Abstract
14-3-3 family members are dimeric phosphoserine-binding proteins that participate in signal transduction and checkpoint control pathways. In this work, dominant-negative mutant forms of 14-3-3 were used to disrupt 14-3-3 function in cultured cells and in transgenic animals. Transfection of cultured fibroblasts with the R56A and R60A double mutant form of 14-3-3ζ (DN–14-3-3ζ) inhibited serum-stimulated ERK MAPK activation, but increased the basal activation of JNK1 and p38 MAPK. Fibroblasts transfected with DN–14-3-3ζ exhibited markedly increased apoptosis in response to UVC irradiation that was blocked by pre-treatment with a p38 MAPK inhibitor, SB202190. Targeted expression of DN–14-3-3η to murine postnatal cardiac tissue increased the basal activation of JNK1 and p38 MAPK, and affected the ability of mice to compensate for pressure overload, which resulted in increased mortality, dilated cardiomyopathy and massive cardiomyocyte apoptosis. These results demonstrate that a primary function of mammalian 14-3-3 proteins is to inhibit apoptosis.
Keywords: apoptosis/cardiac/MAPK/14-3-3 protein/signal transduction
Introduction
14-3-3 proteins are intracellular, dimeric, phosphoserine-binding molecules that have been identified in many eukaryotic organisms, including plants and fungi, and that are found primarily in the cytoplasmic compartment of cells (Aitken et al., 1995; Muslin et al., 1996; Yaffe et al., 1997). In Drosophila melanogaster, genetic screens have established that 14-3-3 proteins are an essential component of the Ras-mediated signaling pathway of the developing embryo, and epistatic analysis has demonstrated that 14-3-3 acts between Ras and MEK (Chang and Rubin, 1997; Kockel et al., 1997; Li et al., 1997). In particular, 14-3-3 proteins play an important role in eye and brain development, and in brain function (Chang and Rubin, 1997; Skoulakis and Davis, 1998).
There are seven mammalian members of the 14-3-3 family encoded by separate genes (β, γ, ɛ, η, σ, τ and ζ) (Aitken et al., 1995; Rittinger et al., 1999). Mammalian 14-3-3 proteins regulate several facets of cell biochemistry, including binding to and promotion of the activation of tyrosine and tryptophan hydroxylases that are important in neurotransmitter synthetic pathways (Furakawa et al., 1993). 14-3-3 proteins bind to the protein kinases Raf-1 (Fantl et al., 1994; Freed et al., 1994; Fu et al., 1994; Irie et al., 1994), KSR-1 (Xing et al., 1997), BCR (Reuther et al., 1994), protein kinase U-α (PKU-α) (S.Zhang et al., 1999), protein kinase C (PKC) (Robinson et al., 1994; Meller et al., 1996) and Ask1 (L.Zhang et al., 1999), and are thought to modulate the activity of these kinases. In the cases of PKC and Ask1, 14-3-3 binding inhibits their activities (Robinson et al., 1994; Meller et al., 1996; L.Zhang et al., 1999). The interaction of 14-3-3 with Raf–1 is required for the Ras-dependent activation of Raf (Muslin et al., 1996; Roy et al., 1998; Thorson et al., 1998; Tzivion et al., 1998). 14-3-3 also interacts with the cell cycle protein phosphatase cdc25c (Peng et al., 1997) and promotes the cytoplasmic localization of cdc25c and PKU-α (Dalal et al., 1999; Kumagai and Dunphy, 1999; Lopez-Girona et al., 1999; S.Zhang et al., 1999). In addition, 14-3-3 binds to the apoptosis-promoting protein BAD, and this interaction prevents BAD from binding to Bcl-XL (Zha et al., 1996). Indeed, one important activity of Akt may be to phosphorylate BAD and thereby create 14-3-3-binding sites (Datta et al., 1997; del Peso et al., 1997). Mutation of the 14-3-3-binding sites of BAD promotes its ability to stimulate apoptosis (Zha et al., 1996). Recently, 14-3-3 was found to bind to and promote the cytoplasmic localization of the apoptosis-promoting forkhead transcription factor FKHRL1 (Brunet et al., 1999). The phosphorylation of FKHRL1 by Akt promotes the association of 14-3-3 with FKHRL1 and inhibits the ability of this transcription factor to stimulate apoptosis (Brunet et al., 1999).
Although many 14-3-3 binding partners have been identified, there is limited information about the biological function of mammalian 14-3-3 proteins. An important advance in this field was the identification of dominant-negative mutant forms of 14-3-3 that were first identified by Chang and Rubin (1997) in a D.melanogaster genetic screen. When mutant forms of mammalian 14-3-3η and ζ were made that were homologous to the dominant-negative forms of DM14-3-3ɛ, they were found to have a modest reduction in their ability to bind to phosphoserine-containing peptides, perhaps due to altered substrate preference (Thorson et al., 1998; Wang et al., 1998; Rittinger et al., 1999). Other mutant forms of mammalian 14-3-3η and ζ were produced that were found to be much more deficient in their ability to bind to phosphoserine-containing peptides (Thorson et al., 1998; Wang et al., 1998). These mutant forms of 14-3-3η and ζ included the point mutant K49E form and the double mutant R56A and R60A form. Transfection of cultured mammalian cells with the R56A and R60A double mutant form of 14-3-3η inhibited the activity of BXB-Raf (Thorson et al., 1998) and also affected the subcellular localization of the 14-3-3 binding partner PKU-α (S.Zhang et al., 1999b). Another mutant form of the fission yeast 14-3-3 homolog Rad24 was identified recently that resulted in the nuclear accumulation of cdc25c (Lopez-Girona et al., 1999). In this mutant, two hydrophobic residues in a putative nuclear export signal motif (NES) were altered. Analysis of the crystal structure of 14-3-3 suggests that these hydrophobic residues lie within the phosphoserine-binding pocket, and the homologous I217A and L220A double mutant form of mammalian 14-3-3ζ is predicted to have dominant-negative activity (Wang et al., 1998).
In this work, we used the double R56A and R60A mutant forms of 14-3-3ζ and 14-3-3η (DN–14-3-3ζ and DN–14-3-3η), and the double I217A and L220A mutant form of 14-3-3ζ (NES–14-3-3ζ) as reagents to evaluate the biological role of 14-3-3 proteins in mammalian cells. We found that a primary function of 14-3-3 proteins is to inhibit apoptosis and that this effect is mediated partially by the differential regulation of MAPK pathways.
Results
14-3-3 is required for serum-stimulated ERK MAPK activation
To evaluate the biological function of 14-3-3 proteins in mammalian cells, we used the double arginine mutant forms (R56A and R60A) of human 14-3-3η and ζ (DN–14-3-3η or DN–14-3-3ζ) and the NES (I217A and L220A) mutant form of human 14-3-3ζ (NES–14-3-3ζ).
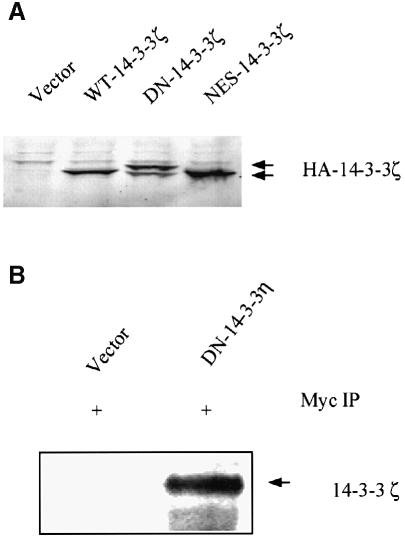
The cDNAs encoding DN–14-3-3ζ, NES–14-3-3ζ and the wild-type 14-3-3ζ (WT–14-3-3ζ), each with hemagglutinin (HA) and FLAG epitope tags, and the cDNA encoding DN–14-3-3η with a Myc-1 epitope tag were used to generate stably transfected NIH 3T3 cell lines. The DN–14-3-3ζ, NES–14-3-3ζ and WT–14-3-3ζ cell lines contained nearly identical transfected protein levels as determined by anti-HA epitope immunoblotting (Figure 1A). Analysis of anti-pan-14-3-3 immunoblots revealed that DN–14-3-3ζ, NES–14-3-3ζ and WT–14-3-3ζ protein levels represent 40% of the total 14-3-3 proteins in the respective cell lines (data not shown). The putative mechanism of action of dominant-negative 14-3-3 is that it forms inactive heterodimers with native 14-3-3 monomers. To determine whether Myc-1 epitope-tagged DN–14-3-3η binds to native 14-3-3ζ protein, we performed co-immunoprecipitation experiments. Anti-Myc-1 immunoprecipitates were analyzed by anti-14-3-3ζ immunoblotting with an isoform-specific antibody that does not recognize DN–14-3-3η (S.Zhang et al., 1999), and this revealed that DN–14-3-3η protein interacts with native 14-3-3ζ (Figure 1B).
Fig. 1. Protein levels of mutant forms of 14-3-3 in NIH 3T3 cells. (A) Immunoblot analysis of protein lysates. NIH 3T3 cells were stably transfected with HA epitope-tagged versions of DN–14-3-3ζ, NES–14-3-3ζ and WT–14-3-3ζ, or with vector alone. Protein lysates from stably transfected NIH 3T3 cells (DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ, Vector) were separated by SDS–PAGE and analyzed by immunoblotting with an anti-HA epitope primary antibody. (B) DN–14-3-3η binds to native WT–14-3-3ζ. NIH 3T3 cells were stably transfected with Myc-1 epitope-tagged DN–14-3-3η or with vector alone. Cytosolic extracts of transfected cells were used to generate anti-Myc-1 immunoprecipitates that were analyzed by immunoblotting with a highly specific anti-14-3-3ζ antibody. The anti-14-3-3ζ antibody does not recognize DN–14-3-3η (S.Zhang et al., 1999).
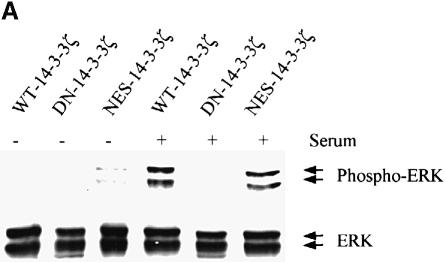
Transfected cell lines were evaluated for serumstimulated ERK MAPK activation by the use of an antibody that is highly specific for the dual phosphorylated, active form of ERK MAPK (Zecevic et al., 1998). Serum-stimulated ERK MAPK activation were markedly inhibited in NIH 3T3 cells transfected with DN–14-3-3ζ (Figure 2A) and DN–14-3-3η (data not shown), but not with NES–14-3-3ζ or WT–14-3-3ζ (Figure 2A). Despite the ability of DN–14-3-3ζ and DN–14-3-3η to inhibit ERK MAPK activation, NIH 3T3 cells that were transfected with these mutants appeared normal, exhibited normal growth rates when cultured in the presence of 10% fetal calf serum (FCS) and had a normal cell cycle distribution as determined by fluorescence cell sorting (data not shown).
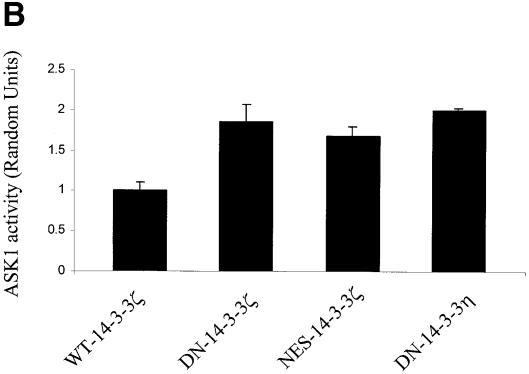
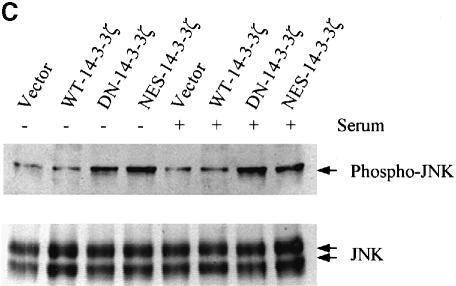
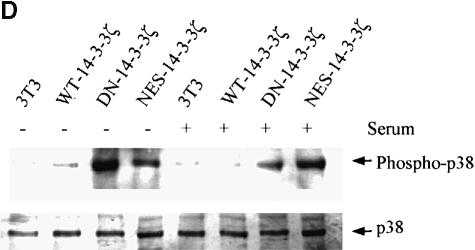
Fig. 2. Mutant forms of 14-3-3 differentially affect MAPK signaling pathways. (A) Transfection of NIH 3T3 cells with DN–14-3-3ζ but not with NES–14-3-3ζ or WT–14-3-3ζ blocks ERK MAP kinase activation. Cells were deprived of serum for 24 h (–) and some (+) were treated with 10% FCS for 10 min. Protein lysates from stably transfected cells (DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ) were separated by SDS–PAGE and analyzed by immunoblotting with a highly specific, anti-phospho-ERK antibody (upper panel). Equal amounts of total protein were loaded in each lane. Lysates were also analyzed in parallel by immunoblotting with an anti-ERK (lower panel) antibody to control for protein content. These immunoblots are representative of five separate experiments. (B) DN–14-3-3ζ, DN–14-3-3η and NES–14-3-3ζ promote Ask1 activation. NIH 3T3 cells stably transfected with DN–14-3-3ζ, DN–14-3-3η, NES–14-3-3ζ or WT–14-3-3ζ were grown in the presence of serum. Anti-Ask1 immunoprecipitates derived from stably transfected NIH 3T3 cells were assayed for kinase activity by the use of MBP as a substrate in the presence of [γ-32P]ATP. Equal amounts of total protein were used for each immunoprecipitate. Proteins were separated by SDS–PAGE and radiolabeled MBP was detected by autoradiography. Bands were quantitated by densitometry using NIH Image software, and each column reflects the mean signal intensity ± SEM of three determinations. (C) DN–14-3-3ζ and NES–14-3-3ζ promote JNK1 activation. Cells were deprived of serum for 24 h and some were treated with 10% FCS for 10 min. Protein lysates from stably transfected cells (DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ, Vector) were separated by SDS–PAGE and analyzed by immunoblotting with a highly specific, anti-phospho-JNK antibody (upper panel). Equal amounts of total protein were loaded in each lane. Lysates were also analyzed in parallel by immunoblotting with an anti-JNK (lower panel) antibody to control for protein content. These immunoblots are representative of three separate experiments. (D) DN–14-3-3ζ and NES–14-3-3ζ promote p38 MAPK activation. Cells were deprived of serum for 24 h and some were treated with 10% FCS for 10 min. Protein lysates from untransfected (3T3) or stably transfected cells (DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ) were separated by SDS–PAGE and analyzed by immunoblotting with a highly specific, anti-phospho-p38 (upper panel) antibody. Equal amounts of total protein were loaded in each lane. Lysates were also analyzed in parallel by immunoblotting with an anti-p38 MAPK (lower panel) antibody to control for protein content. These immunoblots are representative of three separate experiments.
14-3-3 blocks basal JNK and p38 MAP activation
Previous work has demonstrated that 14-3-3 binds to and inhibits the activity of the mitogen-activated protein kinase kinase kinase Ask1 (L.Zhang et al., 1999), a protein that regulates the activation of JNK and p38 MAPK (Ichijo et al., 1997). These findings suggest that 14-3-3 may inhibit signaling by JNK and p38 MAPK and that dominant-negative 14-3-3 could promote the activation of these kinases. To confirm that 14-3-3 regulates Ask1 activity, anti-Ask1 immunoprecipitates derived from unstimulated DN–14-3-3-transfected cell lines were evaluated by in vitro kinase assay with myelin basic protein (MBP) as a substrate. These assays demonstrated that basal Ask1 activity was significantly increased in cells transfected with DN–14-3-3ζ, DN–14-3-3η or NES–14-3-3ζ, but not in cells transfected with WT–14-3-3ζ (Figure 2B).
We next evaluated JNK1 and p38 MAPK activation by the use of antibodies that are highly specific for the dual phosphorylated, active forms of these protein kinases (Chan et al., 1997). Analysis of DN–14-3-3ζ-transfected NIH 3T3 cells revealed that basal levels of JNK1 (Figure 2C) and p38 MAPK (Figure 2D) activation were substantially higher than in untransfected cells or in cells that were transfected with WT–14-3-3ζ. In addition, cells transfected with NES–14-3-3ζ, but not with WT–14-3-3ζ, showed enhanced basal levels of JNK1 and p38 MAPK activation (Figure 2C and D). Therefore, the NES–14-3-3ζ mutant promotes Ask1, JNK1 and p38 MAPK activation, but does not inhibit ERK MAPK activation.
14-3-3 inhibits apoptosis in response to UVC irradiation, serum deprivation and TNF-α treatment
Three binding partners of 14-3-3 are the pro-apoptotic proteins BAD, FKHLR1 and Ask1 (Zha et al., 1996; Brunet et al., 1999; L.Zhang et al., 1999). 14-3-3 is thought to inhibit the ability of these proteins to promote apoptosis by sequestering BAD and FKHRL1 in the cytoplasm, and by inactivating the catalytic activity of the protein kinase Ask1. These results are consistent with the hypothesis that 14-3-3 is a general anti-apoptotic factor in cells. One prediction that can be made on the basis of this hypothesis is that reduction of 14-3-3 activity will promote apoptosis. To test this prediction, we subjected dominant-negative 14-3-3-transfected cells to UVC irradiation or serum deprivation, well-defined stimuli that activate JNK and p38 MAPK and that promote apoptosis (Gunn et al., 1983; Schreiber et al., 1995).
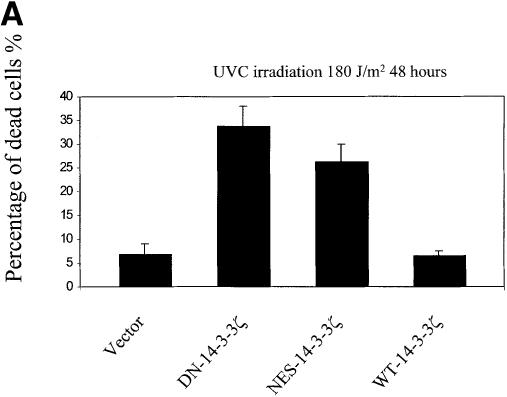
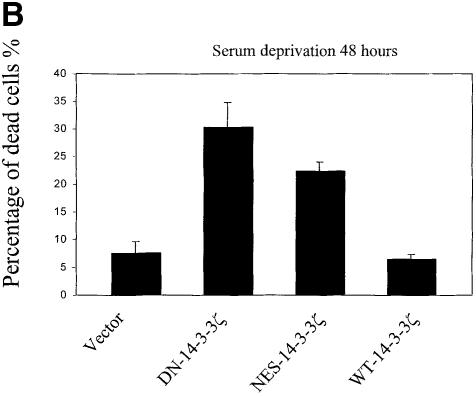
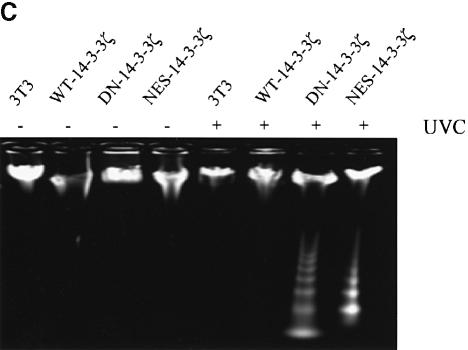
UVC irradiation or serum deprivation of NIH 3T3 cells transfected with DN–14-3-3ζ or NES–14-3-3ζ, but not with WT–14-3-3ζ, caused marked cell death as determined by Trypan blue exclusion (Figure 3A and B). Genomic DNA fragmentation assays confirmed that UV irradiation-induced cell death (Figure 3C) and serum deprivation-induced cell death (data not shown) were secondary to apoptosis.
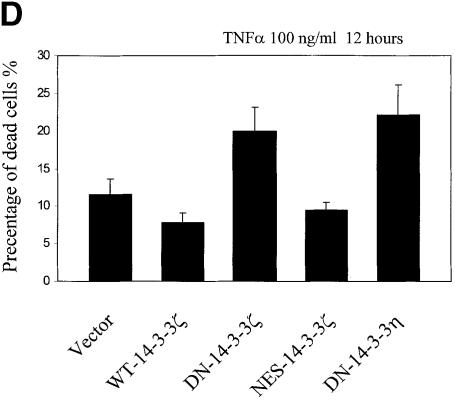
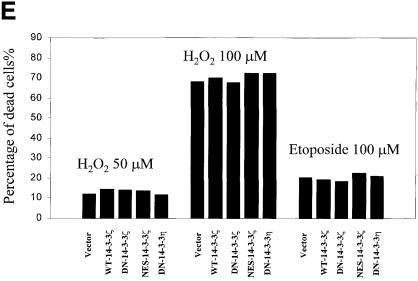
Fig. 3. Increased sensitivity to UVC-, serum deprivation- and TNF-α-induced apoptosis in DN–14-3-3ζ-transfected cells. (A and B) Killing curves of transfected (Vector, DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ) NIH 3T3 cells (A) 48 h after UVC irradiation and (B) after 48 h of serum deprivation. Cells were analyzed by the Trypan blue exclusion method. At least 100 cells were analyzed for each condition. Results are the mean ± SEM of three experiments. (C) DNA fragmentation assays before (–) and after UVC irradiation (+). Untransfected (3T3) or stably transfected (DN–14-3-3ζ, NES–14-3-3ζ, WT–14-3-3ζ) NIH 3T3 cells were irradiated with 180 J/m2 UVC, incubated for 48 h, and then genomic DNA was extracted from cells to assess DNA fragmentation by agarose gel electrophoresis. These results are representative of four separate experiments. (D and E) Killing curves of transfected (Vector, DN–14-3-3ζ, DN–14-3-3η, NES–14-3-3ζ, WT–14-3-3ζ) NIH 3T3 cells after 12 h of stimulation with (D) 100 ng/ml TNF-α and (E) 50–100 μM hydrogen peroxide or 100 μM etoposide. Cells were analyzed by the Trypan blue exclusion method. At least 100 cells were analyzed for each condition.
We next subjected dominant-negative 14-3-3-transfected cells to treatment with the pro-inflammatory cytokine tumor necrosis factor-α (TNF-α), a molecule that activates the TNF receptor I and multiple MAPK cascades (De Cesaris et al., 1999). Interestingly, previous work has suggested that ERK MAPK plays a critical role in suppressing the apoptotic response in HeLa cells to the Fas receptor, a protein that is highly related to TNF receptor I (Holmstrom et al., 1999). TNF-α treatment of NIH 3T3 cells transfected with DN–14-3-3ζ or DN–14-3-3η, but not with NES–14-3-3ζ or WT–14-3-3ζ, caused marked cell death as determined by Trypan blue exclusion (Figure 3D).
Finally, we stimulated cells with pro-apoptotic molecules that are known to activate directly caspases, etoposide and hydrogen peroxide (Kauffman, 1998; DiPietrantonio et al., 1999; Matsura et al., 1999). We found that these agents promoted equivalent amounts of cell death in wild-type 14-3-3- and dominant-negative 14-3-3-transfected NIH 3T3 cells (Figure 3E).
Key role of p38 MAPK in the inhibition of apoptosis by 14-3-3 in response to UVC irradiation
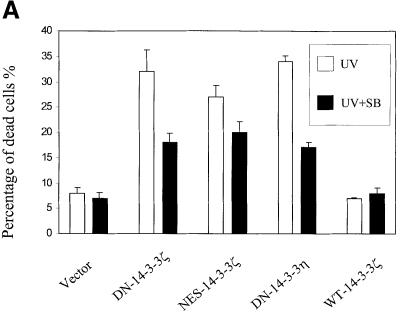
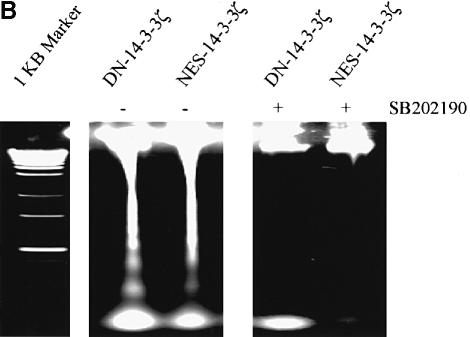
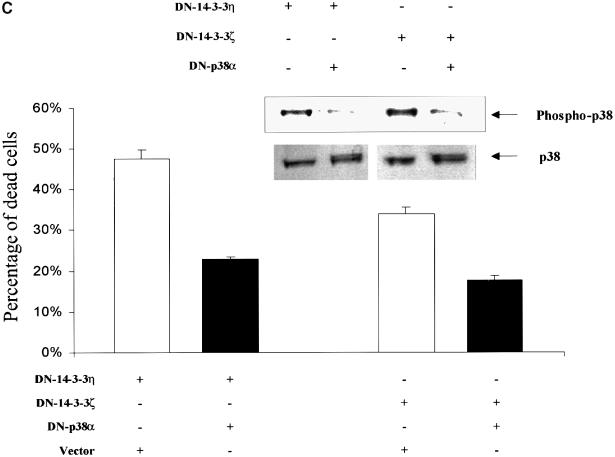
To determine whether UVC-stimulated apoptosis in DN–14-3-3-transfected cells was dependent on p38 MAPK activation, we pre-treated cells with SB202190, a compound that specifically inhibits the activity of p38 MAPK (Horstmann et al., 1998). Indeed, UVC irradiation-induced apoptosis was blocked in DN–14-3-3ζ-, DN–14-3-3η- or NES–14-3-3ζ-transfected NIH 3T3 cells that were pre-treated with SB202190 (Figure 4A and B).
Fig. 4. Treatment with SB202190 or transfection with dominant-negative p38α inhibits UVC irradiation-induced apoptosis in DN–14-3-3ζ-, DN–14-3-3η- and NES–14-3-3ζ-transfected NIH 3T3 cells. NIH 3T3 cells were stably transfected with DN–14-3-3ζ, DN–14-3-3η or NES–14-3-3ζ, and some were treated with SB202190 (20 μM) (+) or with vehicle (–) for 1 h prior to irradiation with 180 J/m2 UVC and then incubated for 48 h. (A) Killing curves of transfected NIH 3T3 cells 48 h after UVC irradiation. Cells were analyzed by the Trypan blue exclusion method. At least 100 cells were analyzed for each condition. Results are the mean ± SEM of three experiments. (B) DNA fragmentation assays performed 48 h after UVC irradiation in transfected NIH 3T3 cells that were treated with SB202190 (+) or with vehicle (–). These results are representative of three separate experiments. (C) Killing curves of dominant-negative p38α-transfected NIH 3T3 cells 48 h after UVC irradiation. At 36 h prior to UVC irradiation, NIH 3T3 cells that were stably transfected with DN–14-3-3ζ or DN–14-3-3η were transiently transfected with 2 μg of vector or with dominant-negative human p38α. Doubly transfected cells were analyzed for total p38 protein levels and p38 activity by immunoblotting with anti-p38 and anti-phospho-p38 antibodies. Cells were analyzed by the Trypan blue exclusion method. At least 100 cells were analyzed for each condition. Results are the mean ± SEM of three experiments.
The role of p38 in the apoptotic response of transfected cells was investigated further by the use of a dominant-negative form of p38α (DN–p38α) that was mutated in the TXY activation loop (Rincon et al., 1998). When NIH 3T3 cells that were stably transfected with DN–14-3-3ζ or DN–14-3-3η were transiently transfected with DN–p38α, the basal p38 MAP kinase activity was reduced by ∼50% (Figure 4C). UVC irradiation-induced apoptosis was blocked in NIH 3T3 cells double-transfected with either DN–p38α and DN–14-3-3ζ or DN–p38α and DN–14-3-3η compared with cells transfected with DN–14-3-3ζ or DN–14-3-3η alone (Figure 4C).
Targeted expression of a dominant-negative form of 14-3-3 to the heart
To determine whether disruption of 14-3-3 function in an intact model system could also promote apoptosis, we generated transgenic mice with a construct that contained the α-myosin heavy chain (α-MHC) promoter that has previously been demonstrated to direct specific postnatal ventricular gene transcription (Subramaniam et al., 1991). The α-MHC promoter was linked to the coding region of DN–14-3-3η that contained a 5′-Myc-1 epitope tag. Dominant-negative 14-3-3 was targeted to cardiac tissue because previous work has demonstrated that pressure overload provokes a modest apoptotic response in cardiomyocytes that can be detected by terminal deoxynucleotidyltransferase (TdT) nicked-end labeling (TUNEL) assay (Condorelli et al., 1999). We hypothesized that in this ‘sensitized’ system, factors that increase or decrease the apoptotic response could be readily identified.
Two transgenic Fo mice were obtained to yield transgenic lines. Slot-blot analysis revealed that there was integration of three copies of the transgene in one line (3×-DN–14-3-3) and 3–4 copies in a second line (4×-DN–14-3-3) (data not shown). All heterozygous F1 and F2 transgenic mice appeared grossly normal at birth and lived for at least 6 months in the absence of experimental intervention.
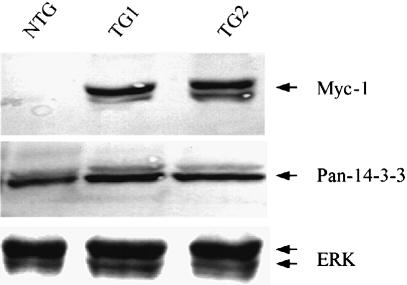
Expression of DN–14-3-3η protein was analyzed in mice by the use of both a polyclonal anti-pan-14-3-3 antibody and a monoclonal anti-Myc-1 epitope antibody. These immunoblots revealed that in ventricular tissue isolated from 3×-DN–14-3-3 mice, DN–14-3-3η protein represented 50% of total 14-3-3 protein (Figure 5). The levels of a control protein, ERK MAPK, were identical in transgenic mice and their non-transgenic littermates (Figure 5). In the absence of experimental intervention, transthoracic echocardiography showed that all 3×–DN–14-3-3 transgenic mice had normal cardiac morphology and basal ventricular systolic function (data not shown). Histological analysis of transgenic ventricular tissue revealed normal cardiomyocyte appearance when compared with non-transgenic littermates (data not shown).
Fig. 5. Targeted expression of DN–14-3-3η to postnatal murine cardiac tissue. Detection of DN–14-3-3η protein in transgenic mouse cardiac tissue. Immunoblotting was performed on ventricular extracts of two 3×-DN–14-3-3 mice (TG1 and TG2) and a non-transgenic (NTG) littermate. Equal amounts of total protein were loaded in each lane. Murine monoclonal anti-Myc-1-epitope antibody, rabbit polyclonal anti-pan-14-3-3 antibody and anti-ERK MAPK primary antibodies were used. Similar results were obtained in three hearts in each group.
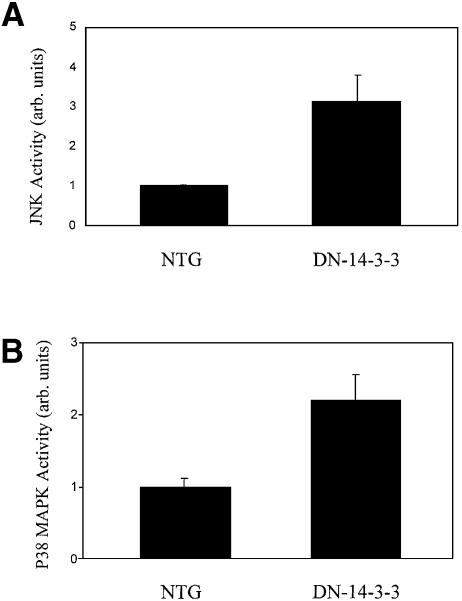
The activation of MAPK family members in transgenic cardiac tissue was evaluated by immunoblotting with phospho-specific antibodies, and these studies revealed that the basal activation of JNK1 and p38 MAPK was enhanced in DN–14-3-3 ventricular tissue when compared with non-transgenic littermates (Figure 6A and B).
Fig. 6. Increased basal JNK1 and p38 MAPK activity in DN–14-3-3 cardiac tissue. (A) JNK1 activity was assessed in the ventricular tissue of three 3×-DN–14-3-3 mice and three non-transgenic littermates. Immunoblots of cytosolic extracts were analyzed by the use of an anti-phospho JNK antibody. Equal amounts of total protein were loaded in each lane. (B) p38 MAPK activity was assessed in the ventricular tissue of three 3×-DN–14-3-3 mice and three non-transgenic littermates. Immunoblots of cytosolic extracts were analyzed by the use of an anti-phospho p38 MAPK antibody. Equal amounts of total protein were loaded in each lane. Densitometric analysis of immunoreactive bands in (A) and (B) was performed by the use of NIH Image software and data are expressed as the mean signal intensity ± SEM.
Abnormal response to pressure overload in transgenic mice
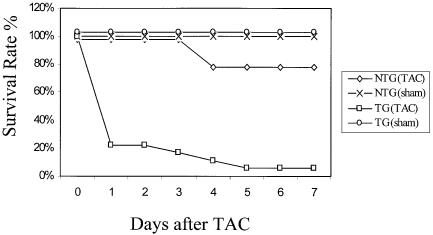
We next examined the ability of transgenic cardiac tissue to compensate for pressure overload induced by transverse aortic constriction (TAC). In this procedure, a 60–70% stenosis in the transverse aorta is created by surgical ligation (Rockman et al., 1991; Rogers et al., 1999). In non-transgenic littermates, TAC was tolerated and seven out of nine animals (78%) survived for at least 7 days (Figure 7). After 1 week, most mice developed significant cardiac hypertrophy that was easily detected by determining the left ventricular weight to body weight ratio.
Fig. 7. Decreased survival of transgenic mice after transverse aortic constriction (TAC). Survival rates in the days following TAC or sham operation (sham) in 3×-DN–14-3-3 mice (TG) and their non-transgenic littermates (NTG).
In contrast, 3×-DN–14-3-3 transgenic mice did not tolerate TAC, and 17 out of 18 animals died within 7 days of the procedure (Figure 7). Many DN–14-3-3 animals deteriorated in the hours following recovery from anesthesia. Although the cause of death was not ascertained in all DN–14-3-3 mice following tight TAC, pre-morbid echocardiography of several animals revealed global left ventricular hypokinesis, left ventricular dilatation and bradycardia in the minutes prior to death. χ2 analysis revealed that the decrease in survival observed in 3×–DN–14-3-3 transgenic mice 7 days after tight TAC was statistically significant when compared with non-transgenic littermates (p = 0.0001). To exclude the possibility that 3×-DN–14-3-3 transgenic mice were uniquely sensitive to anesthesia or thoracotomy, sham operations were performed that were identical to the TAC procedure except that the aorta was not ligated after it was identified by dissection. All five sham-operated 3×-DN–14-3-3 mice tolerated the procedure and survived >7 days.
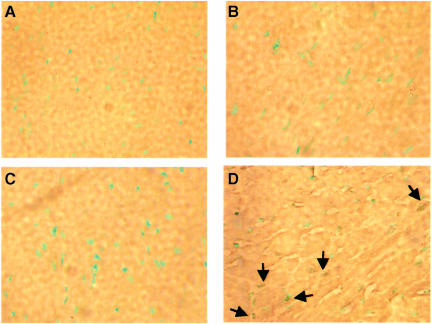
We next investigated the incidence of cardiomyocyte apoptosis in transgenic animals after TAC, and found that there was massive apoptosis in 3×-DN–14-3-3 ventricular tissue obtained from three separate animals 3–5 days after TAC (Figure 8). Indeed, the apoptotic index was 18.9 ± 6.2% in 3×-DN–14-3-3 ventricular tissue obtained 5 days after TAC, but was only 1.6 ± 1.1% in non-transgenic littermates (TAC), and 0.8 ± 1.2% and 1.1 ± 1.4%, respectively, in sham-operated non-transgenic littermates and 3×-DN–14-3-3 mice at this time point (Figure 8). For example, in ventricular tissue obtained from one 3×–DN–14-3-3 mouse 5 days after TAC, 88 out of 437 cardiomyocyte nuclei were TUNEL positive. The profound increase in apoptosis observed in DN–14-3-3 mice following TAC contrasts with our recently reported results with transgenic mice that overexpress RGS4 in cardiac tissue (Rogers et al., 1999). Transgenic mice that overexpress RGS4 in the heart have enhanced mortality after TAC, but do not exhibit any increase in cardiomyocyte apoptosis (Rogers et al., 1999).
Fig. 8. Increased cardiomyocyte apoptosis in transgenic mice after TAC. Hearts were isolated from 3×-DN–14-3-3 transgenic mice and their non-transgenic littermates 5 days after TAC, fixed in paraformaldehyde, embedded in paraffin, sectioned with a microtome and analyzed by TUNEL assay. Sections of ventricular tissue obtained from (A) a sham-operated non-transgenic mouse, (B) a non-transgenic mouse obtained 5 days after TAC, (C) a 3×-DN–14-3-3 transgenic mouse obtained 5 days after sham operation and (D) a 3×-DN–14-3-3 transgenic mouse obtained 5 days after TAC. TUNEL-positive nuclei are indicated by arrows.
Discussion
14-3-3 proteins bind to a variety of signal transduction, checkpoint control and cytoskeletal proteins (Aitken et al., 1995). In this work, we have explored the role of 14-3-3 in cell physiology by the use of dominant-negative forms of 14-3-3ζ and 14-3-3η. We have used mutant forms of 14-3-3 that are unable to bind to phosphoserine-containing peptides (Thorson et al., 1998; Wang et al., 1998). We demonstrated that a dominant-negative form of 14-3-3η can bind to native 14-3-3ζ protein, presumably forming an inactive heterodimer. The use of dominant-negative forms of 14-3-3 permitted us to examine the overall role of these proteins in cell physiology. Previous work has concentrated on the manipulation of 14-3-3 targets, e.g. the expression of mutant forms of BAD (Zha et al., 1996), Ask1 (L.Zhang et al., 1999) or cdc25c (Dalal et al., 1999) that are unable to bind to 14-3-3. In contrast, we have attempted to inhibit all of the actions of 14-3-3 in cells by expression of dominant-negative mutant forms of 14-3-3.
Our results demonstrate that the DN–14-3-3ζ mutant inhibits serum-stimulated ERK MAPK activation in transfected NIH 3T3 cells, but enhances the basal activity of Ask1, JNK and p38 MAPK. In contrast, the NES–14-3-3ζ mutant does not inhibit serum-stimulated ERK MAPK activity in transfected cells, but does enhance basal Ask1, JNK and p38 MAPK activity. One possible explanation for the different biochemical activities of these two mutants is that DN–14-3-3ζ is a more potent inhibitor of wild-type 14-3-3, but Wang et al. (1998) showed that the L220A single mutant form of 14-3-3η is markedly impaired in its ability to interact with Raf-1 by yeast two-hybrid assay.
Transfection of NIH 3T3 cells with DN–14-3-3ζ, DN–14-3-3η or NES–14-3-3ζ did not affect the phenotypic appearance of cultured cells, nor did it have an effect on cell proliferation, but it markedly increased the sensitivity of cells to apoptotic stimuli, such as UVC irradiation, serum deprivation and TNF-α stimulation. The ability of cells to proliferate at normal rates when grown in the presence of serum despite transfection with dominant-negative forms of 14-3-3 may imply that 14-3-3-independent pathways can mediate the proliferative response. Alternatively, transfected cells may still retain low but sufficient levels of 14-3-3 activity to permit cell proliferation.
Transfection of NIH 3T3 cells with DN–14-3-3ζ, DN–14-3-3η or NES–14-3-3ζ, but not with WT–14-3-3ζ, markedly increased the sensitivity of cells to apoptotic stimuli, such as UVC irradiation and serum deprivation. Furthermore, administration of a pharmacological inhibitor of p38 MAPK, SB202190, or transfection with dominant-negative p38α blocked apoptosis in DN–14-3-3-transfected cells. These findings demonstrate that activation of the p38 MAP kinase pathway by DN–14-3-3 sensitizes cells to apoptotic stimuli, but it is possible that other pathways may also be involved in this response.
The ability of DN–14-3-3 to lower the apoptotic threshold of cells was validated by experiments with transgenic mice. The α-MHC promoter was linked to DN–14-3-3 and this construct was used to generate transgenic mice. These mice appeared normal at birth, had normal cardiac function and morphology, and lived normal lifespans in the absence of experimental manipulation. One well-established trigger of apoptosis in cardiac tissue is pressure overload caused by TAC. When DN–14-3-3 transgenic mice were subjected to TAC, they exhibited a marked increase in mortality in comparison with non-transgenic littermates. Examination of cardiac tissue by TUNEL assay revealed that DN–14-3-3 transgenic cardiac tissue had a profound increase in cardiomyocyte apoptosis after TAC when compared with control cardiac tissue. These results are similar to the ventricular-restricted GP130 knockout mouse where massive cardiomyocyte apoptosis (>30%) was observed in the 3–4 days following TAC (Hirota et al., 1999). These results are also complementary to recently described experiments with isolated perfused rabbit hearts, where it was found that myocardial ischemia followed by reperfusion led to cardiomyocyte apoptosis in >30% of cells and that this apoptotic response was blocked by pre-treatment with SB203580, another inhibitor of p38 MAPK (Ma et al., 1999). Therefore, DN–14-3-3 promotes the apoptotic pathway in cultured cells and in cardiac tissue in response to provocative stimulation because of its ability to activate p38 MAPK.
Our findings establish that 14-3-3 is a critical anti-apoptotic factor in cells. 14-3-3 blocks apoptosis by inhibiting the activation of p38 MAPK. The ability of 14-3-3 to inhibit the action of BAD and FKHLR1 and to promote the activation of Raf-1 also probably contributes to this effect on apoptosis. These findings suggest that chemical agents that inhibit 14-3-3 activities will promote apoptosis.
Materials and methods
Plasmid construction
The cDNA encoding the R56A and R60A mutant form of 14-3-3η (DN–14-3-3η) has been described previously (S.Zhang et al., 1999). The cDNA encoding the T180A and Y182F dominant-negative mutant form of human p38α MAPK in pCMV5 was a gift from Dr Aubrey R.Morrison (Washington University, St Louis, MO).
An N-terminal FLAG epitope tag and a C-terminal HA epitope tag were added to the cDNA encoding human WT–14-3-3ζ by PCR, and the PCR product was inserted into pCINeo (Promega), a mammalian expression vector that contains a cytomegalovirus (CMV) promoter. PCR was used to generate the 14-3-3ζ point mutants using primers as described below. All mutants were verified by DNA sequencing.
Primers used to generate WT–14-3-3ζ were: 5′-CAGAATTCATGGACTACAAGGACGACGATGACAAGATGGATAAAAATGAGCTG-3′ and 5′-GAATTCTTAGAGGCTAGCATAATCAGGAACATCATACGGATAATTTTCCCCTCCTTCTCC-3′. Primers used to generate the R56A and R60A double mutant form of 14-3-3ζ (DN–14-3-3ζ) were: 5′-CTTATAAAAATGTTGTAGGAGCCCGTGCTAGCTCTTGGGCCGTCGTCTCAAGTATTGAAC-3′ and 5′-GTTCAATACTTGAGACGACGGCCCAAGAGCTAGCACGGGCTCCTACAACATTTTTATAAG-3′.Primers used to generate the I217A and L220A double mutant form of 14-3-3ζ (NES–14-3-3ζ) were: 5′-CATACAAAGACAGCACGCTAGCAATGCAAGCACTGAGAGACAACTTGACA-3′ and 5′-TGTCAAGTTGTCTCTCAGTGCTTGCATTGCTAGCGTGCTGTCTTTGTATG-3′.
Cell culture, transfection and reagents
NIH 3T3 fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS and antibiotics. For transient transfections, 5 × 105 cells were plated in 6-well culture dishes 12 h before standard lipofectamine transfection. For stable transfections, cells were replated in selective medium containing 0.4 mg/ml geneticin. After 2–3 weeks, distinct colonies were trypsinized and transferred to multiwell plates for further propagation in the presence of selective medium. For serum stimulation studies, cells were serum starved for 24 h and treated with DMEM plus 10% FCS for 10 min.
Protein analysis and antibodies
NIH 3T3 cells were lysed using NP-40 lysis buffer (0.5% NP-40, 137 mM NaCl, 50 mM NaF, 5 mM EDTA, 10 mM Tris–HCl pH 7.5, 2 mM phenylmethylsulfonyl fluoride, 25 μM leupeptin, 0.2 U/ml aprotinin). Lysates were cleared by low-speed centrifugation and stored at –80°C. Murine ventricular cytosolic lysates were obtained as described previously (Zhang et al., 1998). For Western blot experiments, proteins were separated by SDS–PAGE and electrophoretically transferred to nitrocellulose filters. Filters were blocked with either 5% non-fat dried milk or 5% bovine serum albumin (BSA) in TBST (10 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween-20). Anti-HA epitope rabbit polyclonal antibody (Santa Cruz Biotech) was used at a dilution of 1:600, anti-Myc-1 mouse monoclonal antibody (Santa Cruz Biotech) at 1:600, anti-phospho-ERK MAPK mouse monoclonal antibody (New England Biolabs) at 1:1000, anti-ERK MAP kinase rabbit polyclonal antibody (Santa Cruz Biotech) at 1:400, anti-phospho-JNK mouse monoclonal antibody (Santa Cruz Biotech) at a dilution of 1:300, anti-JNK kinase rabbit polyclonal antibody (Santa Cruz Biotech) at 1:300, anti-phospho-p38 MAPK rabbit polyclonal antibody (New England Biolabs) at 1:1000, anti-p38 MAP kinase rabbit polyclonal antibody (New England Biolabs) at 1:1000, anti-14-3-3ζ rabbit polyclonal antibody (Santa Cruz Biotech) at 1:500 and anti-14-3-3β (anti-pan-14-3-3) rabbit polyclonal antibody (Santa Cruz Biotech) at 1:500. After incubation in primary antibody, bound antibody was visualized with alkaline phosphatase or horseradish peroxidase-coupled secondary antibody and color-developing agents (Promega) or chemiluminescence-developing agents (ECL, Amersham).
For co-immunoprecipitation assays, protein A/G–agarose (Santa Cruz Biotech) was used to immobilize antibody-bound proteins. Immunoprecipitates were washed three times with NP-40 lysis buffer, and analyzed by SDS–PAGE as above.
Ask1 activity assays
Transfected NIH 3T3 cells were treated with 1000 μM hydrogen peroxide for 10 min or 100 ng/ml TNF-α for 30 min. Treated and untreated NIH 3T3 cells were lysed in NP-40 lysis buffer, cleared by low-speed centrifugation, and anti-Ask1 immunoprecipitates were obtained by the use of a rabbit polyclonal anti-Ask1 antibody (Santa Cruz Biotech). Immunoprecipitates were immobilized on protein A/G–agarose beads, washed three times with NP-40 lysis buffer, and then in vitro kinase reactions were performed. In brief, 2 μg of MBP (UBI) were incubated with the immune complexes for 20 min at 30°C in buffer containing 40 mM HEPES pH 8.0, 5 mM magnesium acetate, 2 mM dithiothreitol, 1 mM EGTA, 50 μM ATP and 1 μCi of [γ-32P]ATP. The kinase reaction was terminated with SDS loading buffer, proteins were separated by SDS–PAGE and bands were visualized by autoradiography and quantitated by densitometry using NIH Image software.
UVC irradiation, serum deprivation and TNF-α treatment
For UVC irradiation studies, cells were washed once with phosphate-buffered saline (PBS) and irradiated with 180 J/m2 UVC (UV Stratalinker 2400, Stratagene). SB202190 (Calbiochem) was added to culture media of some cells at a concentration of 20 μM 1 h prior to UV irradiation and cells were maintained in media supplemented with SB202190.
For serum deprivation studies, cultured cells were washed once with PBS, then grown in DMEM and antibiotics without added FCS for 48 h.
For TNF-α stimulation studies, NIH 3T3 cells were plated in 12-well plates at a density of 1 × 105 cells/well 12 h prior to ligand stimulation. Cells were treated with 100 ng/ml TNF-α (Sigma) for 12 h and then harvested. Other cells were treated with 100 μM etoposide (Sigma) or 50–100 μM hydrogen peroxide (Sigma) for 12 h and then harvested.
Trypan blue staining
At selected time points after UVC irradiation or serum deprivation, NIH 3T3 cells in 6-well plates were harvested with trypsin/EDTA and washed in PBS. Trypan blue (Gibco) was added to suspended cells at a concentration of 0.4% w/v. After 10 min, cells were transferred to a hemocytometer and Trypan blue dye uptake (dead cells) was detected by the use of a compound microscope.
DNA laddering assays
At 40–48 h after UVC irradiation, 5 × 106 cells were harvested and lysed overnight at room temperature in 545 μl of lysis buffer [10 mM Tris–HCl pH 8.0, 100 mM NaCl, 25 mM EDTA, 0.5% SDS, 1.0 mg/ml proteinase K (Sigma)]. Protein was removed by precipitation in 1.2 M NaCl. After phenol–chloroform extraction, genomic DNA was precipitated with ethanol, washed in 70% ethanol and resuspended in TE with added RNase A for 30 min at 37°C. Equal amounts of each sample (30 μg) were subjected to electrophoresis in 1.4% agarose gels.
Generation of DN–14-3-3 transgenic mice
The coding region of the human DN(R56A and R60A)-14-3-3η cDNA with a 5′-Myc-1-epitope tag was subcloned into a vector (clone 26; gift of Dr Jeffrey Robbins) containing the α-MHC promoter and an SV40 polyadenylation site (Subramaniam et al., 1991). Linearized DNA was injected into the pronuclei of one-cell C57BL/6 × SJL embryos at the Neuroscience Transgenic Facility at Washington University School of Medicine. Progeny were analyzed by PCR to detect transgene integration (Rogers et al., 1999). Two founder mice were obtained.
All research involving the use of mice was performed in strict accordance with the Recommendation from the Declaration of Helsinki.
Transverse aortic constriction
TAC was performed largely as previously described (Rockman et al., 1991; Rogers et al., 1999). In brief, 12-week-old mice were anesthetized with a mixture of xylazine (16 mg/kg) and ketamine (80 mg/kg). The chest was opened and, following blunt dissection through the intercostal muscles, the thoracic aorta was identified. A 7-0 silk suture was placed around the transverse aorta and tied around a 26 gauge blunt needle that was subsequently removed. The chest was closed with a purse-string suture. At the end of the procedure, the incision was closed in two layers with an interrupted suture pattern. The mouse was kept on a heating pad until responsive to stimuli. The surgeon was blinded to the transgenic status of the mice. Sham-operated animals underwent an identical surgical procedure except that the aortic constriction was not placed. After 7 days, surviving animals were killed and hearts were dissected out and weighed.
Echocardiography
Transthoracic echocardiography was performed in anesthetized mice (intraperitoneal injection of 0.01 ml of 2.5% avertin/g body weight) by the use of an Acuson Sequoia 256 Echocardiography System equipped with a 15 MHz (15L8) transducer as described previously (Rogers et al., 1999). The echocardiographer was blinded to the transgenic status of the mice.
Statistical analysis
Statistical analysis was performed by χ2 analysis. A value of p <0.05 was considered to be statistically significant.
TUNEL assay
Animals were killed, and the heart was excised, fixed overnight at 4°C in 10% formalin in PBS, embedded in paraffin and sectioned with a microtome. TUNEL was performed on 5 μm sections (TdT-FragEL DNA Fragmentation Detection Kit; Oncogene, Cambridge, MA) as described previously (Rogers et al., 1999). Sections were mounted on coverslips and evaluated by light microscopy. For each animal, five sections from the left ventricle free wall were scored for apoptotic nuclei. Only nuclei that were clearly located in cardiac myocytes were scored.
Acknowledgments
Acknowledgements
We thank Andrey S.Shaw for the DN(R56A and R60A)-14-3-3η cDNA, Dan Kelly, Helen Piwnica-Worms and Andrey Shaw for technical advice and critical reading of the manuscript, and Mia Nichol and the Neuroscience Transgenic Facility at Washington University School of Medicine for technical assistance. This work was supported by grants from the National Institutes of Health (GM54670) and the Barnes-Jewish Hospital Foundation.
References
- Aitken A., Jones, D., Soneji, Y. and Howell, S. (1995) 14-3-3 proteins: biological function and domain structure. Biochem. Soc. Trans., 23, 605–611. [DOI] [PubMed] [Google Scholar]
- Brunet A. et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]
- Chan E.D., Winston, B.W., Jarpe, M.B., Wynes, M.W. and Riches, D.W.H. (1997) Preferential activation of the p46 isoform of JNK/SAPK in mouse macrophages by TNF. Proc. Natl Acad. Sci. USA, 94, 13169–13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.C. and Rubin, G.M. (1997) 14-3-3ɛ positively regulates Ras-mediated signaling in Drosophila. Genes Dev., 11, 1132–1139. [DOI] [PubMed] [Google Scholar]
- Condorelli G. et al. (1999) Increased cardiomyocyte apoptosis and changes in proapoptotic and antiapoptotic genes bax and bcl-2 during left ventricular adaptations to chronic pressure overload in the rat. Circulation, 99, 3071–3078. [DOI] [PubMed] [Google Scholar]
- Dalal S.N., Schweitzer, C.M., Gan, J. and DeCaprio, J.A. (1999) Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol., 19, 4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S.R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y. and Greenberg, M.E. (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell, 91, 231–241. [DOI] [PubMed] [Google Scholar]
- De Cesaris P., Starace, D., Starace, G., Filippini, A., Stefanini, M. and Ziparo, E. (1999) Activation of jun N-terminal kinase/stress-activated protein kinase pathway by tumor necrosis factor α leads to intercellular adhesion molecule-1 expression. J. Biol. Chem., 274, 28978–28982. [DOI] [PubMed] [Google Scholar]
- del Peso L., Gonzalez-Garcia, M., Page, C., Herrera, R. and Nunez, G. (1997) Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science, 278, 687–689. [DOI] [PubMed] [Google Scholar]
- DiPietrantonio A.M., Hsieh, T. and Wu, J.M. (1999) Activation of caspase 3 in HL-60 cells exposed to hydrogen peroxide. Biochem. Biophys. Res. Commun., 255, 477–482. [DOI] [PubMed] [Google Scholar]
- Fantl W.J., Muslin, A.J., Kikuchi, A., Martin, J.A., MacNicol, A.M., Gross, R.W. and Williams, L.T. (1994) Activation of Raf-1 by 14-3-3 proteins. Nature, 371, 612–614. [DOI] [PubMed] [Google Scholar]
- Freed E., Symons, M., Macdonald, S.G., McCormick, F. and Ruggieri, R. (1994) Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science, 265, 1713–1716. [DOI] [PubMed] [Google Scholar]
- Fu H., Xia, K., Pallas, D.C., Cui, C., Conroy, K., Narsimhan, R.P., Mamon, H., Collier, R.J. and Roberts, T.M. (1994) Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science, 266, 126–129. [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Ikuta, N., Omata, S., Yamauchi, T., Isobe, T. and Ichimura, T. (1993) Demonstration of the phosphorylation-dependent interaction of tryptophan hydroxylase with the 14-3-3 protein. Biochem. Biophys. Res. Commun., 194, 144–149. [DOI] [PubMed] [Google Scholar]
- Gunn A., Scrimgeour, D., Potts, R.C., Mackenzie, L.A., Brown, R.A. and Beck, J.S. (1983) The destruction of peripheral-blood lymphocytes by extracorporeal exposure to ultraviolet radiation. Immunology, 50, 477–485. [PMC free article] [PubMed] [Google Scholar]
- Hirota H., Chen, J., Betz, U.A.K., Rajewsky, K., Gu, Y., Ross, J.,Jr, Muller, W. and Chien, K.R. (1999) Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell, 97, 189–198. [DOI] [PubMed] [Google Scholar]
- Holmstrom T.H., Tran, S.E.F., Johnson, V.L., Ahn, N.G., Chow, S.C. and Eriksson, J.E. (1999) Inhibition of mitogen-activated kinase signaling sensitizes HeLa cells to Fas receptor-mediated apoptosis. Mol. Cell. Biol., 19, 5991–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann S., Kahle, P.J. and Borasio, G.D. (1998) Inhibitors of p38 mitogen-activated protein kinase promote neuronal survival in vitro. J. Neurosci. Res., 52, 483–490. [DOI] [PubMed] [Google Scholar]
- Ichijo H. et al. (1997) Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science, 275, 90–92. [DOI] [PubMed] [Google Scholar]
- Irie K., Gotoh, Y., Yashar, B.M., Errede, B., Nishida, E. and Matsumoto, K. (1994) Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science, 265, 1716–1719. [DOI] [PubMed] [Google Scholar]
- Kaufmann S.H. (1998) Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim. Biophys. Acta, 1400, 195–211. [DOI] [PubMed] [Google Scholar]
- Kockel L., Vorbruggen, G., Jackle, H., Mlodzik, M. and Bohmann, D. (1997) Requirement for Drosophila 14-3-3ζ in Raf-dependent photoreceptor development. Genes Dev., 11, 1140–1147. [DOI] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy, W.G. (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer cdc25. Genes Dev., 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Skoulakis, E.M., Davis, R.L. and Perrimon, N. (1997) The Drosophila 14-3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras 1-dependent manner. Development, 124, 4163–4171. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A., Furnari, B., Mondesert, O. and Russell, P. (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature, 397, 172–175. [DOI] [PubMed] [Google Scholar]
- Ma X.L. et al. (1999) Inhibition of p38 mitogen-activated protein kinase decreases cardiomyocyte apoptosis and improves cardiac function after myocardial ischemia and reperfusion. Circulation, 99, 1685–1691. [DOI] [PubMed] [Google Scholar]
- Matsura T., Kai, M., Ito, H. and Yamada, K. (1999) Hydrogen peroxide-induced apoptosis in HL-60 cells requires caspase-3 activation. Free Radic. Res., 30, 73–83. [DOI] [PubMed] [Google Scholar]
- Meller N., Liu, Y.C., Collins, T.L., Bonnefoy-Berard, N., Baier, G., Isakov, N. and Altman, A. (1996) Direct interaction between protein kinase C θ (PKC θ) and 14-3-3 τ in T cells: 14-3-3 overexpression results in inhibition of PKC θ translocation and function. Mol. Cell. Biol., 16, 5782–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A.J., Tanner, J.W., Allen, P.M. and Shaw, A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Peng C.Y., Graves, P.R., Thoma, R.S., Wu, Z., Shaw, A.S. and Piwnica-Worms, H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Reuther G.W., Fu, H., Cripe, L.D., Collier, R.J. and Pendergast, A.M. (1994) Association of the protein kinases c-Bcr and Bcr-Abl with proteins of the 14-3-3 family. Science, 266, 129–133. [DOI] [PubMed] [Google Scholar]
- Rincon M., Enslen, H., Raingeaud, J., Recht, M., Zapton, T., Su, M.S., Penix, L.A., Davis, R.J. and Flavell, R.A. (1998) Interferon-γ expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J., 17, 2817–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Budman, J., Xu, J., Volinia, S., Cantley, L.C., Smerdon, S.J., Gamblin, S.J. and Yaffe, M.R. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Robinson K. et al. (1994) Mechanism of inhibition of protein kinase C by 14-3-3 isoforms. 14-3-3 isoforms do not have phospholipase A2 activity. Biochem. J., 299, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman H.A., Ross, R.S., Harris, A.N., Knowlton, K.U., Steinhelper, M.E., Field, L.J., Ross, J., Jr and Chien, K.R. (1991) Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl Acad. Sci. USA, 88, 8277–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Tamirisa, P., Kovacs, A., Weinheimer, C., Blumer, K.J., Kelly, D.J. and Muslin, A.J. (1999) RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J. Clin. Invest., 104, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., McPherson, R.A., Apolloni, A., Yan, J., Lane, A., Clyde-Smith, J. and Hancock, J.F. (1998) 14-3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol. Cell. Biol., 18, 3947–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M., Baumann, B., Cotten, M., Angel, P. and Wagner, E.F. (1995) Fos is an essential component of the mammalian UV response. EMBO J., 14, 5338–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis E.M. and Davis, R.L. (1998) 14-3-3 proteins in neuronal development and function. Mol. Neurobiol., 16, 269–284. [DOI] [PubMed] [Google Scholar]
- Subramaniam A., Jones, W.K., Gulick, J., Wert, S., Neumann, J. and Robbins, J. (1991) Tissue-specific regulation of the α-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem., 266, 24613–24620. [PubMed] [Google Scholar]
- Thorson J.A., Yu, L.W., Hsu, A.L., Shih, N.Y., Graves, P.R., Tanner, J.W., Allen, P.M., Piwnica-Worms, H. and Shaw, A.S. (1998) 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell. Biol., 18, 5229–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G., Luo, Z. and Avruch, J. (1998) A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature, 394, 88–92. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang, L., Liddington, R. and Fu, H. (1998) Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3ζ disrupt its interaction with Raf-1 kinase. J. Biol. Chem., 273, 16297–16304. [DOI] [PubMed] [Google Scholar]
- Xing H., Kornfeld, K. and Muslin, A.J. (1997) The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr. Biol., 7, 294–300. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger, K., Volinia, S., Caron, P.R., Aitken, A., Leffers, H., Gamblin, S.J., Smerdon, S.J. and Cantley, L.C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Zecevic M., Catling, A.D., Eblen, S.T., Renzi, L., Hittle, J.C., Yen, T.J., Gorbsky, G.J. and Weber, M.J. (1998) Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J. Cell Biol., 142, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J., Harada, H., Yang, E., Jockel, J. and Korsmeyer, S.J. (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X. Cell, 87, 619–628. [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen, J. and Fu, H. (1999) Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl Acad. Sci. USA, 96, 8511–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Watson, N., Zahner, J., Blumer, K.J. and Muslin, A.J. (1998) RGS3 and RGS4 are G protein inhibitors in the heart. J. Mol. Cell. Cardiol., 30, 269–276. [DOI] [PubMed] [Google Scholar]
- Zhang S., Xing, H. and Muslin, A.J. (1999) Nuclear localization of PKU–α regulated by 14-3-3. J. Biol. Chem., 275, 24865–24872. [DOI] [PubMed] [Google Scholar]