Abstract
Liver-specific ablation of cytochrome P450 reductase in mice (LCN) results in hepatic steatosis that can progress to steatohepatitis characterized by inflammation and fibrosis. The specific cause of the fatty liver phenotype is poorly understood but is hypothesized to result from elevated expression of genes encoding fatty acid synthetic genes. Since expression of these genes is known to be suppressed by polyunsaturated fatty acids, we performed physiological and genomics studies to evaluate the effects of dietary linoleic and linolenic fatty acids (PUFA) or arachidonic and decosahexaenoic acids (HUFA) on the hepatic phenotypes of control and LCN mice by comparison with a diet enriched in saturated fatty acids. The dietary interventions with HUFA reduced the fatty liver phenotype in livers of LCN mice and altered the gene expression patterns in these livers to more closely resemble those of control mice. Importantly, the expression of genes encoding lipid pathway enzymes were not different between controls and LCN livers, indicating a strong influence of diet over POR genotype. These analyses highlighted the impact of POR ablation on expression of genes encoding P450 enzymes and proteins involved in stress and inflammation. We also found that livers from animals of both genotypes fed diets enriched in PUFA had gene expression patterns more closely resembling those fed diets enriched in saturated fatty acids. These results strongly suggest only HUFA supplied from an exogenous source can suppress hepatic lipogenesis.
Keywords: cytochrome P450 reductase, functional genomics, gene set analysis
present u.s. society benefits, and suffers, from the great technological advances made that have increased our ability to produce an abundance of food while decreasing the amount of time and physical energy spent obtaining food. At the same time, we have stressed our environment and introduced into it chemicals that can alter physiological parameters and influence gene expression. Many nutrients and environmental toxins are metabolized by the cytochrome P450 system. In the liver, the P450 system plays a critical role in bile acid synthesis on the one hand and detoxification of environmental chemicals, drugs, and xenobiotics on the other, functions that are much appreciated and the subject of intense investigations. However, the role of P450 enzymes in metabolism of nutrients and natural metabolites remains poorly understood. Among these are P450 enzymes that catalyze the conversion of long chain highly unsaturated fatty acids to bioactive compounds.
The liver-specific cytochrome P450 reductase (NADPH-hemoprotein reductase, EC 1.6.2.4, CPR, or POR) knockout (LCN) mouse, a model in which the gene for POR, the master enzymatic regulator of microsomal P450 metabolism, has been selectively deleted from the liver, was observed to display a fatty liver phenotype with characteristics in common with nonalcoholic fatty liver disease (NAFLD) (13, 16). These characteristics include steatosis involving accumulation of triglycerides and cholesterol esters, eventual development of fibrosis, and evidence of inflammation. Previously we had provided evidence that the fatty liver phenotype was coincident with an accumulation of the C18 polyunsaturated fatty acids C18:2ω6 and C18:3ω3 [linoleic (LA) and linolenic (LN) acid, respectively; collectively called PUFA] when mice were fed a standard chow diet (45). This PUFA accumulation occurs at the expense of the synthesis of the very long chain highly unsaturated fatty acids C20:4ω6, C20:5ω3, and C22:6ω3 (AA, EPA, and DHA, respectively; collectively called HUFA) and levels of these fatty acids are decreased in livers from LCN mice compared with livers from control animals. Transcription profiling demonstrated that this probably occurred due to downregulation of specific elongase and desaturase enzymes required for HUFA synthesis. These results were unanticipated because previously there was no known connection between the P450 system and fatty acid elongation and desaturation. The reductase activity associated with the fatty acid desaturase enzymes (Fads1 and Fads2) is not POR. However, it is known that members of several cytochrome P450 enzyme (CYP) families including CYP1, CYP2, CYP3, CYP4, and CYP5 function in the metabolism of C20:4ω6 (arachidonic acid) to form bioactive regulatory eicosanoids (reviewed in Ref. 30). The role of these enzymes in the metabolism of C20:5ω3 and C22:6ω3 fatty acids is less well studied. Therefore, the importance of the LCN model lies in the opportunity to uncover important integrative relationships between lipid homeostasis and other metabolic pathways including the detoxification of drugs and elimination of toxic environmental factors.
In the present study, we explored the hypothesis that the fatty liver phenotype and associated gene expression changes that occur with the specific deletion of the Por gene in adult mouse liver could be abrogated by supplementation of the mouse diet with the HUFA, arachidonic acid (AA, C20:4ω6), eicosapentaenoic acid (EPA, C20:5ω3) and docosahexaenoic acid (DHA, C22:6ω3). As we expected the fatty liver phenotype was not reduced by the PUFA linoleic (LA, C18:2ω6) and linolenic (LN, C18:3ω3), since these accumulated in the fatty livers of LCN animals. However, we also made two surprising observations. First, control animals fed a diet enriched in PUFA had fatty livers and gene expression profiles similar to animals fed a lard diet, which was deficient in both PUFA and HUFA. Second, while a diet enriched in HUFA did result in reduced steatosis in livers of the LCN animals, fat accumulation was still elevated relative to controls. Array analyses indicated that most differences in gene expression that were related to fatty acid metabolism in the LCN livers could be attenuated by modification of the fat composition of the diet.
MATERIALS AND METHODS
Animal husbandry and dietary protocols.
Generation of the founder line with hepatic ablation of Por is described in Wu et al. (48). Male test animals used in this study had the genotype Alb-Cre+/−/Porlox/lox and were designated LCN. The control animals were co-isogenic male mice with the genotype Alb-Cre−/−/Porlox/lox. The test and control animals were derived from intercrosses between Alb-Cre+/−/Porlox/lox and Alb-Cre−/−/Porlox/lox mice; the breeders had been backcrossed for at least 10 generations onto the C57BL/6 background. All animal procedures were approved by the Wadsworth Center Institutional Animal Care and Use Committee of the New York State Department of Health.
Upon weaning (4 wk after birth) 6–12 male mice of each genotype were placed on one of the test diets for 8 wk. Siblings of the same genotype were randomized between dietary groups. Synthetic diets used base diet AIN-93G and contained 18.7% protein, 64.7% carbohydrate, 5% fiber, 6% fat, 0.025% cholesterol, 0.008% sodium cholate, and 3.97 kcal per gram. The added fats were as follows: 6% lard (lard diet), 6% canola oil (canola diet), or 2.73% menhaden oil (fish/fungal diet) and 3.27% ARASCO oil (Martek Biosciences). The synthetic chows were stored in evacuated and sealed plastic bags at −80°C until use. The complete fatty acid composition of the diets is given in Sealls et al. (36). Weight gain and body length (from the nose to anus) were monitored once a week, and no differences were detected between genotypes or diets. Food was changed frequently and fatty acid analysis by gas chromatography/mass spectrometry (GC-MS) was repeated periodically to assure the unsaturated fats were not oxidized and or otherwise modified.
Sample collection.
After 8 wk on the test diets (12 wk of age) the animals were fasted for 4 h prior to death. Venous blood was drawn from the heart and collected in lithium heparin-coated tubes; 50 μl of whole blood was saved for analysis and plasma was obtained by centrifugation of the remaining blood. Both whole blood and plasma were immediately stored at −20°C for further analysis. Liver samples were placed in RNAlater for mRNA analysis, in OCT tissue-Tex for histology, or were immediately frozen and stored at −80°C.
Blood chemistry.
One week before the mice were killed they were fasted for 12 h and blood was obtained by tail clipping. Glucose was measured using a glucometer and test strips (ACCU-CHECK Active, Roche). Plasma cholesterol and triglyceride (Tg) levels were determined after terminal bleed with commercially available kits (Wako Diagnostics) following the manufacturer's instructions. Liver function was measured by analysis of 4 h fasted whole blood using a mammalian liver profile rotor in a VetScan classic hematological analyzer (Abaxis).
Lipid analyses.
Total lipid extraction from liver samples was performed according to Folch's method (11). All the lipid extractions were carried out in the presence of the antioxidant β-hydroxy-toluene (0.05%, wt/vol). Heptadecanoic and nonadecanoic acid (100 μg each) were added to all samples to estimate the recovery of fatty acids. For fatty acid analysis, complex lipids were hydrolyzed by adding 0.5 ml of methanol containing 1% (wt/vol) sulfuric acid and 0.25 ml of toluene to each sample. The samples were incubated at 60°C overnight. After this period 1.25 ml of water containing sodium chloride (5%, wt/vol) was added and the required esters were extracted with hexane (2 × 1.25 ml). The hexane layer was washed with water (1 ml) containing potassium bicarbonate (2%, wt/vol) and dried over anhydrous sodium sulfate. The solvent was removed under a stream of nitrogen and the fatty acid methyl esters were analyzed by GC/MS using an Agilent 6890 series gas chromatograph equipped with a 5873 mass-selective detector. For assessment of sterols, 100 μg 5α-cholestane was added to samples as an internal standard. Sterols were also separated and detected by GC/MS using a DB-17ms column (Agilent).
RNA isolation and quantitative RT-PCR.
RNA was extracted using a Qiagen RNeasy mini kit and reverse transcribed to cDNA using Bio-Rad iScript cDNA synthesis kit for production of a template for quantitative (Q)-PCR. Reactions were performed in a 96-well plate format using a Bio-Rad iCycler. Each Q-PCR reaction contained the following: 12.5 μl of Bio-Rad iQ SYBR Green Supermix, 500 nM of each primer (Supplemental Table S1)1, 1–6 μl of cDNA, and milliQ H2O up to a total volume of 25 μl. For all genes studied, standard curves were generated using serial dilutions of the corresponding PCR product using the primers specified. The PCR products had been previously purified using a Qiagen PCR purification kit and the concentration measured using a NanoDrop. Total copy number/μl was calculated with the following equation: {[(DNA)/molecular weight of fragment] × 6.023 × 1023}. Q-PCR amplification was set using the following parameters: a denaturing cycle of 3 min at 95°C; 30 cycles of PCR 95°C for 30 s, annealing temperature specific to each primer set for 30 s, and 72°C for 30 s; a melt cycle consisting of 95°C for 1 min, 55°C for 1 min, and a step cycle starting at 55°C and ending at 95°C increasing by 5°C increments for 10 s each. Standard curve generation, quantification of cDNA, and melt curve generation were performed with the Bio-Rad iQ-PCR cycler. Each hepatic gene quantified for the lard, canola, and fish/fungal diets was then corrected for varying cDNA microgram inputs (each was set equivalent to 100 ng of input RNA). Results for each gene is presented relative to the values obtained for livers from control mice fed the lard diet.
Microarray analysis.
Total RNA was isolated from the livers of mice as described above for Q-PCR. To reduce variability between arrays, three RNA samples from individual mice of the same genotype/diet group were pooled using equivalent RNA amounts for each animal (3 biological replicates for each diet/genotype combination). The quality of RNA was analyzed using an Agilent 2100 Bioanalyzer. Three micrograms of RNA was reverse transcribed to make cDNA. Hybridization of cDNA, washing, and scanning of the Affymetrix GeneChip Mouse Genome 430 2.0 (containing 45,101 probe sets representing over 34,000 mouse genes) was performed according to the standard Affymetrix protocol (performed by The Applied Genomics Technologist Core at the New York State Department of Health, Wadsworth Center). Data analysis was performed, in part, using GeneSpring GX 9.0 software. Probe data from the gene chips were corrected for background, normalized, and summarized using the robust multiarray analysis (RMA) method (18). This was followed by linear models of gene expression corrected with empirical Bayes models as implemented in the new limma package extended from reference (37). Both RMA and limma are packages of the Bioconductor Suite (12). Raw data were transformed to base-2 logarithms for better visualization and to give equal emphasis on both up- and downregulated genes. The array data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database with accession number GSE20944. For the main effect of genotype and diet, log2 fold change was calculated relative to the genotype or diet indicated. Genes whose expression changed relative to both diet and genotype interactions were expressed as the raw log2 transformed data. In other calculations, the main effect of genotype was calculated using a t-test (using an asymptotic P value computation) with a P value cutoff of P < 0.05. Main effect of diet was calculated first by ANOVA followed by Tukey's honestly significant difference (HSD) (using an asymptotic P value computation) with a P value cutoff of P < 0.05. Interaction between genotype and diet was calculated using a two-way ANOVA with a P value cutoff of P < 0.05. False discovery rate (FDR) was calculated according to Benjamini and Hochberg and applied to the independent main effects (genotype or diet) but not the interaction effect (3, 34).
Pathway and gene ontology studies by gene set enrichment analysis.
To identify biochemical/signaling pathways or Gene Ontology (1) categories that were significantly differentially regulated, we used rigorous Gene Set Enrichment Analysis (GSEA) (40). GSEA calculates enrichment on the basis of all genes on the chip as opposed to ad hoc methods based on the biased samples of up- or downregulated genes. GSEA estimates the statistical significance of enrichment in the upregulated and downregulated categories. For example, gene sets as a whole may be significantly upregulated even when the upregulation of most individual genes is not significant. We performed 10,000 random permutations of all genes for the permutation tests. Multiple test correction was performed by FDR using a threshold of FDR ≤ 0.05 (3, 34).
Most biochemical and signaling pathways were obtained from the Curated Dataset (C2) of the Molecular Signature Database created by the GSEA Team that we mapped from human genes to their mouse orthologs (40). We also used the Kyoto Encyclopedia of Genes and Genomes Pathways Database (KEGG, 31). Gene Ontology sets for biological processes, molecular functions, and cellular localizations (1) were downloaded from the Gene Ontology Consortium (http://www.geneontology.org/GO.downloads.shtml). Gene set databases were adapted for GSEA using our PERL libraries (University of Nebraska - Lincoln, Department of Statistics). GSEA was performed on a command-line interface at a 16 gb RAM LINUX server.
Western blot analysis.
Livers were mechanically disrupted by homogenization on ice in 20 mM Tris·HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA pH 7.4, 1 mM EGTA pH 7.5, 0.1% Triton, 2.5 mM Na pyrophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and one protease inhibitor cocktail tablet (Roche) per 50 ml. For fatty acid synthase (FASN) 20 μg of total protein was separated on an 8% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was blotted using anti-FASN at 0.5 μg/μl. Secondary anti-rabbit horseradish peroxidase (HRP) was used at a 1:5,000 dilution. β-Actin (antibody dilution of 1:10,000) was used as a loading control. For Scd1, microsomes were prepared according to Coon et al. (7). In brief, liver homogenates (above) were centrifuged at 10,000 g for 20 min at 4°C. Supernatants were recovered and centrifuged at 110,000 g for 1 h, at 4°C. Pellets containing the microsomes were washed twice with cold pyrophosphate buffer (100 mM sodium pyrophosphate pH 7.4, 1 mM EDTA) and then resuspended and sonicated in cold storage buffer (50 mM Tris-acetate pH 7.4, 20% glycerol, 0.1 mM EDTA). To detect Scd1 30 μg of microsomal protein was separated on a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was blotted using anti-Scd1 antibody from Santa Cruz Biotechnology (sc-14719) at a 1:200 dilution. Secondary anti-goat HRP antibody from Santa Cruz Biotechnology was used at a 1:5,000 dilution. Proteins were visualized using an ECL kit (Pierce) according to the manufacturer's instructions. Protein concentrations were determined using the Bio-Rad protein assay kit according to manufacturer's instructions. BSA was used as a standard.
Statistical analysis.
Unless otherwise indicated lipid, plasma chemistry and Q-PCR data were analyzed by two-way ANOVA followed by Tukey's HSD or t-test (JMP statistical analysis software). Gene expression array data was analyzed as described above.
RESULTS
Physiological and histological examination of control and LCN mice fed diets varying in lipid composition.
Livers of LCN mice are steatotic when mice are reared on standard rodent chow (13, 16). To determine if steatosis could be reduced and differences in gene expression for lipid metabolic enzyme normalized despite POR deficiency, we performed a dietary study in which the mice were reared from weaning until 12 wk of age on three defined diets differing only in fat composition, which included lard (limited in PUFA and HUFA), canola oil (enriched in PUFA), or a mixture of Menhaden and ARASCO oils (fish/fungal, enriched in HUFA) (36). The diets were not high in calories, fat, or carbohydrate. We expected the high percentage of HUFA in the fish/fungal diet would dampen fatty acid and triglyceride synthesis to limit steatosis in the LCN animals (4, 23). Both LCN and control mice grew well on each diet, and there were no significant differences in growth between genotypes or diets (Table 1). Liver size was not affected by dietary treatment, but livers of LCN mice were significantly larger than their control counterparts, and this was reflected in a slightly higher liver to body weight ratio.
Table 1.
Body weights, organ weights, and blood and plasma chemistry parameters
| Genotype | Lard | Canola | Fish/Fungal | Significance, P | ||
|---|---|---|---|---|---|---|
| Body and Organ Weights | ||||||
| Body, g | Ctrl | 26.6 (1.1) | 25.8 (4.4) | 25.6 (0.6) | Diet | NS |
| LCN | 24.98 (1.0) | 25.1 (0.5) | 24.7 (0.8) | Geno | NS | |
| D*G | NS | |||||
| Liver, g | Ctrl | 1.17 (0.07) | 1.18 (0.9) | 1.15 (0.04) | Diet | NS |
| LCN | 1.48 (0.09) | 1.33 (0.07) | 1.39 (0.03) | Geno | 0.0002 | |
| D*G | NS | |||||
| Liver: Body | Ctrl | 0.04 (0) | 0.05 (0) | 0.05 (0) | Diet | NS |
| LCN | 0.06 (0) | 0.05 (0) | 0.06 (0) | Geno | <0.0001 | |
| D*G | NS | |||||
| Adipose | Ctrl | 0.59 (0.12) | 0.74 (0.1) | 0.45 (0.06) | Diet | NS |
| Tissue, g | LCN | 0.67 (0.08) | 0.58 (0.07) | 0.50 (0.06) | Geno | NS |
| D*G | NS | |||||
| Blood and Plasma Chemistry | ||||||
| ALP, U/l | Ctrl | 68.7 (6.4) | 79.5 (5.2) | 73.3 (0.9) | Diet | NS |
| LCN | 93.8 (7.9) | 92.3 (4.2) | 109.3 (18.1) | Geno | 0.0040 | |
| D*G | NS | |||||
| ALT, U/l | Ctrl | 34.7 (9.3) | 20.0 (3.2) | 16.8 (2.5) | Diet | NS |
| LCN | 154.3 (79.0) | 97.0 (37.0) | 34.0 (9.3) | Geno | 0.0192 | |
| D*G | NS | |||||
| TBIL, mg/dl | Ctrl | 0.35 (0.02) | 0.35 (0.03) | 0.35 (0.03) | Diet | NS |
| LCN | 0.30 (0.00) | 0.30 (0.00) | 0.38 (0.03) | Geno | NS | |
| D*G | NS | |||||
| ALB, g/dl | Ctrl | 3.9 (0.2) | 4.0 (0.2) | 4.2 (0.2) | Diet | NS |
| LCN | 3.9 (0.2) | 3.6 (0.1) | 3.6 (0.1) | Geno | 0.0332 | |
| D*G | NS | |||||
| BUN, mg/dl | Ctrl | 17.0 (0.9) | 19.5 (1.0) | 19.5 (0.9) | Diet | NS |
| LCN | 19.3 (2.8 | 15.3 (0.3) | 19.0 (2.6) | Geno | NS | |
| D*G | NS | |||||
| CHOL, mg/dl | Ctrl | 132.2 (8.3) | 122.9 (10.9) | 88.3 (5.2) | Diet | 0.0020 |
| LCN | 81.5 (5.4) | 74.0 (7.1) | 73.0 (5.8) | Geno | <0.0001 | |
| D*G | 0.0245 | |||||
| TG, mg/dl | Ctrl | 33.8 (5.6) | 35.3 (5.1) | 19.9 (4.6) | Diet | 0.0237 |
| LCN | 16.0 (6.2) | 8.3 (4.4) | 4.7 (4.1) | Geno | 0.001 | |
| D*G | NS | |||||
| GLUC, mg/dl | Ctrl | 187.7 (7.3) | 181.7 (15.4) | 136.0 (11.5) | Diet | 0.0007 |
| LCN | 171.0 (7.7) | 174.4 (8.6) | 139.4 (11.4) | Geno | NS | |
| D*G | NS | |||||
Values are the mean for 6–9 mice; values in parentheses indicate SE. Significance of difference was evaluated by 2-way ANOVA (P < 0.05; t-test). ALP, alkaline phosphatase; ALT, alanine aminotransferase; TBIL, total bilirubin; ALB, albumin; BUN, blood urea nitrogen; CHOL, total cholesterol; TG, triglyceride; GLUC, fasting (10 h) glucose; Geno, genotype; Ctrl, control; D*G, diet × genotype; NS, not significant.
Plasma chemistry parameters, glucose, and lipid levels were assessed to evaluate the effect of different dietary fat compositions on animals of each genotype (Table 1). Blood ALP and ALT varied significantly with genotype whereby the LCN animals had higher levels than controls possibly indicative of hepatic stress. Fasting blood glucose levels measured after a 12 h fast were not different between LCN and control animals but were significantly lower when animals were fed the fish/fungal diet (36). Plasma cholesterol levels were low in LCN animals compared with controls as previously reported (16), and this was only partially alleviated by dietary fat treatment. Animals on the lard based diet had the highest plasma cholesterol levels followed by canola and fish/fungal fed animals. The levels for the LCN mice were more similar to the controls maintained on the fish/fungal diet. The plasma triglyceride (Tg) levels were low in the LCN animals compared with controls and partially affected by diet. In each case animals fed the fish/fungal diet had lower levels of plasma Tg than animals on lard or canola based chows.
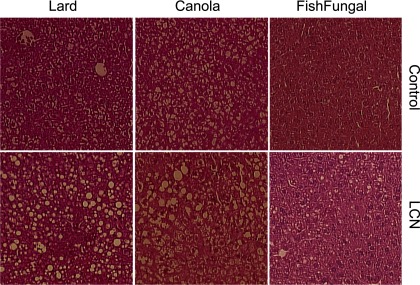
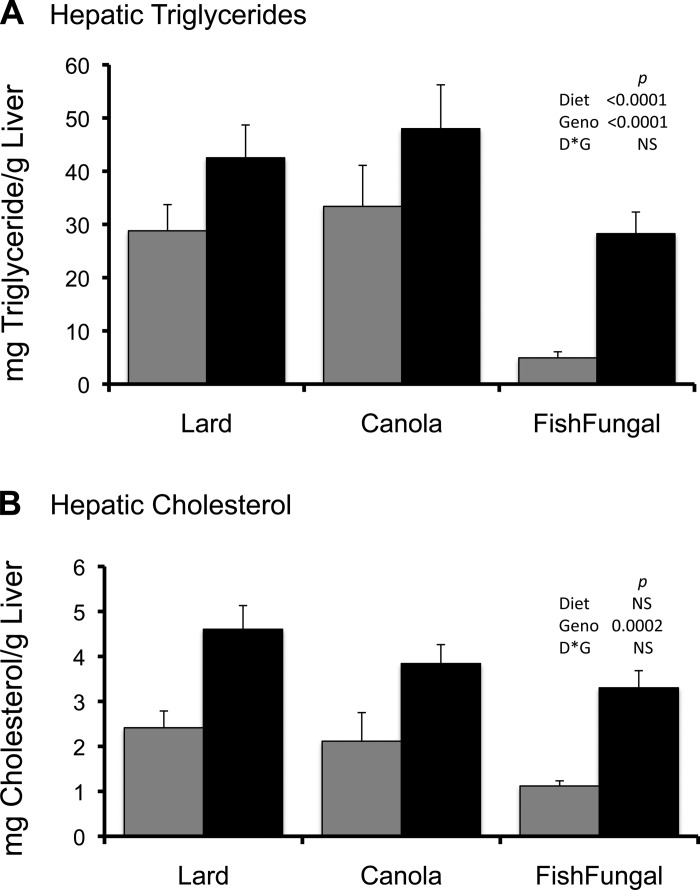
A notable phenotype of the LCN model is hepatic steatosis. One of the goals of the present study was to alter fat accumulation by treatment with different dietary fats. Since we had previously noted livers of these mice were relatively enriched in the PUFAs over HUFAs, we hypothesized feeding a HUFA-enriched diet would suppress fatty acid synthesis and excessive accumulation of Tg. Histological examination revealed extensive lipid droplet accumulation in livers of control animals fed lard and canola, which was even more exaggerated in livers from the LCN mice (Fig. 1). We found hepatic lipid accumulation was favored and was similar when mice were fed either the lard or canola diets regardless of genotype (Fig. 2A). The amount of Tg that accumulated in LCN livers from animals fed the fish/fungal diets was about half the amount for livers of animals of the same genotype fed either the lard or canola diets. However, the amount of Tg that accumulated in livers of the LCN mice on any diet was still higher than that of control livers. This was surprising since, as reported below, expression of most fatty acid synthetic genes were suppressed by fish/fungal feeding regardless of genotype.
Fig. 1.
Steatosis in livers of controls and LCN mice fed lard or canola diets. Representative liver micrographs at ×10 magnification from control or LCN mice fed each of the diets as indicated. Sections were stained with hematoxylin and eosin.
Fig. 2.
Hepatic triglyceride and cholesterol levels in control and LCN mice fed the 3 test diets. Triglycerides (A) and cholesterol (B) were extracted from livers as detailed in the text. Gray bars indicate data for controls and black bars for LCN; n = 7 mice per group. The error bar gives SE. Data were analyzed using 2-way ANOVA and the Student's t-test; significance values are as indicated.
Hepatic cholesterol synthesis is largely dependent upon POR function since pathway enzymes leading from lanosterol to cholesterol and bile acids are dependent upon P450 enzymes. Therefore both synthesis and clearance of cholesterol from liver is compromised in the LCN animals. The levels of cholesterol were measured using a standard kit (Fig. 2B) and by GC/MS analysis to identify cholesterol and other sterol precursors and metabolites. LCN livers had higher levels of hepatic cholesterol than control animals. The greater sequestration of cholesterol in the LCN livers than in the control livers was probably because cholesterol was not eliminated as bile.
Due to the defect in P450 reductase, it was expected that the de novo synthesis of cholesterol would be halted or compromised at steps beyond squalene due to insufficient squalene epoxidase (CYP51) activity. As predicted we found squalene to accumulate to high levels in the LCN livers (0.34 ± 0.07 mg/g liver) compared with controls (0.05 ± 0.01 mg/g liver) (P = 0.002). In contrast, there was no significant difference specifically due to diets (data not shown).
Liver fatty acid profiles are modified by genotype and diet.
Liver synthesizes saturated and monounsaturated fatty acids de novo from acetyl-CoA usually derived from dietary carbohydrate. Preformed fatty acids may also be acquired from the blood stream, and these are generally thought to be derived from lipoproteins during the fed state and from free fatty acids originating from adipose tissue lipolysis during fasting. Hepatocytes elongate and desaturate the essential PUFA, C18:2ω6 (LA) and C18:3ω3 (LN), to form the HUFA, C20:4ω6 (AA), C20:5ω3 (EPA), and C22:6ω3 (DHA). One of the questions arising from the present study was: what is the interrelationship between dietary fatty acids and hepatic gene expression, particularly with regard to lipid synthesis? Our previous analyses, comparing fatty acid profiles of animals of each genotype fed a standard lab chow, suggests that liver specific deletion of Por resulted in suppression of elongation and desaturation of PUFA, resulting in low levels of HUFA in these animals compared with controls (45). We therefore analyzed hepatic fatty acids from livers of animals of both genotypes on each of the synthetic test diets. The present data were analyzed in two ways. First the total amount of fatty acid was measured and expressed in μmol/g wet weight of tissue (Supplemental Table S2). This verified total lipid accumulated in the LCN livers was higher than in control livers. Second, these data were analyzed as mol% of each class of fatty acid relative to the total fatty acid in the sample to facilitate assessment of the relative amount of any fatty acid acquired from the diet or synthesized de novo (Table 2). Monounsaturated fatty acids made up 50–60 mol% of all fatty acids when mice of either genotype were fed the lard- or canola-based diets. Since this is higher than the amount provided in the diets, it suggests efficient conversion of the saturated fatty acids to the monounsaturated and/or net synthesis. The relative abundance of monounsaturated fatty acids was much lower in the fish/fungal diet groups, than in the other two diet groups, for either genotype. Furthermore, the control animals maintained on the fish/fungal diet had twofold higher levels of saturated fatty acids than monounsaturated fatty acids, while the fish/fungal-fed LCN animals had essentially equivalent levels for the two groups of fatty acids. This indicates the fish/fungal diet suppresses the expression and/or activity of the Δ9-acyl-CoA desaturase encoded within the Scd1 gene (see below).
Table 2.
Hepatic fatty acid content (mol %) for controls and LCNs fed each of the experimental diets
| Fatty Acid | Genotype | Lard mol% (SE) | Canola mol% (SE) | Fish/Fungal mol% (SE) | FA Class Significance, P | |
|---|---|---|---|---|---|---|
| C14:0 | Ctrl | 0.7 (0.0) | 0.6 (0.1) | 0.5 (0.0) | ||
| LCN | 0.6 (0.0) | 0.6 (0.0) | 0.5 (0.1) | |||
| C16:0 | Ctrl | 21.5 (0.3) | 21.9 (0.9) | 26.4 (0.4) | ||
| LCN | 19.0 (0.5) | 18.5 (0.7) | 21.1 (0.5) | |||
| C18:0 | Ctrl | 5.8 (1.2) | 5.6 (0.6) | 12.8 (0.8) | ||
| LCN | 4.8 (0.2) | 4.4 (0.3) | 7.3 (0.4) | |||
| C20:0 | Ctrl | 0.3 (0.0) | 0.2 (0.0) | 0.3 (0.1) | ||
| LCN | 0.5 (0.1) | 0.4 (0.0) | 0.4 (0.1) | |||
| C22:0 | Ctrl | 0.1 (0.0) | 0.0 (0.0) | 0.2 (0.1) | ||
| LCN | 0.2 (0.0) | 0.1 (0.0) | 0.2 (0.1) | |||
| Σ SFA | Ctrl | 28.7 (1.5) | 28.4 (1.0) | 40.4 (1.1) | Diet | <0.0001 |
| LCN | 25.2 (0.6) | 24.0 (0.8) | 29.7 (0.8) | Geno | <0.0001 | |
| D*G | 0.0006 | |||||
| C16:1 | Ctrl | 8.3 (1.0) | 7.1 (0.7) | 3.0 (0.5) | ||
| LCN | 9.3 (0.3) | 8.3 (0.6) | 4.4 (0.5) | |||
| C18:1 | Ctrl | 44.2 (2.8) | 39.7 (2.1) | 14.2 (1.5) | ||
| LCN | 47.4 (0.9) | 46.4 (1.0) | 25.1 (2.0) | |||
| C20:1 | Ctrl | 1.9 (0.3) | 1.2 (0.2) | 0.0 (0.0) | ||
| LCN | 2.9 (0.2) | 2.1 (0.1) | 0.7 (0.1) | |||
| C22:1 | Ctrl | 0.2 (0.1) | 1.5 (0.1) | 0.4 (0.2) | ||
| LCN | 0.5 (0.1) | 0.2 (0.0) | 0.2 (0.1) | |||
| Σ MUFA | Ctrl | 54.6 (4.1) | 48.2 (3.0) | 17.7 (2.0) | Diet | <0.0001 |
| LCN | 60.0 (1.0) | 57.2 (1.4) | 30.5 (2.5) | Geno | <0.0001 | |
| D*G | NS | |||||
| C18:2ω3 | Ctrl | 7.4 (1.0) | 10.6 (1.0) | 5.6 (0.6) | ||
| LCN | 8.3 (0.3) | 11.0 (0.7) | 6.4 (0.5) | |||
| C18:3ω6 | Ctrl | 0.1 (0.0) | 1.3 (0.3) | 0.1 (0.0) | ||
| LCN | 0.1 (0.1) | 1.9 (0.2) | 0.8 (0.1) | |||
| C20:3ω9 | Ctrl | 1.1 (0.3) | 0.7 (0.2) | 0.8 (0.2) | ||
| LCN | 1.0 (0.1) | 0.7 (0.1) | 1.4 (0.1) | |||
| Σ PUFA | Ctrl | 7.5 (1.0) | 11.9 (1.2) | 5.7 (0.6) | Diet | <0.0001 |
| LCN | 8.4 (0.3) | 12.7 (0.9) | 7.2 (0.5) | Geno | NS | |
| D*G | NS | |||||
| C20:4ω6 | Ctrl | 6.0 (1.4) | 4.5 (0.8) | 19.6 (0.8) | ||
| LCN | 3.4 (0.1) | 2.4 (0.2) | 15.4 (0.5) | |||
| C20:5ω3 | Ctrl | 0.0 (0.0) | 0.8 (0.1) | 0.9 (0.1) | ||
| LCN | 0.0 (0.0) | 0.5 (0.0) | 1.4 (0.1) | |||
| C22:6ω3 | Ctrl | 2.0 (0.4) | 5.5 (1.0) | 15.0 (1.0) | ||
| LCN | 1.7 (0.1) | 2.2 (0.5) | 14.2 (1.9) | |||
| Σ HUFA | Ctrl | 8.0 (1.8) | 10.7 (1.9) | 35.4 (1.2) | Diet | <0.0001 |
| LCN | 5.1 (0.2) | 5.0 (0.7) | 31.1 (2.1) | Geno | 0.0009 | |
| D*G | NS | |||||
The percentage of the fatty acids in each class (SFA, MUFA, PUFA, HUFA) is noted. Data shown are means (± SE) of 6–7 animals per group. Significance of difference was evaluated by 2-way ANOVA; significance values are as indicated.
The PUFA cannot be synthesized de novo in mammals and must be obtained in the diet. Therefore, tissue PUFA content reflects accumulation and retention of these fatty acids from a dietary source. The canola oil diet contained 37% PUFA at a ratio of ∼2:1, C18:2ω6 to C18:3ω3. However, the livers of control and LCN mice fed the canola diet accumulated only 12–13% PUFA. Since there was very little HUFA (approx. 0.1%) provided in the canola diet, any amount accumulated in liver should be derived from anabolism of dietary PUFA. While livers of animals of both genotypes fed the canola diet had measurable amounts of HUFA, the LCN livers only accumulated half as much as the control livers, suggesting a reduction in metabolic synthesis compared with controls. The fish/fungal diet contained about 37% HUFA at a ratio of ∼2:1 C20:4ω6 to C20:5ω3 and C22:6ω3. Fatty acid profiles in livers from animals of both genotypes fed the fish/fungal diet each accumulated a high level of HUFA (35.4 vs. 31.1 mol%). Therefore, we had achieved one goal in elevating liver HUFA levels in both the control and LCN livers. This was necessary to assess impact on the liver transcriptome as detailed below.
Manipulation of dietary fat alters gene expression profiles irrespective of POR genotype.
Previous expression analysis of livers from LCN and control mice fed standard rodent chow diets identified numerous differences particularly with respect to genes encoding cytochrome P450 enzymes and genes involved in fatty acid metabolism (29, 45). Since HUFA are known to suppress hepatic lipid synthesis, we evaluated gene expression array results for livers from mice of each genotype on the three test diets. Data were compared using two-way ANOVA to assess differences between diets and genotypes and interactions between diets and genotype.
The largest number of gene expression differences, 80 in total, was detected by comparisons between diets, regardless of genotype (Table 3). Surprisingly, expression of 57 of these was similar between lard and canola by comparison with fish/fungal, while only seven were found to be in common between canola and fish/fungal compared with lard. In all but eight cases the gene expression level was lower in livers of mice fed the fish/fungal diets. Notable among these eight was Acsm2, encoding a medium chain acyl-CoA synthetase found in the mitochondria (5), and Gpcpd1, a putative glycerophosphocholine phosphodiesterase (32). These enzymes may be involved in maintaining fatty acid and glycerolipid homeostasis when animals are fed diets enriched in HUFA.
Table 3.
Hepatic genes differentially expressed in response to dietary fat
| Gene | C/L | L/FF | C/FF | Protein/Function |
|---|---|---|---|---|
| Fatty Acid Metabolism and Transport | ||||
| Acacb | ns | 5.33 | 3.46 | acetyl-Coenzyme A carboxylase beta |
| Acsl3 | 0.74 | 3.73 | 1.37 | acyl-CoA synthetase long-chain family member 3 |
| Acsl4 | 0.65 | 1.48 | ns | acyl-CoA synthetase long-chain family member 4 |
| Acsm2 | ns | 0.61 | 0.43 | acyl-CoA synthetase medium-chain family member 2 |
| Acly | 0.53 | 3.73 | 1.98 | ATP citrate lyase |
| Acbp/ Dbi | ns | 1.30 | 1.22 | Acyl-CoA binding protein |
| Echdc1 | 0.83 | 1.80 | 1.50 | enoyl Coenzyme A hydratase domain containing 1 |
| Elovl2 | 0.83 | 2.03 | 1.68 | FA elongase family member 2 |
| Elovl5 | ns | 5.81 | 4.70 | FA elongase family member 5 |
| Fabp2 | ns | 1.98 | 2.21 | Fatty acid binding protein 2 |
| Fabp5 | 0.39 | 5.62 | 2.17 | Fatty acid binding protein 5 |
| Fads1 | ns | 6.43 | 4.94 | Fatty acid desaturase 1 |
| Fads2 | ns | 5.05 | 3.92 | Fatty acid desaturase 2 |
| Fasn | ns | 6.56 | 3.68 | Fatty acid synthase |
| Tecr/Gpsn2 | ns | 1.49 | 1.28 | Trans-2,3-enoyl-CoA reductase |
| Pcyt2 | ns | 1.67 | 1.57 | CTP:phosphoethanolamine cytidylyltransferase 2 |
| Gpcpd1/Prei4 | ns | 0.49 | 0.61 | Glycerophosphocholine phosphodiesterase GDE1 homolog |
| Sterol Metabolism | ||||
| Cyp51 | 0.66 | 4.17 | 2.03 | Cytochrome P450 51 |
| Dhcr7 | ns | 2.27 | 1.84 | 7-dehydrocholesterol reductase |
| Fdps | ns | 6.17 | 4.10 | Farnesyl diphosphate synthetase; |
| Hmgcs1 | ns | 2.19 | 1.79 | HMG CoA synthase |
| Idi1 | ns | 7.07 | 4.59 | Isopentenyl-diphosphate delta isomerase |
| Insig1 | ns | 2.34 | 1.87 | Insulin-induced gene 1 |
| Lss | ns | 2.03 | 1.61 | Lanosterol synthase |
| Nsdhl | ns | 2.50 | 1.76 | NAD(P) dependent steroid dehydrogenase-like |
| Pmvk | ns | 2.15 | 1.60 | Phosphomevalonate kinase |
| Rdh11 | ns | 3.64 | 2.56 | Retinol dehydrogenase 11 |
| Retsat | 1.58 | 0.62 | ns | Retinol saturase |
| Sc4mol | 0.75 | 2.18 | 1.58 | Sterol-C4-methyl oxidase-like |
| Sc5d | 0.77 | 1.87 | 1.44 | Sterol-C5-desaturase |
| Sqle | 0.49 | 3.40 | ns | Squalene epoxidase |
| Stard4 | ns | 2.77 | 2.27 | Steroidogenic acute regulatory-related lipid transfer (START) domain containing 4 |
| Carbohydrate metabolism | ||||
| Dlat | ns | 1.34 | 1.28 | dihydrolipoamide S-acetyltransferase |
| Gpsn2 | ns | 1.49 | 1.28 | Glycoprotein |
| Pgm3 | ns | 1.62 | 1.48 | phosphoglucomutase 3 |
| Membrane transport | ||||
| Aqp4 | ns | 2.26 | 4.15 | aquaporin 4 |
| Mfsd2 | ns | 9.75 | 10.9 | major facilitator superfamily domain containing 2 |
| Slc10a2 | 0.74 | 1.63 | ns | solute carrier family 10 |
| Slc30a10 | ns | 1.77 | 1.59 | solute carrier family 30 |
| Apolipoproteins | ||||
| Apoa4 | ns | 8.58 | 8.10 | Apolipoprotein A-IV |
| Apoa5 | ns | 1.21 | 1.22 | Apolipoprotein A-V |
| Apoc2 | ns | 1.49 | 1.62 | Apolipoprotein C-II |
| Cell Adhesion/Cytoskeletal | ||||
| Olfm3 | ns | 1.69 | 1.59 | olfactomedin 3 |
| Pstpip2 | 0.71 | 2.34 | 1.90 | Pro-ser-thr-phosphatase-interacting protein 2 |
| Tjp3 | ns | 1.52 | 1.51 | Tight junction protein 3 |
| Arhgap24/Filgap | ns | 1.59 | 1.41 | Rho GTPase activating protein 24 |
| Regulation | ||||
| Atf5 | ns | 2.08 | 1.92 | Activating transcription factor 5 |
| Bace2 | 0.83 | 0.88 | 0.72 | Beta-site APP-cleaving enzyme 2 |
| Camk1d | ns | 2.96 | 2.25 | calcium/calmodulin-dependent protein kinase ID |
| Rassf6 | ns | 1.54 | ns | Ras association (RalGDS/AF-6) domain family 6 |
| Rgs3 | ns | 3.01 | 2.34 | regulator of G protein signaling 3 |
| Sema4a | 0.88 | 1.25 | 1.11 | sema domain/Semaphorin-4A |
| Vegfb | ns | 1.57 | 1.42 | vascular endothelial growth factor B |
| Others | ||||
| Car14 | ns | 0.68 | 0.63 | carbonic anhydrase 14 |
| Cxadr | 0.70 | 2.05 | 1.45 | coxsackievirus and adenovirus receptor |
| Ddx58 | ns | 1.43 | 1.22 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| Dntt | ns | 1.67 | 1.37 | deoxynucleotidyltransferase |
| Enpep | ns | 1.41 | 1.31 | glutamyl aminopeptidase |
| Fkbp14 | 0.80 | 1.46 | 1.17 | FK506 binding protein 14 |
| Frmd4b | 0.78 | 2.12 | 1.62 | FERM domain containing 4B |
| H2-K1 | 0.89 | 1.30 | 1.17 | histocompatibility 2 |
| H2-Q10 | ns | 1.43 | 1.38 | histocompatibility 2 |
| H2-Q6 | 0.79 | 1.52 | 1.19 | histocompatibility 2 |
| Limd2 | ns | 0.69 | 0.72 | LIM domain containing 2 |
| Lonp2 | 1.20 | 0.75 | ns | lon peptidase 2 |
| Mfsd2 | ns | 9.75 | 10.89 | major facilitator superfamily domain containing 2 |
| Oprs1 | ns | 1.33 | 1.24 | opioid receptor |
| Paqr9 | ns | 2.20 | 2.33 | progestin and adipoQ receptor family member IX |
| Pnpla7 | ns | 1.56 | 1.50 | patatin-like phospholipase domain containing 7 |
| Pter | ns | 1.67 | 1.48 | phosphotriesterase related |
| Tm7 sf2 | ns | 1.80 | 1.71 | transmembrane 7 superfamily member 2 |
| Ube2i | ns | 1.24 | 1.17 | ubiquitin-conjugating enzyme E2I |
| Unknowns | ||||
| ns | 1.59 | 1.35 | RIKEN cDNA 1110059G02 gene | |
| 0.75 | 1.63 | 1.22 | RIKEN cDNA 1600029D21 gene | |
| ns | 2.67 | 1.88 | RIKEN cDNA 1810008I18 gene | |
| 1.20 | 0.82 | ns | RIKEN cDNA 2410001C21 gene | |
| ns | 0.74 | 0.68 | RIKEN cDNA 6430514L14 gene | |
| ns | 4.84 | 2.61 | RIKEN cDNA 9130221J18 gene | |
| ns | 1.82 | 2.00 | RIKEN cDNA C730029A08 gene | |
| ns | 1.97 | 1.47 | RIKEN cDNA D130043K22 gene | |
| ns | 1.43 | 1.32 | RIKEN cDNA D730039F16 gene | |
Data are expressed as ratios when identified as significant at q < 0.05 using 1-way ANOVA with Benjamini-Hochberg false discovery rate (FDR) correction; ns, not significant; L, lard; C, canola; FF, fish fungal. Protein/function identification is from the UniProtKB/Swiss-Prot Database.
Many of the genes whose expression was found to be similarly high between livers of mice fed the lard and canola diets were fatty acid and sterol synthetic genes (Table 3). Among these were Acly, encoding the ATP-citrate lyase, which provides cytosolic acetyl-CoA for both fatty acid and cholesterol synthesis; acetyl-CoA carboxylase subunit gene (Acacb); Fasn; the fatty acid elongase genes fatty acid elongase (Elovl) 2 and Elovl 5; and the fatty acid desaturase genes Fads1 and Fads2. Two fatty acid binding protein genes, Fabp2 and Fabp5, required for intracellular fatty acid transport and trafficking, were highly expressed in lard and canola versus fish/fungal fed livers. However, Fabp5 expression was unique in that expression in livers of canola fed mice was only 40% of that in lard-fed mice, indicating PUFA as well as HUFA may suppress this gene's expression in liver. Fifteen genes involved in sterol metabolism were identified as significantly different. Again, most were expressed in a similar manner when comparing livers from animals fed the lard or canola diets, and most were low when animals were fed the fish/fungal diets. Among these was Insig1, which regulates the processing of both SREBP1 and SREBP2 to the mature active form of these transcription factors. Overall, this pattern of fatty acid and sterol gene expression indicates both SREBP1 and SREBP2 are highly active in livers of animals fed either the lard or canola diets. These findings emphasize the impact of dietary HUFA over dietary PUFA in regulating gene expression through SREBP1 in particular.
Expression of three apolipoprotein genes was high in livers from lard and canola fed mice compared with fish/fungal. The ApoA4 gene was remarkable in that it was very strongly expressed (8- to 8.5-fold) in livers of lard- and canola-fed mice compared with the fish/fungal diet. This gene is found in a cluster with ApoC3 and ApoA5 (8). ApoA4 is a dynamic protein that has been found to be associated with both chylomicrons and high-density lipoproteins and is implicated in diseases associated with elevated lipid levels (33, 46). It is an activator of lecithin-cholesterol acyltransferase (LCAT) and has also been implicated as a satiety signal (39). ApoA5 was more highly expressed in liver of mice fed lard or canola diets compared with fish/fungal. Allelic variants of this protein have been associated with hypertriglyceridemia (9, 15). Elevation of genes encoding these lipoproteins by the lard and canola diets is consistent with high levels of lipid synthesis and trafficking that would result in elevated plasma lipids.
The primary source of calories in the base diet was from carbohydrate (66.5% kcal/g) and did not differ between the three test diets. Few genes were identified as significantly different that were involved in carbohydrate metabolism, and those identified were not necessarily indicative of control by ChREBP, the major carbohydrate-responsive transcription factor. Neither were the identified genes suggestive of coordinate control between SREBP1 and ChREBP. These observations suggest the differences in the type of fatty acid supplied in our test diets did not have a significant impact on carbohydrate responsive gene expression.
Among the genes involved in regulation that were identified as significantly different due to diet, were several that have been implicated in Type 2 diabetes and cardiovascular disease, including Vegf-B and calmodulin-dependent kinase 1d (Cdc123/Camk1d). Vegf-B has been implicated in the regulation of genes involved in fatty acid uptake and mitochondrial metabolism (14). Camk1d was indentified in genome-wide association studies for Type 2 diabetes susceptibility loci (50).
For the biostatistical analyses at the level of metabolic and regulatory pathways we used GSEA, which evaluates microarray expression datasets not at the level of individual genes but at the level of gene sets based on published information of biochemical function and coexpression patterns (40). GSEA is a statistically robust method because a normalized enrichment score is computed based on all members of a gene set compared with all nonmembers measured. Supplemental Table S3 lists the gene sets that were significantly different using a 5% FDR threshold for at least one comparison between diets. As expected, among the six top pathways identified were five sets related to lipid synthesis. Interestingly, oxidative phosphorylation was identified as significantly different only for the canola versus lard comparison.
GSEA identified two peroxisome proliferator-activated receptor (PPAR)α pathways as strongly influenced by dietary fatty acid. This is plausible since both PPARα and sterol regulatory element binding protein (SREBP) 1 activities are regulated by DHA (reviewed in Ref. 19). SREBP1 and PPARα co-regulate, among others, genes encoding elongases and desaturases required to synthesize HUFA from PUFA. For most other regulatory pathways identified, there was a general pattern of downregulation by the fish/fungal diet. Notable exceptions included prostaglandin synthesis regulation and G protein signaling, which were only significantly different for the canola versus lard comparison. In the nuclear receptor category, only the fish/fungal versus canola comparison was identified.
Many inflammation- and infection-related pathways emerged by GSEA. At least 12 of the identified pathways were significantly different for both fish/fungal versus lard and canola versus lard. Interesting exceptions included the complement and coagulation cascades and the related blood clotting cascade gene sets, each of which was significantly different only for the canola versus lard comparison. These distinctions are likely to indicate unique regulatory cascades are responsible for the observed regulatory patterns due to dietary PUFA versus HUFA.
Other GSEA pathways identified could be generally grouped into those involved in cell cycle and division or cell structure and adhesion. There is evidence in the literature for control of each process by dietary fatty acids and/or fatty acid regulated transcription factors such as PPARα and SREBP1 (e.g., see Ref. 22).
Taken together, these data indicate the HUFA in the fish/fungal diets, particularly DHA have a dominant effect on gene expression compared with the lard and canola diets, which have low levels of HUFA and differ between each other in the relative amount of PUFA. However, as noted, some regulatory patterns could not be explained by HUFA effects alone and were observed to either be influenced by the PUFA alone or by both PUFA and HUFA.
Differences in gene expression between LCN and control livers irrespective of diet.
We estimated that 71 genes were differentially expressed specifically due to genotype at P ≤ 0.05 (Table 4). Of these 63 were higher in LCN livers compared with controls and eight were lower. It is important to note that feeding the synthetic diets differing only in fat composition eliminated many of the apparent differences in expression of genes involved in fatty acid and sterol metabolism that we previously reported for control and LCN mice fed a standard chow diet (compare data in Ref. 45 with Table 4). In fact, the only gene related to fatty acid metabolism identified as significantly different due to genotype was Lpl encoding lipoprotein lipase (2.5-fold higher for LCN livers than control livers). This protein is required for hydrolysis of Tg from lipoproteins in the bloodstream and its activity is a prerequisite for subsequent fatty acid uptake into cells.
Table 4.
Genes whose expression is different due to genotype
| Gene | KO/C | Protein Name/Function |
|---|---|---|
| Regulation | ||
| Rarb | 1.66 | retinoic acid receptor beta |
| Metabolism | ||
| Acp2 | 1.51 | acid phosphatase 2 |
| Anxa2 | 1.60 | annexin A2; calpactin 1 |
| Anxa5 | 1.60 | annexin A5 |
| Ces6 | 2.11 | carboxylesterase 6 |
| Idh2 | 1.56 | isocitrate dehydrogenase |
| Lpl | 2.53 | lipoprotein lipase |
| Rarres1 | 1.63 | retinoic acid receptor responder 1 |
| Sdro | 0.53 | orphan short chain dehydrogenase |
| Smpd3 | 1.85 | sphingomyelin phosphodiesterase 3 |
| Membrane Transport | ||
| Abcb1a | 2.38 | ATP-binding cassette |
| Abcc3 | 2.28 | ATP-binding cassette |
| Slc16a7 | 1.62 | solute carrier family 16 transporter |
| Slc3a1 | 0.63 | solute carrier family 3 a1 transporter |
| Slco1a4 | 3.10 | solute carrier family 1a4 transporter |
| Oxidative Stress | ||
| Gpx7 | 1.76 | glutathione peroxidase 7 |
| Gsta1 | 2.91 | glutathione S-transferase a1 |
| Gsta4 | 1.56 | glutathione S-transferase m4 |
| Gstm1 | 1.53 | glutathione S-transferase m1 |
| Gstm3 | 8.46 | glutathione S-transferase m3 |
| Inflammatory Response | ||
| Akr1b3 | 1.51 | aldo-keto reductase family1b3 |
| Akr1b7 | 1.97 | aldo-keto redcutase family 1b7 |
| Il1rn | 2.01 | interleukin 1 receptor antagonist |
| Hspb1 | 1.51 | heat shock protein 1 |
| Lcn2 | 3.93 | lipocalin 2 |
| Mmp12 | 3.00 | matrix metallopeptidase 12 |
| Orm3 | 2.16 | orosomucoid 3 |
| Serpinb1a | 2.96 | serpin peptidase inhibitor b1a |
| Serpinb6b | 2.08 | serpin peptidase inhibitor 6b |
| Serpina4 | 0.36 | serpin peptidase inhibitor a4 |
| Tgfbr2 | 1.56 | transforming growth factor |
| Tnfrs19 | 2.23 | tumor necrosis factor receptor 19 |
| Cytochrome P450 Enzymes | ||
| Cyp1a2 | 1.56 | cytochrome P450 1a2 |
| Cyp26a1 | 2.69 | cytochrome P450 26a1 |
| Cyp2b1 | 127.69 | cytochrome P450 2b10 |
| Cyp2c54 | 1.67 | cytochrome P450 2c54 |
| Cyp2c55 | 17.87 | cytochrome P450 2c55 |
| Cyp2u1 | 0.65 | cytochrome P450 2u1 |
| Cyp3a41a | 1.86 | cytochrome P450 3a41a |
| Ubiquitination | ||
| Usp18 | 1.57 | ubiquitin-specific peptidase 18 |
| Ubd | 2.38 | ubiquitin D |
| Yod1 | 0.65 | YOD1 deubiquitinating enzyme 1 |
| Others | ||
| Basp1 | 1.52 | brain abundant |
| Casc4 | 1.51 | cancer susceptibility candidate 4 |
| Cml4/Nat8 | 0.56 | camello-like 4; N-acetyl tranferase |
| Cotl1 | 1.64 | coactosin-like 1 (Dictyostelium) |
| D1Ertd471e | 1.82 | DNA segment |
| Eid1 | 1.50 | EP300 interacting inhibitor of differentiation 1 |
| Entpd5 | 1.65 | ectonucleoside triphosphate diphosphohydrolase 5 |
| Flvcr2 | 1.55 | feline leukemia virus subgroup C cellular receptor family |
| Gdf15 | 1.91 | growth differentiation factor 15 |
| H2-Aa | 1.54 | histocompatibility 2 |
| Lgals7 | 1.67 | lectin |
| Ndrg1 | 1.82 | N-myc downstream regulatory gene 1 |
| Nebl | 4.79 | nebulette; thin filament protein |
| Ptpla | 1.50 | protein tyrosine phosphatase-like |
| Pvr | 1.59 | poliovirus receptor |
| Pvt1 | 1.70 | plasmacytoma variant translocation 1 |
| Rhbg | 1.66 | Rhesus blood group-associated B glycoprotein |
| Rragd | 1.91 | Ras-related GTP binding D |
| Snx17 | 0.60 | sorting nexin 17 |
| Sulf2 | 1.54 | sulfatase 2 |
| Tlr2 | 1.52 | Toll-like receptor 2 |
| Tmed3 | 1.51 | transmembrane emp24 domain containing 3 |
| Tmem120a | 0.47 | transmembrane protein 120A |
| Tmem176a | 1.53 | transmembrane protein 176A |
| Tmem51 | 1.53 | transmembrane protein 51 |
| Ugt2b37 | 2.73 | UDP glucuronosyltransferase 2 family |
| Unknowns | ||
| 1810023F06Rik | 1.61 | RIKEN cDNA 1810023F06 |
| 4931406C07Rik | 1.71 | RIKEN cDNA 4931406C07 |
| ? | 2.29 | 0 day neonate lung cDNA |
Data are expressed as ratio of LCN vs. control (KO/C) when identified as significant at q < 0.05 using the t-test with Benjamini-Hochberg FDR correction. Protein/function identification is from the UniProtKB/Swiss-Prot Database.
At least 17 identified genes were associated with oxidative stress and inflammation probably reflecting the compensation for hepatic dysfunction associated with POR ablation. Six genes encoding cytochrome P450 enzymes were also upregulated, while expression of one, Cyp2u1, was low in the LCN livers. A number of phase II drug-metabolizing enzymes were also identified including Gstm3, which has been identified as a cancer susceptibility gene (24, 49). Many of these gene changes correlate with those previously observed for the POR knockout animals and were not unexpected (43, 45). Our unique finding, as noted above, was the elimination of the lipid metabolic pathway genes as significantly different between genotypes.
We also performed GSEA pathway analysis by including all array datasets without consideration of diet (9 each for control and LCN) using an FDR q-value of ≤ 0.25. Although relatively high, this threshold is considered useful to provide biological insights (40). Employing these criteria, 53 pathway gene sets were identified (Supplemental Table S4). Only four of the identified pathways were coincident with those identified as different due to diet: prostaglandin synthesis regulation, HSA05220 chronic myeloid leukemia, HSA04110 cell cycle, and cell cycle Kegg. Interestingly, in the Molecular Signature Databases (Broad Inst.) used, four pathway sets are included from our previous report on the LCN gene expression array data (45). However, none of these were identified in our GSEA as significantly enriched even at an FDR q-value of ≤ 0.25. These results point to the dominant effect of diet over genotype uncovered in the present studies.
Diet-gene interactions affect liver transcriptome.
Array data for livers from animals of each genotype fed each of the three diets was analyzed using two-way ANOVA to examine interactive effects between genotype and diet (GeneSpring software). However, we found that if FDR was applied for multiple test correction, no genes were identified as significantly different suggesting the interactive effects between genotype and diet were subtle. Without applying a correction but using a P value of < 0.01, 22 genes were identified whose expression was influenced by both diet and genotype (Table 5). Among these, one gene was identified that is required for fatty acid elongation, Elovl 1. Also identified were the genes encoding lipoprotein lipase (Lpl) and lipocalin 13 (Lcn13), two proteins implicated in fatty acid transport. Lcn13 expression was highest in the control livers from mice fed the fish/fungal diet, while Lpl expression was highest in the LCN livers on the fish/fungal diet. Both genes had low expression in livers when the animals had been fed the lard or canola diets. While the function of Lpl is very well defined, that of Lcn 13 is not. Lipocalins are members of a family of secreted proteins that bind and transport small hydrophobic molecules. One family member Lcn2/NGAL has been implicated in inflammatory response (41, 42, 44) and is suggested as a biomarker of kidney disease (26, 38). Identification of Lcn13 as a factor affected by both dietary fat and hepatic POR indicates it should be explored as a sensitive biomarker of liver function. Fewer genes associated with inflammation and P450 function were identified as being affected by both genotype and diet as were identified by comparisons of genotype effects alone.
Table 5.
Hepatic genes whose expression is significantly different due to both diet and genotype
| Gene | Ctl L | KO L | Ctl C | KO C | Ctl FF | KO FF | P Value | Protein Name/Function |
|---|---|---|---|---|---|---|---|---|
| Fatty Acid Metabolism | ||||||||
| Elovl1 | 0.02 | 0.30 | −0.20 | 0.08 | −0.64 | −0.01 | 0.011 | FA elongation |
| Lcn13 | −0.15 | −0.81 | −0.14 | −0.58 | 2.81 | 0.13 | 0.003 | lipocalin 13 |
| Lpl | −0.20 | 0.89 | −0.59 | 0.43 | −0.35 | 1.55 | 0.003 | lipoprotein lipase |
| Ppt1 | −0.02 | 0.01 | −0.07 | −0.07 | 0.18 | 0.04 | 0.014 | palmitoyl-protein thioesterase 1 |
| Inflammation/Inflammatory Response | ||||||||
| Mmp12 | −0.44 | 2.43 | −0.15 | 0.83 | −0.23 | 0.68 | 0.012 | matrix metallopeptidase 12 |
| Regulation | ||||||||
| Vegfb | 0.20 | 0.18 | 0.17 | −0.07 | −0.62 | −0.30 | 0.007 | vascular endothelial growth factor B |
| Creb3l2 | −0.25 | 0.21 | −0.16 | 0.02 | 0.01 | 0.01 | 0.002 | cAMP responsive element binding protein 3-like 2 |
| Others | ||||||||
| Adamts16 | 0.17 | −0.02 | 0.01 | 0.00 | 0.00 | −0.09 | 0.011 | A disintegrin-like and metallopeptidase |
| Clcn6 | 0.01 | 0.20 | −0.03 | −0.01 | −0.33 | 0.04 | 0.006 | chloride channel 6 |
| Col23a1 | 0.00 | −0.27 | 0.07 | 0.03 | 0.00 | −0.02 | 0.001 | procollagen |
| Dsn1 | −0.11 | 0.27 | −0.14 | 0.06 | 0.03 | 0.00 | 0.002 | MIND1 kinetochore complex component |
| Gpc1 | 0.44 | −0.07 | 0.56 | −0.14 | −0.25 | −0.10 | 0.000 | glypican 1 |
| Gpr137b | −0.38 | 0.94 | −0.25 | 0.12 | 0.04 | 0.20 | 0.008 | G protein-coupled receptor 137B |
| Neu1 | −0.08 | 0.05 | 0.00 | 0.07 | −0.24 | 0.13 | 0.009 | neuraminidase 1 |
| Odf1 | 0.01 | 0.04 | 0.12 | −0.21 | 0.02 | −0.15 | 0.002 | outer dense fiber of sperm tails 1 |
| Rbpms | −0.35 | 0.03 | −0.32 | 0.02 | 0.08 | 0.13 | 0.014 | RNA binding protein gene with multiple splicing |
| Sgcg | 0.14 | −0.19 | 0.18 | −0.16 | 0.01 | −0.04 | 0.002 | sarcoglycan |
| Tmem120a | 0.23 | −0.39 | 0.49 | −0.33 | 0.52 | −1.27 | 0.003 | transmembrane protein 120A |
| Tmem97 | 0.05 | 0.24 | 0.01 | 0.13 | −0.65 | −0.02 | 0.015 | transmembrane protein 97 |
| Wdr51b | 0.24 | 0.11 | −0.16 | −0.06 | −0.07 | 0.02 | 0.004 | WD repeat domain 51B |
| Wrnip1 | −0.24 | 0.11 | −0.11 | 0.05 | −0.10 | 0.35 | 0.014 | Werner helicase interacting protein 1 |
| Unknown | ||||||||
| −0.01 | −0.01 | 0.06 | −0.12 | 0.21 | −0.03 | 0.003 | RIKEN cDNA 9030224M |
Log2 transformed fold change and significance were calculated by 2-way ANOVA, q < 0.01. Ctl, control; KO, LCN; L, lard; C, canola; FF, fish/fungal.
It should be pointed out that many of the genes identified as differentially expressed due to interaction between genotype and diet did not cluster into easily identifiable categories. Among these were several regulatory proteins including Vegf-B, Creb family member Creb3l2, and G protein-coupled receptor 137b. Previous studies showed that Vegf-B overexpression in cardiomyocytes results in accumulation of ceramide, decreased triglyceride, increased mitochondrial lysis, and accumulation of intracellular lipid membrane vacuoles (21). A recent paper demonstrated Vegf-B targets fatty acids to endothelial cells by upregulating expression of fatty acid transport proteins (14). Creb3l2 has been implicated in regulating the unfolded protein response (2, 25).
Examination of gene expression patterns for selected genes by Q-PCR and Western analysis.
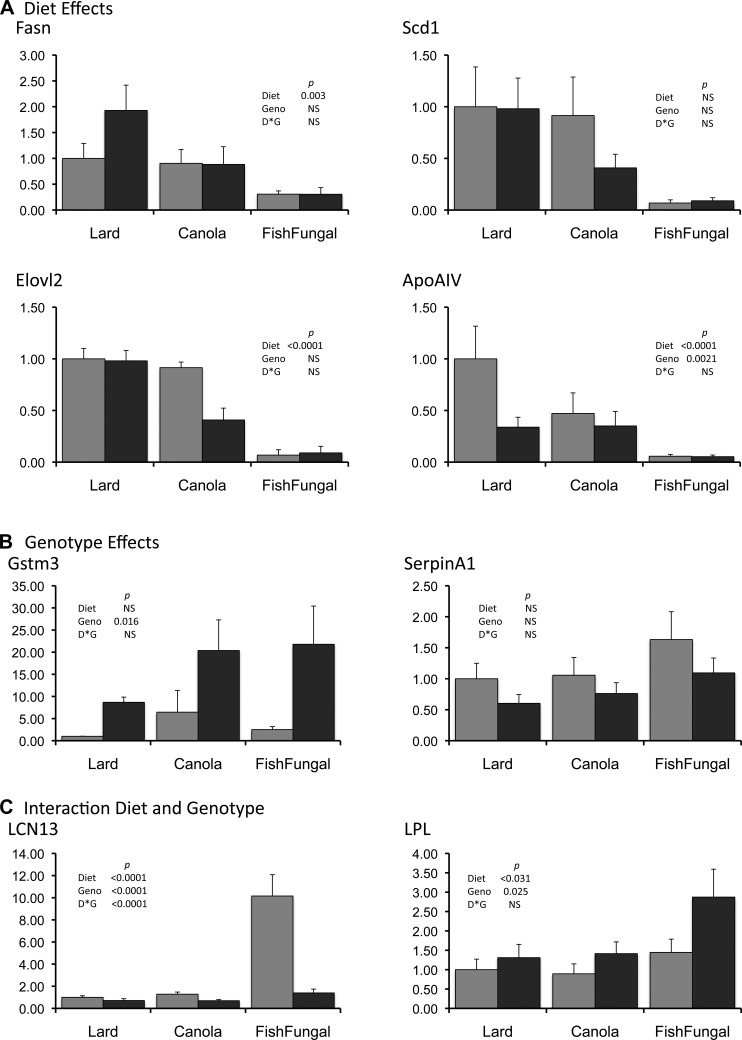
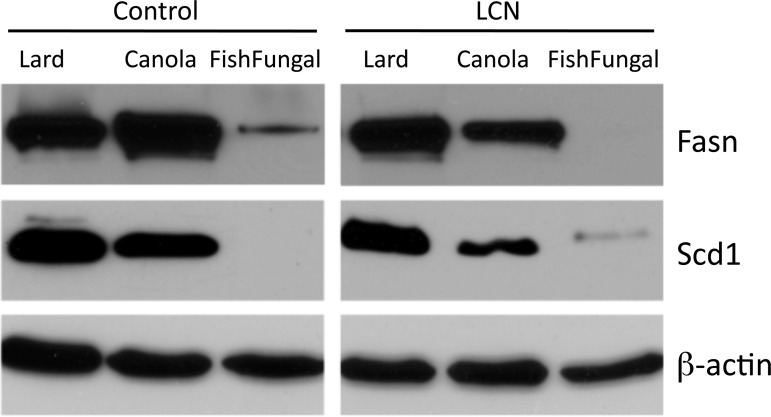
Q-PCR of selected genes was performed using RNA from individual liver samples from six to eight separate mice maintained on each diet to further evaluate apparent expression changes due to diet, genotype, and the interaction between diet and genotype. Among the genes identified as most significantly sensitive to dietary fatty acid by expression array analysis, we focused on those encoding Fasn, fatty acid elongase 2 (Elovl 2) and ApoAIV (Fig. 3A). As expected each was significantly different for the diet comparison. ApoAIV was also determined to be significant at the level of genotype but no interactions were detected. Expression of the Δ9-stearyl CoA desaturase gene Scd1 is often reported to be sensitive to dietary fat. Because it was not identified in our array analyses, we examined its expression by Q-PCR as well. We found there was substantial between-animal variation in Scd1 expression that precluded establishment of statistically significant differences. However, there was a strong trend for higher expression in livers of mice fed the lard and canola diets compared with those of mice fed the fish/fungal diet. Protein levels of Fasn and Scd1 followed the same QPCR patterns (Fig. 4).
Fig. 3.
Quantitation of expression of specific genes using Q-PCR. Genes were selected among those that showed significant differences in expression on arrays due to diet (A), genotype (B), or both diet and genotype (C). Gene designations are as indicated in the text. Gray bars indicate data for controls and black bars for LCN. Data are expressed relative to the value for control lard within each experiment; n = 6–8 mice per group. The error bar gives SE. Data analyzed using 2-way ANOVA and the Student's t-test; significance values are as indicated.
Fig. 4.
Protein expression in control and LCN livers from mice fed the 3 test diets. Representative images are for fatty acid synthase (Fasn) and Δ9-stearyl-CoA desaturase (Scd1) as indicated. β-Actin served as a loading control.
Two genes, Gstm3 and SerpinA1, that were found to be significantly different between genotypes by array analysis, were also selected for Q-PCR (Fig. 3B). Expression of Gstm3 was significantly higher in the LCN livers compared with controls. SerpinA1 was lower in the knockout livers but this difference was not statistically significant (P = 0.078). Neither was significantly different when diets were compared, and there were no interaction effects.
To examine genes whose expression was suggested to be sensitive to both diet and genotype, we selected Lcn13 and Lpl (Fig. 3C). In the case of Lcn13 all three parameters tested were significantly different. For lipoprotein lipase, genotype and diet were significantly different but no interaction was confirmed.
DISCUSSION
The liver-specific deletion of the Por gene results in a complex phenotype that affects P450 pathways and sterol and fatty acid metabolism. In the present study, we provide evidence that dietary fatty acids alter hepatic Tg accumulation and the gene expression profile in LCN mice as well as controls. The fish/fungal diet, high in the HUFA AA, EPA, and DHA, reduced the total amount of Tg and cholesterol in liver and had a substantial, primarily suppressive, effect on hepatic gene expression. Surprisingly, the impact of the canola diet, high in the PUFA LN and LA, on gene expression and lipid accumulation was more similar to the lard diet, which was low in PUFA and HUFA but high in SFA. The impact on LCN gene expression was such that most fatty acid and sterol metabolic genes identified were primarily influenced by diet over genotype. The relatively subdued effects of Por deletion on the expression of fatty acid and sterol metabolic genes in mice fed with the three synthetic diets are in sharp contrast to the large genotype effects seen previous by us and others in mice fed standard lab chow (43, 45). Analysis of gene expression changes clearly distinguished effects related to diet and effects related to genotype with few overlapping or interactive differences. Thus, the synthetic diets in which only the fat component was different were useful to scrutinize the impact of POR on hepatic metabolic genes and resulted in distinguishing those involved in P450-related pathways, oxidative stress, and inflammation apart from lipid metabolism.
Most evidence to date specifically implicates DHA as the fatty acid that is responsible for modulation of genes encoding enzymes of fatty acid metabolism by SREBP1, as well as for many PPAR-dependent effects (20). In supplying diets enriched in the PUFA, LA and LN, we were able to distinguish the precursor-product relationship between PUFA and HUFA. Liver is able to efficiently convert LN to DHA as evidenced by the low levels of LN that accumulated in livers of animals fed the canola diet. However, this did not result in the same regulatory impact as the fish/fungal diet containing preformed DHA. In fact, when estimating significant differences between diets, we found the expression patterns in livers of mice fed the canola diet were more similar to those fed the lard diet than to the fish/fungal diet. The lard and canola diets also caused steatosis in both control and LCN livers, while lipid accumulation in livers of animals fed the fish/fungal diet was more limited. These results are important given the very low HUFA content of the traditional western diet and preponderance of LA as the essential PUFA in that diet. It also points to the importance of differentiating between dietary intake of the various omega 3 fatty acids for apparent health benefits.
Apart from individual gene analysis, further insights were provided by performing GSEA. As expected, sterol and fatty acid metabolic pathways were identified by the comparison between diets. In addition, we also noted the consistent occurrence of signaling pathways related to insulin-dependent regulation and sensitivity to insulin. These pathways share genes including: AKT1/PKB, AKT2, AKT3, MAPK1, MAPK3, MAPK8/JNK, INSR, INS1, INS2, INS4, SOS1, SOS2, PIK3CA, and RAF1 (reviewed in Refs. 35, 47). For example, AKT1 is included in 18 identified pathways, PIK3CA in 22, and MAPK8/JNK in 9. The AKT signaling pathway has also been implicated in a number of processes required to maintain cellular homeostasis by regulating protein synthesis and survival processes (17, 35, 47).
Also identified by GSEA with genes in common with the insulin signaling pathways were the PTEN, growth hormone, and several MAPK and signaling pathways. Similar signaling events involving the MAPKs occur in inflammation, immune function, and cell growth and adhesion (17, 35, 47). For this reason, these insulin-related signaling molecules are also associated with cancer and are identified here in several cancer pathways (e.g., HAS05211_RENAL_CELL_CARCINOMA and HSA05212_PANCREATIC CANCER) (6, 17, 27). Overall, the coordinate identification of the lipid metabolic pathways, insulin, growth hormone and some cancer/cell growth pathways reflects the strong association between nutrient fatty acids and cellular homeostasis that is so critical to health maintenance (17).
Among the 53 pathway sets selected by GSEA using genotype as the criterion was one (HSA00980) that included genes involved in the metabolism of xenobiotics by cytochrome P450. Another group of genes was repeated in seven sets: GSTM1, GSTM2, and GSTM3. The GSTM gene products are glutathione S-transferase M enzymes involved in phase II drug metabolism and have been implicated in oxidative stress pathways. Also repeated in seven other identified pathways were CALM1, CALM2, and CALM3. The Ca2+/calmodulin pathway has been implicated in induction of the P450 pathway genes and enzymes by various drugs and xenobiotics such as phenobarbital (28). While these gene associations are not as strong as those identified for the diet effects, they point to important regulatory and metabolic interrelationships between these pathways and liver POR activity.
Very few studies have been conducted evaluating the impact of essential nutrients on the P450 system. However, recently the Wolf lab reported a study in which they placed liver specific POR-deficient mice on diets with either no fat or added saturated, monounsaturated, or polyunsaturated fat enriched in C18:2ω6 (10). Similar to this report, they found the PUFA were very potent effectors of Tg accumulation in these livers. Only a diet completely deficient in fat prevented the steatosis generally associated with LCN mice. These investigators examined the expression of certain CYP genes and found a connection between fat accumulation and CYP induction that was dependent upon the transcription factor CAR. However, because they did not evaluate any expression changes for fatty acid metabolic genes, nor did they conduct an expression array analysis, it is unclear whether CAR contributes to steatosis by regulating these metabolic genes as well. Given the patterns of gene expression we have uncovered as differentially expressed due to dietary fat it seems more likely that at least SREBP1 and PPARα are playing a regulatory role in this complex phenotype as well.
LCN animals accumulated more PUFA than controls (Supplemental Table S2), indicating conversion of PUFA to HUFA was dampened. However, it was not completely suppressed and the LCN livers accumulated more Tg than controls even when they were fed the fish/fungal diet rich in HUFA. This suggests that the HUFA-dependent regulatory processes are not optimal in the absence of P450 reductase. We speculate this is due to a reduced capacity to synthesize the proximal ligand(s) necessary for maximal suppression of the fatty acid synthetic genes. A focus of future work will be to investigate whether or not the effectors are CYP-dependent DHA metabolites. Taken together, our results demonstrate that the liver P450 systems have a major impact on the subsequent metabolism of lipid nutrients and this may result in downstream regulatory effects.
GRANTS
This work was supported in part by grants from the Charitable Leadership Foundation (Clifton Park, NY), a program of excellence award from the University of Nebraska-Lincoln, National Institute of Environmental Health Sciences Grant ES-007462 (to X. Ding), and the USDA through the Hatch Act. W. Sealls was supported in part by a graduate student award from the Kappa Gamma Fraternity.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
This work was initiated at the Ordway Research Institute in Albany, NY, and completed at the University of Nebraska-Lincoln, NE. All animal husbandry, breeding, and tissue sample collections were carried out in Albany, NY, within the animal research facilities of the Wadsworth Center of the NY State Department of Health. Special thanks are due to Christopher Petty and Zhigang Wang for expert technical assistance.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Audas TE, Li Y, Liang G, Lu R. A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response. Mol Cell Biol 28: 3952–3966, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284, 2001. [DOI] [PubMed] [Google Scholar]
- 4. Berger A, Mutch DM, German JB, Roberts MA. Dietary effects of arachidonate-rich fungal oil and fish oil on murine hepatic and hippocampal gene expression. Lipids Health Dis 1: 2, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boomgaarden I, Vock C, Klapper M, Doring F. Comparative analyses of disease risk genes belonging to the acyl-CoA synthetase medium-chain (ACSM) family in human liver and cell lines. Biochem Genet 47: 739–748, 2009. [DOI] [PubMed] [Google Scholar]
- 6. Chaussade C, Rewcastle GW, Kendall JD, Denny WA, Cho K, Gronning LM, Chong ML, Anagnostou SH, Jackson SP, Daniele N, Shepherd PR. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem J 404: 449–458, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coon MJ, van der Hoeven TA, Dahl SB, Haugen DA. Two forms of liver microsomal cytochrome P-450, P-450lm2 and P-450LM4 (rabbit liver). Methods Enzymol 52: 109–117, 1978. [DOI] [PubMed] [Google Scholar]
- 8. Dorfmeister B, Brandlhofer S, Schaap FG, Hermann M, Furnsinn C, Hagerty BP, Stangl H, Patsch W, Strobl W. Apolipoprotein AV does not contribute to hypertriglyceridaemia or triglyceride lowering by dietary fish oil and rosiglitazone in obese Zucker rats. Diabetologia 49: 1324–1332, 2006. [DOI] [PubMed] [Google Scholar]
- 9. Dorfmeister B, Zeng WW, Dichlberger A, Nilsson SK, Schaap FG, Hubacek JA, Merkel M, Cooper JA, Lookene A, Putt W, Whittall R, Lee PJ, Lins L, Delsaux N, Nierman M, Kuivenhoven JA, Kastelein JJ, Vrablik M, Olivecrona G, Schneider WJ, Heeren J, Humphries SE, Talmud PJ. Effects of six APOA5 variants, identified in patients with severe hypertriglyceridemia, on in vitro lipoprotein lipase activity and receptor binding. Arterioscler Thromb Vasc Biol 28: 1866–1871, 2008. [DOI] [PubMed] [Google Scholar]
- 10. Finn RD, Henderson CJ, Scott CL, Wolf CR. Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J 417: 43–54, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509, 1957. [PubMed] [Google Scholar]
- 12. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278: 25895–25901, 2003. [DOI] [PubMed] [Google Scholar]
- 14. Hagberg CE, Falkevall A, Wang X, Larsson E, Huusko J, Nilsson I, van Meeteren LA, Samen E, Lu L, Vanwildemeersch M, Klar J, Genove G, Pietras K, Stone-Elander S, Claesson-Welsh L, Yla-Herttuala S, Lindahl P, Eriksson U. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature 464: 917–921, 2010. [DOI] [PubMed] [Google Scholar]
- 15. Hagerty BP, Schaap FG, Hermann M, Krenn B, Eder C, Dorfmeister B, Stangl H, Patsch W, Strobl W. Changes in hepatic ApoAV expression are not required for the rapid triglyceride lowering effect of fish oil diet in rats. Horm Metab Res 40: 69–71, 2008. [DOI] [PubMed] [Google Scholar]
- 16. Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278: 13480–13486, 2003. [DOI] [PubMed] [Google Scholar]
- 17. Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43: 389–410, 2009. [DOI] [PubMed] [Google Scholar]
- 18. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jump DB. N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr Opin Lipidol 19: 242–247, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jump DB, Botolin D, Wang Y, Xu J, Demeure O, Christian B. Docosahexaenoic acid (DHA) and hepatic gene transcription. Chem Phys Lipids 153: 3–13, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karpanen T, Bry M, Ollila HM, Seppanen-Laakso T, Liimatta E, Leskinen H, Kivela R, Helkamaa T, Merentie M, Jeltsch M, Paavonen K, Andersson LC, Mervaala E, Hassinen IE, Yla-Herttuala S, Oresic M, Alitalo K. Overexpression of vascular endothelial growth factor-B in mouse heart alters cardiac lipid metabolism and induces myocardial hypertrophy. Circ Res 103: 1018–1026, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller MP, Attie AD. Physiological insights gained from gene expression analysis in obesity and diabetes. Annu Rev Nutr 30: 341–364, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim HJ, Takahashi M, Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mRNAs. J Biol Chem 274: 25892–25898, 1999. [DOI] [PubMed] [Google Scholar]
- 24. Koutros S, Berndt SI, Sinha R, Ma X, Chatterjee N, Alavanja MC, Zheng T, Huang WY, Hayes RB, Cross AJ. Xenobiotic metabolizing gene variants, dietary heterocyclic amine intake, and risk of prostate cancer. Cancer Res 69: 1877–1884, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang Y, She P, Wang X, Demarest K. The messenger RNA profiles in liver, hypothalamus, white adipose tissue, and skeletal muscle of female Zucker diabetic fatty rats after topiramate treatment. Metabolism 55: 1411–1419, 2006. [DOI] [PubMed] [Google Scholar]
- 26. Liangos O, Tighiouart H, Perianayagam MC, Kolyada A, Han WK, Wald R, Bonventre JV, Jaber BL. Comparative analysis of urinary biomarkers for early detection of acute kidney injury following cardiopulmonary bypass. Biomarkers 14: 423–431, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene 29: 3033–3043. [DOI] [PubMed] [Google Scholar]
- 28. Marc N, Galisteo M, Lagadic-Gossmann D, Fautrel A, Joannard F, Guillouzo A, Corcos L. Regulation of phenobarbital induction of the cytochrome P450 2b9/10 genes in primary mouse hepatocyte culture. Involvement of calcium- and cAMP-dependent pathways. Eur J Biochem 267: 963–970, 2000. [DOI] [PubMed] [Google Scholar]
- 29. Mutch DM, Temanni MR, Henegar C, Combes F, Pelloux V, Holst C, Sorensen TI, Astrup A, Martinez JA, Saris WH, Viguerie N, Langin D, Zucker JD, Clement K. Adipose gene expression prior to weight loss can differentiate and weakly predict dietary responders. PLoS One 2: e1344, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet 360: 1155–1162, 2002. [DOI] [PubMed] [Google Scholar]
- 31. Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 36: W423–W426, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patton-Vogt J. Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim Biophys Acta 1771: 337–342, 2007. [DOI] [PubMed] [Google Scholar]
- 33. Qi L, Liu S, Rifai N, Hunter D, Hu FB. Associations of the apolipoprotein A1/C3/A4/A5 gene cluster with triglyceride and HDL cholesterol levels in women with type 2 diabetes. Atherosclerosis 192: 204–210, 2007. [DOI] [PubMed] [Google Scholar]
- 34. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375, 2003. [DOI] [PubMed] [Google Scholar]
- 35. Salminen A, Kaarniranta K. Insulin/IGF-1 paradox of aging: regulation via AKT/IKK/NF-kappaB signaling. Cell Signal 22: 573–577, 2010. [DOI] [PubMed] [Google Scholar]
- 36. Sealls W, Gonzalez M, Brosnan MJ, Black PN, DiRusso CC. Dietary polyunsaturated fatty acids (C18:2 omega6 and C18:3 omega3) do not suppress hepatic lipogenesis. Biochim Biophys Acta 1781: 406–414, 2008. [DOI] [PubMed] [Google Scholar]
- 37. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Soni SS, Cruz D, Bobek I, Chionh CY, Nalesso F, Lentini P, de Cal M, Corradi V, Virzi G, Ronco C. NGAL: a biomarker of acute kidney injury and other systemic conditions. Int Urol Nephrol 42: 141–150, 2010. [DOI] [PubMed] [Google Scholar]
- 39. Steinmetz A, Schneider WJ. Evolving functions of apolipoprotein AIV: from birds to mammals. Trends Cardiovasc Med 9: 153–157, 1999. [DOI] [PubMed] [Google Scholar]
- 40. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sunil VR, Patel KJ, Nilsen-Hamilton M, Heck DE, Laskin JD, Laskin DL. Acute endotoxemia is associated with upregulation of lipocalin 24p3/Lcn2 in lung and liver. Exp Mol Pathol 83: 177–187, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Dam RM, Hu FB. Lipocalins and insulin resistance: etiological role of retinol-binding protein 4 and lipocalin-2? Clin Chem 53: 5–7, 2007. [DOI] [PubMed] [Google Scholar]
- 43. Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (POR) null mice. Biochem J 388: 857–867, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y, Lam KS, Kraegen EW, Sweeney G, Zhang J, Tso AW, Chow WS, Wat NM, Xu JY, Hoo RL, Xu A. Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem 53: 34–41, 2007. [DOI] [PubMed] [Google Scholar]
- 45. Weng Y, Dirusso CC, Reilly AA, Black PN, Ding X. Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene: mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J Biol Chem 280: 31686–31698, 2005. [DOI] [PubMed] [Google Scholar]
- 46. Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wing SS. The UPS in diabetes and obesity. BMC Biochem 9, Suppl 1: S6, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, Zhang QY, Zhuo X, Xie Q, Ding X. Conditional knockout of the mouse NADPH-cytochrome p450 reductase gene. Genesis 36: 177–181, 2003. [DOI] [PubMed] [Google Scholar]
- 49. Yu KD, Fan L, Di GH, Yuan WT, Zheng Y, Huang W, Chen AX, Yang C, Wu J, Shen ZZ, Shao ZM. Genetic variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat 121: 485–496, 2010. [DOI] [PubMed] [Google Scholar]
- 50. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40: 638–645, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.