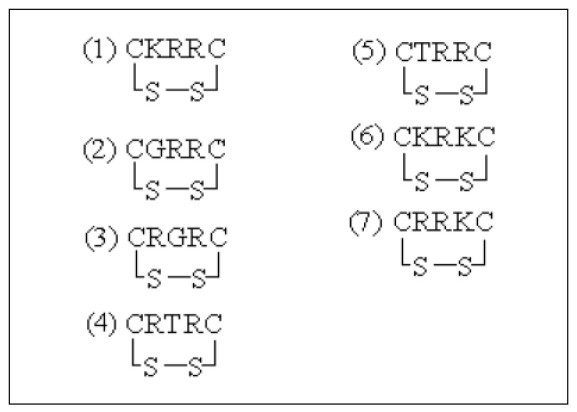
The seven cyclopentapeptides that we designed are illustrated in Figure 1. Despite the low stability, the peptides were suitable for design of drug inhibitors (ligands) because of their high activity and specificity. We cyclized our ligands by use of an S-S disulfide bridge in order to increase their stability [18]. This cyclic formation is likely to increase the hydrophobic interaction within the peptides by lowering the hydrogen interactions with water (solvent), which as a result is likely to increase the total entropy and stability of the peptide. Cyclic peptide with a small size will be more stable to the enzymatic activity from lumen secretion in stomach and duodenum that can degrade the peptide ligand.
Figure 1.
Amino acid sequences of seven designed cyclopentapeptides.
Estimated docking energy
Overall, the free energy of binding (ΔGbind) for all the designed ligands were lower than the standard ligand with significant lowest value of -8.39 kcal/mol, obtained for the ligand CKRKC (Table 1, see supplementary material). The negative and low value of ΔGbind indicates strong favorable bond between enzyme and ligand. The standard ligand showed the highest ΔGbind because the standard ligand has more rotatable bonds, thus increasing its torsional free energy and lowering its binding affinity. These ΔGbind values are parallel to the Ki values observed in Table 1 (see supplementary material). The ligand CKRKC showed the lowest Ki value, indicating that the reaction equilibrium shifted to the formation of complex. In general, all calculated Ki values were reasonably small (within micromolar range), indicating the formation of stable enzyme-ligand complexes. All the ligands showed reasonable low internal energy, indicating that the ligands were in their most favourable conformations.
Binding interaction between enzyme and ligand
Ligand CKRKC
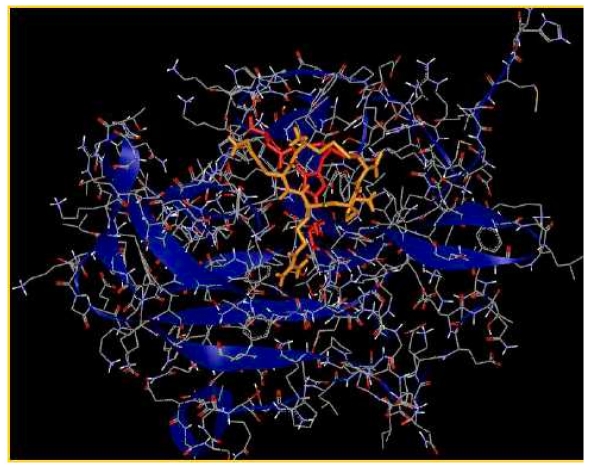
Figure 2 provides a view of ligand CKRKC at the binding site of enzyme. The cumulative effect of many weak interactions within a complex enzyme-ligand could be very significant to the stability of the complex. Table 2 (see supplementary material) shows that there are 9 residues in the active site of NS3/NS2B protease that are involved in hydrogen bonding with the ligand CKRKC, in comparison to only 8 residues with the standard ligand, therefore contributing to the stronger bond of complex enzyme-ligand for CKRKC compared to the complex enzyme-standard ligand. The hydrogen bonding with His51 and Asp75 to ligand CKRKC could disrupt the enzyme activity. Henceforth, the ligand could act as a potential competitive inhibitor for NS3-NS2B protease.
Figure 2.
View of ligand CKRKC (orange) at the binding site of enzyme (blue ribbons). Catalytic triads are shown as red sticks.
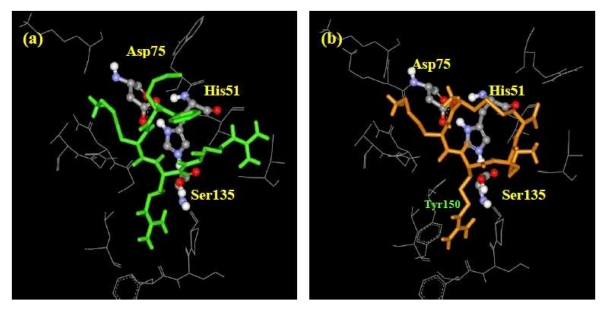
The contact residues of the enzyme to ligand CKRKC and the standard ligand are shown in (see Table 3supplementary material). As Table 2(see supplementary material) only accounts for hydrogen bond, Table 3 provides all weak interactions within the complex enzyme-ligand, such as hydrogen bond, ionic bond, van der Waals interaction, and hydrophobic interaction. Both the CKRKC and the standard ligands interacted with the enzyme's catalytic triad (His51, Asp75, and Ser135). These interactions could interrupt electron transfer between the carboxyl group of Asp75 and nitrogen atom on imidazole group of His51, possibly disrupting the ability of His51 to activate nucleophilic attack of hydroxyl group (β-OH) of Ser135, which is essential for the initiation of proteolysis. Between the standard ligand and ligand CKRKC, the weak interaction of Tyr150 residue was only observed to CKRKC (Figure 3). We propose that π electron of aromatic group Tyr150 interacted with side chain of arginine residue of ligand CKRKC that has a conjugate π bond, so it could form π-π interaction.
Figure 3.
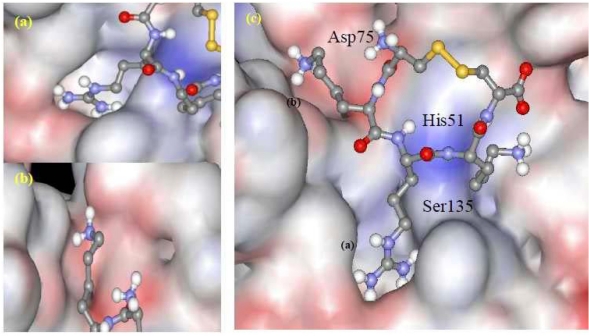
View of ligand conformation at the binding site. (a) Standard ligand, (b) Ligand CKRKC. Contact residues of the enzyme that interact with the ligands are shown as grey sticks. Catalytic triads are shown as balls and sticks.
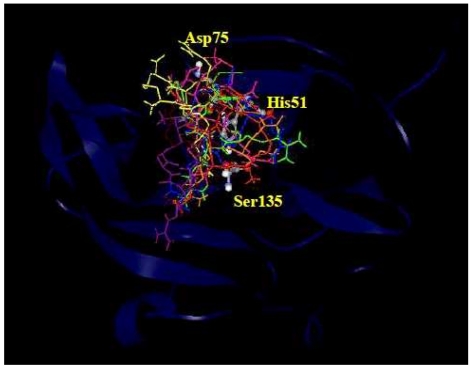
The orientations of all the ligands in the binding site of NS3-NS2B protease are provided in Figure 4. The shape of the binding pocket of enzyme was observed to complement the shape or pose of the ligand CKRKC (Figure 5). The side chain of the arginine and lysine residues of ligand CKRKC which had positive charge and long chain were located at the cleft of the binding pocket (Figure 5a, 5b), which was narrow and had negative charge. The different charge may result in electrostatic interaction between the negative charge of the binding pocket and the ‐NH3+ group of the arginine and lysine residues of the ligand, thus increasing affinity of interaction and complex enzyme-ligand stability.
Figure 4.
All the ligands are superimposed at the catalytic site. DEN-2 NS3-NS2B protease is represented as blue ribbons. The catalytic triads are labeled as His51, Asp75, and Ser135 and are shown as balls and sticks.
Figure 5.
(a) & (b) View of ligand's side chain in the binding pocket from a different angle. (c) Connolly surface representation of ligand CKRKC at the binding site.
Surprisingly, the backbone pose of the ligand CKRKC (Figure 5c) was also in complement to the enzyme's binding site. The peptide backbone was protruded out toward the surface of the enzyme, while the side chains were embedded into the pocket of the enzyme. This orientation was contributed by the low free energy for binding, indicating the formation of a favourable stable enzyme-ligand complex. Overall, this showed that the conformation of the complex complemented the specificity of enzyme's binding pocket (lock-and-key model), therefore the ligand cyclopentapeptide CKRKC could be proposed as a potential inhibitor to the serine protease activities of DEN2 better than the standard ligand
Despite the significant sequence diversity between the dengue serotypes [19], the binding pocket of the NS3-NS2B protease for the four serotypes share similar substrate specificities, suggesting that the ligand is likely to act as an inhibitor for NS3 proteases of the four serotypes [10]. While these preliminary results are promising, further experimental investigation are necessary to validate the docking result.
Conclusions
In this study, we designed seven cyclopentapeptide inhibitors against DEN-2 NS3-NS2B protease: CKRRC, CGRRC, CRGRC, CRTRC, CTRRC, CKRKC and CRRKC. Docking studies on these inhibitors against the protease showed that the free energy binding and estimated Ki values for all the inhibitors were small (within micromolar range), indicating that the ligands bind favourably to the binding site. The cyclopentapeptide CKRKC was observed as the best peptide inhibitor candidate, with value of free energy binding -8.39 kcal/mol and estimated inhibition constant Ki of 0.707 µM compared to the standard inhibitor. Several modes of weak interactions were observed between the cyclopentapeptide CKRKC and the active site of DEN-2 NS3-NS2B protease. Thus, the cyclopentapeptide CKRKC is a potential inhibitor to the NS3-NS2B protease activities of DEN-2.
Supplementary material
Acknowledgments
We are grateful to Ridla Bakri PhD, Chairman of Department of Chemistry, Faculty of Mathematics and Natural Science, University of Indonesia for his support of this research.
Footnotes
Citation:Tambunan & Alamudi, Bioinformation 5(6): 250-254 (2010)
References
- 1.Melino S, Paci M. FEBS J. 2007;274:2986. doi: 10.1111/j.1742-4658.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JR, Rico-Hesse R. Am J Trop Med Hyg. 2006;75:886. [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. Novartis Found Symp. 2006;277:3. doi: 10.1002/0470058005.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Bennett SN, et al. J Gen Virol. 2006;87:885. doi: 10.1099/vir.0.81309-0. [DOI] [PubMed] [Google Scholar]
- 5.Kliks SC, et al. Am J Trop Med Hyg. 1989;42:444. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 6.Irie K, et al. Gene. 1989;75:197. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 7.Brinkworth RI, et al. J Gen Virol. 1999;80:1169. doi: 10.1099/0022-1317-80-5-1167. [DOI] [PubMed] [Google Scholar]
- 8.Chambers TJ, et al. Proc Natl Acad Sci. 1990;87:8898. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers TJ, et al. Annu Rev Microbiol. 1990;44:649. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 10.Li J, et al. J Biol Chem. 2005;280:28766. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- 11.Keller TH, et al. Novartis Found Symp. 2006;277:102. [PubMed] [Google Scholar]
- 12.Leung D, et al. J Biol Chem. 2001;276:45762. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- 13.Yin Z, et al. Bioorg Med Chem Lett. 2006;16:40. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 14.Kiat TS, et al. Bioorg Med Chem Lett. 2006;16:3337. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 15.Annamala MK, et al. Bioinformation. 2008;9:339. doi: 10.6026/97320630001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sousa SF, et al. Proteins. 2006;65:15. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 17.Hetenyi C, Spoel VD. Protein Sci. 2002;11:1729. doi: 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell AJ, et al. Pharm Res. 2009;26:2186. doi: 10.1007/s11095-009-9933-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan AM, et al. PLoS Negl Trop Dis. 2008;2:e272. doi: 10.1371/journal.pntd.0000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.