Abstract
The atomic force microscope (AFM) is widely used in materials science and has found many applications in biological sciences but has been limited in use in vision science. The AFM can be used to image the topography of soft biological materials in their native environments. It can also be used to probe the mechanical properties of cells and extracellular matrices, including their intrinsic elastic modulus and receptor-ligand interactions. In this review, the operation of the AFM is described along with a review of how it has been thus far used in vision science. It is hoped that this review will serve to stimulate vision scientists to consider incorporating AFM as part of their research toolkit.
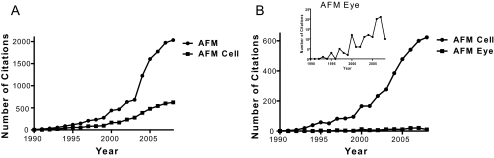
Although atomic force microscopy (AFM) has found increasing use in the physical and biological sciences, it has been underused in vision science. Since 1990, the number of citations of AFM in the physical sciences has dramatically increased (Fig. 1A). The number of research publications making use of AFM to investigate biological processes has also increased, though not at the same rate as in the physical sciences. The number of publications using AFM in vision science has seen only a very slight increase in the past 15 years, suggesting an unrealized potential of AFM in this field (Fig. 1B). AFM is a very powerful technique for the biological sciences, allowing samples to be imaged in situ in physiological conditions. Although it is outside the scope of this review to provide a comprehensive review of AFM in the biological sciences, many reviews exist that describe the uses and advantages of AFM for biological materials, many of which are referenced here.1–25 The AFM has several advantages over electron microscopy in the study of biological materials, including the ability to image in liquid with minimal sample preparation (no labeling, fixing, or coating). The AFM also allows the topographic characterization of surfaces at resolutions not achievable by optical microscopy. An optical microscope is limited by the diffraction limit of light; the achievable lateral resolution of the AFM is limited by the tip size and shape and is typically on the order of a few nanometers. The height (z) resolution of the AFM is approximately 1 Å, limited only by electronic and thermal noise in the system. High-resolution imaging, therefore, allows molecular scale features to be identified in the native environment of the sample and in real time. High-resolution images of normal and diseased states can be compared, providing insight into molecular reorganizations not observable by other techniques. In the vision sciences, AFM high-resolution imaging has been used in the imaging of the ommatidium surface of Diptera compound eyes, the imaging of rhodopsin in the native membrane, and the imaging of aquaporin in both a normal and a cataractous lens capsule.26–32
Figure 1.
Plots showing the incidence of publications using the term AFM since 1990. (A) PubMed search terms “AFM” and “AFM and cell.” (B) PubMed search terms “AFM and cell” and “AFM and eye or cornea or sclera or lens or retina or RPE.”
In addition to high-resolution imaging, the AFM is able to exert and measure forces on the order of piconewtons. This feature provides the ability to probe mechanical properties, including obtaining a local elastic modulus of a surface, measuring modulus variations across a sample surface, and measuring ligand-receptor interactions. The ability to explore the mechanical properties of biological materials and surfaces in their native state is essential for a complete characterization of the material and an understanding of how biophysical cues, such as substrate modulus, influence cell behaviors. It has been demonstrated that cells respond to the mechanical properties of the underlying substrate.33–35 In particular, fibroblasts and osteoblasts change their stiffness by cytoskeletal reorganization to adapt to changes in substrate modulus.36,37 Substrate modulus also affects cellular orientation and alignment, migration, proliferation and differentiation.35,38–46 A change in the substrate modulus is thought to play a role in disease development and, in particular, may have implications for vascular disease,34 muscle diseases,39,47–50 osteoarthritis,51 liver fibrosis,52,53 and tumor cell migration.54
Several techniques have been used to measure mechanical properties of biological materials. As an example, in the vision sciences, tensile testing and bulge testing have been used to determine the elastic modulus of the cornea. These techniques measure the properties of a sample in its entirety. AFM, however, has the force sensitivity and the spatial resolution necessary to measure the local modulus of a very soft sample surface and to investigate sample heterogeneities across small size scales. AFM has been successfully used for determining the mechanical properties of many tissues and cells,19,55–65 imaging of biological membranes,3,5,21 analyzing protein structure,66 and probing molecular interactions.9,57,67–70 This review will describe how mechanical properties measurements have impacted vision sciences, including studies on the lens, the corneal basement membranes, the trabecular meshwork and its role in glaucoma (Russell P, et al. IOVS. 2010;50:ARVO E-Abstract 3205), mucin interactions, melanosomes of the human retinal pigment epithelium, and recoverin interactions with lipid bilayers.71–75 The purpose of this review is to describe the operation of AFM as it applies to vision science and to examine the studies that have used AFM to explore fundamental questions in vision research.
AFM Operation
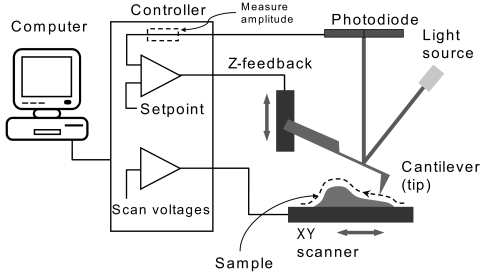
AFM was developed in 1986 to eliminate the need for a conductive sample, as was required with the scanning tunneling microscope.76 Although many reviews, as well as tutorials provided by instrument manufacturers, have provided an in depth discussion of the operation of the AFM and the available imaging modes, a brief description will be provided here.4,10,77,78 The AFM can be used for both topographical imaging and force measurements. Topographical imaging involves scanning the cantilever/tip across the sample surface (Fig. 2). A laser beam is reflected off the back of the cantilever, and small changes in cantilever deflection are detected with a position-sensitive photodiode detector. This deflection is processed by the system electronics to determine topological height changes on the sample surface. The achievable lateral resolution depends on the details of the tip geometry but can approach sub-nanometer levels, whereas the height resolution is sub-angstrom. Imaging can be accomplished with either contact mode or tapping mode, and both modes can be used in either air or fluid. The tip is in constant contact with the sample surface in contact mode, whereas in tapping mode the cantilever is oscillated as it is rastered across the sample surface, reducing the lateral forces on the sample and consequently reducing sample damage. The tapping mode is, therefore, more commonly used when imaging very soft materials. With the introduction of the tapping mode fluid cell in 1994,79 allowing the tapping mode to be used with samples under fluid, the AFM was increasingly used for the imaging of soft biological materials and for application to answer questions in the biological sciences.
Figure 2.
Schematic of the operation of the AFM. To obtain an image, a cantilever is scanned over the sample surface. A laser beam is deflected off the back of the cantilever, and changes in deflection are monitored with a photodiode detector. Reprinted with permission from Asylum Research (Santa Barbara, CA).
An AFM force curve is a plot of the deflection of the cantilever as the tip is brought into contact with the sample. The force curve is taken at a single location on the sample surface. Force curves can be used to measure mechanical properties such as elastic modulus, which is a measure of the stiffness of a material or its tendency to deform elastically under an applied force, or adhesion. Force measurements have been used extensively in the characterization of biological materials.1,8–10,23,55,63–65,80,81 In addition, many force curves can be obtained within a set scan size to obtain a force map of the surface.8,82 Force maps allow for discovery of surface heterogeneities in mechanical properties and can correlate those features with topographic features. Ligand-receptor interactions can also be measured with this technique.
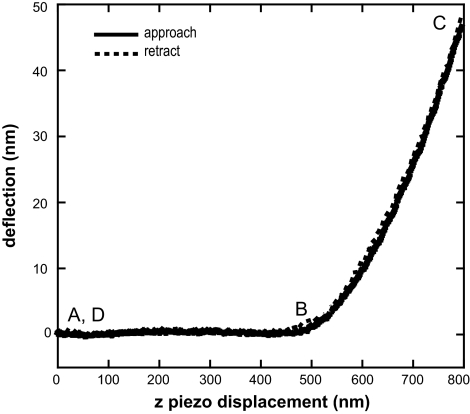
A typical force curve on a soft sample is shown in Figure 3. When the tip is not in contact with the surface, the force curve shows a straight, horizontal line approach (A in Fig. 3). When the tip comes into contact with a soft sample the force curve shows a gradual increase in the cantilever deflection (B in Fig. 3). The cantilever deflection will continue to increase until the tip is moved away from the sample (C in Fig. 3). When the tip is no longer in contact with the sample, the force curve is again a horizontal line (D in Fig. 3). Any adhesion or interaction of the tip with the surface will be observed in the retraction force curve. The interaction force associated with this pull-off can be calculated using the equation F = k(d − do), where k is the cantilever spring constant and d − do is the cantilever deflection.
Figure 3.
A typical force curve obtained from a soft sample. The z position of the cantilever is plotted on the x-axis and the cantilever deflection is plotted on the y-axis. Solid line: cantilever deflection as the cantilever approaches the surface. Dashed line: cantilever deflection on retraction from the surface. When the tip is not in contact with the surface the force curve shows a straight, horizontal line approach (A). When the tip comes into contact with a soft sample the force curve shows a gradual increase in the cantilever deflection (B). The cantilever deflection will continue to increase until the tip is moved away from the sample (C). When the tip is no longer in contact with the sample, the force curve is again a horizontal line (D). Values for the initial contact point of the tip with the sample (zo, do) and the cantilever deflection and z piezo position (z, d) are obtained from the force curve and used in the Hertz analysis to determine the elastic modulus.
On a hard surface, the cantilever deflection on approach will be equal to the movement of the piezo in the z direction (denoted as z), with z = d. On a soft sample, however, the cantilever will both deflect and indent into the sample surface as z is increased. Values obtained from the force curve are z, zo, d, and do, where z is the piezo displacement, d is the cantilever deflection, and zo and do are the values at initial contact of the tip with the sample. These values can be used to calculate the indentation depth, which is given by:
The elastic modulus of the biological sample can be calculated by fitting the force curve with an appropriate theoretical model. Most biological materials can be modeled as perfectly elastic at typical indentation rates and depths, and the Hertz model is most often used to determine modulus. The assumptions and concerns associated with the use of this equation are discussed in the following section. The Hertz model provides a relationship between the loading force and the indentation and has a specific form for each tip shape.83,84 The two most common tip geometries used for AFM nanoindentation are conical (equation 2) and spherical (equation 3):
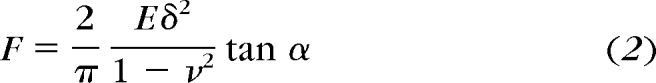 |
 |
where F is the loading force, ν is Poisson's ratio (assumed to be 0.5), δ is the indentation depth, and E is the elastic modulus. The half- opening angle of a conical indenter is α, and for spherical indenters R is the radius of the tip. With these equations and the knowledge that F = k (d − do), the equations can be solved for the elastic modulus E. The value of E can then be plotted for each point on the force curve as a function of indentation depth. The elastic modulus will be constant as a function of indentation depth when there is no contribution from the substrate or a stiffening of the sample. The advantages and disadvantages of the different tip geometries will be discussed in a later section.
Assumptions and Challenges
There are many assumptions that must hold for an accurate modulus determination with the Hertz model, including that the material is linear elastic, homogeneous, and infinitely thick, with no adhesion or attraction between the tip and the sample. Care must be taken to ensure that the sample in question can be described within these assumptions or that the deviations from these assumptions are considered. Although most biological materials have viscoelastic properties, at the indentation rates (2 μm/s) and depths (<100 nm) typically used in AFM nanoindentation, the force curves can be consistent with a linear elastic material and an elastic modulus can be defined. If the force curves show any indication of viscoelasticity (hysteresis in the loading and unloading curves in the force curve) or sample adhesion, it may not be appropriate to use the Hertz model or to define an elastic modulus. The range of indentation depths that can be accurately modeled is also dependent on the sample thickness. For soft or thin samples (and larger indentation depths), the influence of the substrate on the measured elastic modulus must be considered. The elastic modulus can be plotted as a function of indentation depth, and it can be verified that the elastic modulus is independent of indentation depth for at least small indentation depths. An increase in modulus with indentation depth may be indicative of an increasing contribution from the substrate.
It is also important to note that the AFM measures the modulus at the surface of the material and may be more accurately termed local modulus. Measuring a local modulus can be advantageous for certain experiments. For example, this allows a determination of the substrate elastic modulus a cell would sense on the surface of the substrate. Additionally, any heterogeneity in modulus across the substrate can be observed by obtaining a force map. It should also be noted that with the AFM, the modulus is measured in compression as the AFM tip is indented into the sample surface. The modulus measured with AFM may, therefore, be different from that obtained from other techniques, such as tensile testing, because of the differences in the direction of applied strain and any anisotropy of the sample. In addition, tensile testing measures the modulus of the sample in its entirety, whereas the AFM will be sensitive to changes in the mechanical properties between the surface and the bulk.
The Hertz equation also requires accurate knowledge of the initial contact point of the tip with the sample. This value can be difficult to determine from the force curves, particularly when there is tip-sample attraction or there is water or a contamination layer on the sample surface that the tip senses before it comes in contact with the surface. The Hertz equation can be fitted to the experimental data with two unknowns in the equation, fitting for both E and zo, the initial contact point. This eliminates the need to choose the value of zo manually. The user must still be aware that changes in the value of zo can significantly affect the calculated modulus value.
Accurate determination of the elastic modulus requires accurate calibration of the system, including the scanner and the cantilever. Calibration of the scanner must be checked periodically with an appropriate calibration grid for the size of the scanner. Each cantilever must also be calibrated so that the deflection sensitivity and the spring constant are known. The deflection sensitivity (nm/V) is found by taking a force curve on a hard surface, with no indentation into the surface. In this case d = z, and the sensitivity is the slope of the deflection (d) versus z force curve. The deflection sensitivity must be calculated each time the laser is realigned on the cantilever. The spring constant must also be determined for each cantilever because the actual value may vary significantly from the nominal value provided by the manufacturer. There are several techniques that have been used to measure spring constants.85–96 The advantages and disadvantages of each available method have been reviewed by Clifford and Seah.97 Two popular methods are the Sader method, an equation that relates the spring constant to the length and width of the cantilever and the cantilever's resonant frequency, and the thermal tune method, which measures the cantilever's response to thermal noise.90,92–94 In some instruments, the thermal tune method has been incorporated into the instrument hardware and software.
Tip and Cantilever Selection and Tip Functionalization
Proper choice of both cantilever stiffness and geometry of the incorporated tip are necessary for successful application of the AFM. Commercially available cantilevers have spring constants ranging from 0.006 N/m to 200 N/m. To avoid surface damage of delicate biological samples, imaging in contact mode, in either air or fluid, requires cantilevers with low spring constants, typically <0.2 N/m. As mentioned previously, tapping mode is more generally used when imaging soft, biological samples. A low spring constant cantilever is also used for tapping mode in fluid, whereas a stiffer cantilever (typically approximately 45 N/m) is needed for tapping mode in air to reduce noise. Low spring constant cantilevers also provide better deflection resolution, with a larger deflection for a given force, necessary for measuring intermolecular interactions, which typically range from 10−12 N to 10−7 N. The tip can be functionalized with molecules, proteins, or cells, and the interactions of these tips with a surface of interest can be obtained. Several reviews are available describing these types of experiments.1–3,5,98–100
AFM cantilevers with varying tip shapes are available. The standard geometry is of an integrated pyramidal tip with an end radius of 20 to 30 nm; sharper tips are also available. Sharper tips have the advantage of producing images with higher resolution. Sharper tips, however, have been shown to quickly deteriorate during scanning of cells, most likely because of tip contamination and blunting. Additionally, sharper tips have been associated with inducing increased damage to the cell surface.101 Spheres can be attached to the end of the cantilever and used as the tip. Although not useful for imaging because of poor resolution, these types of tip-cantilever configurations are often used for nanoindentation experiments to acquire information on the mechanical properties of the surface being probed, with a larger sample area included in the analysis. The sphere allows for nanoindentation with a well-defined tip geometry, simplifying analysis. The sphere size can also be chosen to probe the desired surface area. For example, in one report, a sphere with a 1-μm radius was chosen to probe the corneal basement membranes to include the contributions from several collagen fibrils and to avoid probing the properties of a single fiber as well as to mimic the dimensions of a typical focal adhesion with the contact area of the tip.73
AFM Combined with Optical or Spectroscopic Techniques
The AFM has been integrated with various optical and spectroscopic techniques, such as bright-field microscopy, epifluorescence, surface interference microscopy, scanning confocal laser microscopy (SCLM), Raman spectroscopy, total internal reflection fluorescence, and fluorescence lifetime imaging microscopy.26,101–112 Integration of these techniques allows the high-resolution imaging capabilities of the AFM to be used in conjunction with the chemical specificity available with optical techniques. This area of research is very active; a large supporting literature and an in-depth review lies outside the scope of this paper. It is obvious, however, that the integration of AFM with complementary techniques will find increasing use in the characterization of biological molecules and interfaces and will impact many areas of biological research. In a recent example, combined AFM and SCLM were used to detect the tyrosine kinase B (TrkB) receptors in hippocampus neurons.105 The location of the fluorescently labeled TrkB receptors could be correlated with AFM images acquired of the cellular structure. It was determined in this study that the TrkB receptors were highly populated in the center of the neuron but less populated along the edges. This type of correlation would not be possible with either technique alone. Such an approach could be used in spatially mapping the distribution of focal adhesions during corneal epithelial cell migration and correlation to spatial differences in cell compliance.
In addition to fluorescence labeling, receptors have also been labeled with superparamagnetic microbeads and imaged with magnetic AFM (MFM).113 MFM provides many of the same advantages of optical techniques, presenting a means to correlate surface topographical imaging with chemical specificity. The magnetic interactions, however, are long-ranged, allowing the microbeads to be imaged up to 150 nm from the surface. This technique may find future application in the imaging of receptors after a process in which they are internalized in the membrane.
In an example from vision science research, an instrument with integrated AFM-Raman spectroscopy was used to image the ommatidial surfaces of compound eyes.26 This combined approach was used to characterize the differences in topography among species and to probe compositional variations. This example will be described in more detail in the following section.
AFM Studies of the Eye
Ocular Surface.
One of the most studied tissues of the eye from a number of species has been the ocular surface. In one study, the integration of AFM with Raman spectroscopy and surface-enhanced Raman spectroscopy (SERS) has provided a means to differentiate topographic and chemical differences in the ommatidial surfaces between species.26 For example, AFM was used to determine that the ommatidial surface of the species Chrysopilus testaceipes consists of a cerebral cortex-like ridge structure. Other species, however, have vastly different topographies, including a smooth surface, a surface with nodule-like bumps, a relatively flat surface with pits and grooves, and a surface with series of bumps arranged to form ridges.
The integration with Raman spectroscopy reveals that there is also variation in surface composition between the tops of the ridges and the troughs of C. testaceipes ommatidial surface, with the ridges showing more hydrophobicity and more protein while the troughs have more carbonyl functionality. These results may provide insight into how differences in topography and composition contribute to differences in function, such as in anti-reflection and surface wetting.
Mucins.
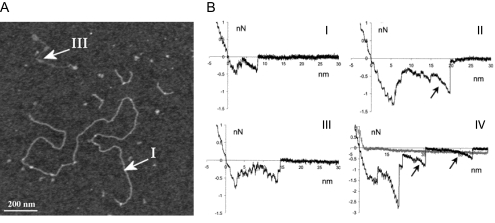
The ability of the AFM to provide both topological and force data is elegantly shown in the study of mucins.71,114–116 Mucin samples have been characterized as polydispersive, making it difficult to separate and describe the varying conformations with a small amount of sample. AFM, however, is capable of distinguishing individual mucins and acquiring information on the sample heterogeneity. In one study, the AFM has allowed investigation of the intramolecular conformational heterogeneity of gene products from one specific mucin gene (MUC5AC, a gel-forming mucin), revealing three distinct conformational populations (Fig. 4A).115,116 From the acquired images, it was possible to identify heterogeneity in polymer length and in flexibility. The three conformations observed were a stiff, extended polymer (I in Fig. 4A), a long, thick, coiled polymer, and a small, thin, flexible polymer (III in Fig. 4A).
Figure 4.
(A) AFM image of the affinity-purified mucins highlighting examples of two polymer types, labeled I and III. 1200 × 1200 nm scan size, z-range 3 nm. Reprinted with permission from Round AN, McMaster TJ, Miles MJ, Corfield AP, Berry M. The isolated MUC5AC gene product from human ocular mucin displays intramolecular conformational heterogeneity. Glycobiology. 2007;17:578–585. The Society for Glycobiology. (B) Examples of force-distance retract curves for mucin-mica interactions. Force curves in I and III were taken without added cations, with dwell times of 0 s and 5 seconds, respectively. Force curves II and IV were taken with the addition of Ni(II) cations and with dwell times of 0 seconds and 5 seconds, respectively. Both detachment (short jumps) and single molecule stretching (arrows) were observed in the force curves. The number of detachment events and the prevalence of the stretching events increased with the addition of the cations. Reprinted with permission from Berry M, McMaster TJ, Corfield AP, Miles MJ. Exploring the molecular adhesion of ocular mucins. Biomacromolecules. 2001;2:498–503. Copyright 2001 American Chemical Society.
The involvement of mucins in both adhesive and nonadhesive functions prompted the use of AFM to study mucin-mica and mucin-mucin interactions.71 Force curves obtained with a mucin-coated tip on a mica surface showed many adhesion events, with adhesion force increasing with the addition of cations (Fig. 4B). Details obtained from the force curves, such as mucin detachment distances from the surface, suggest that the mucins exist as aggregates on the tip. The attraction of the mucin with the mica surface was suggested to be due to the hydrophobic interactions between the peptide core and the mica and through a cation bridge between the negatively charged mucins and the negatively charged mica surface. However, no mucin-mucin interactions were observed when the mucin-coated tip was brought into contact with a mucin gel. It was suggested that this observation may be due to the electrostatic repulsion of the negatively charged oligosaccharides on the mucins. The lack of adhesion between mucins is observed during blinking, with the mucin on the upper lid gliding over the mucin covering the cornea.
Cornea.
Early work of AFM on the mammalian cornea focused on the collagen fibrils of the cornea and fibrils from the sclera.117–124 The AFM was able to resolve individual collagen fibrils and provide structural details, such as diameter and the axial D-periodicity. Although this information can be obtained from electron microscopy, the sample preparation for AFM is less destructive and does not require sample coating or drying. Comparing the structures of the corneal collagen fibrils with the scleral collagen fibrils reveals a difference in the structures that may be necessary for the transparency of the cornea.
Corneal epithelial and endothelial cells and surfaces have been characterized with AFM.125–128 The AFM again has the advantage that structural details can be observed under physiological conditions without destructive sample preparation, which may affect the result. The AFM, therefore, provides a complementary technique with which to verify the accuracy of measurements taken with electron microscopy. The importance of imaging these samples without fixation is highlighted in the following studies. An early study examined rabbit corneal epithelium in both fixed and unfixed tissue. It was concluded that fixed tissue gave sharper images with AFM, but surface features were sometimes lost as a result of fixation.128 In an in vitro study, conjunctival epithelial cells and corneal fibroblasts and epithelial cells were compared.127 This study showed that structural differences were observed between the fixed and unfixed cells, with a loss of surface features on the fixed cells.
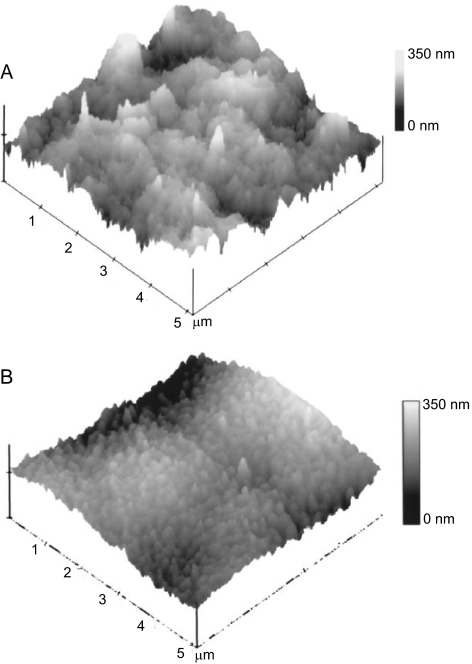
The human corneal anterior basement membrane and Descemet's membrane are the surfaces directly in contact with the epithelial and endothelial cell layers, respectively. Our laboratory has characterized the topography of these corneal basement membranes and related AFM results to images captured with scanning electron microscopy (SEM) and transmission electron microscopy (TEM; Fig. 5).129 All three complementary imaging modalities provided similar measurements of feature dimensions. Obtaining data with AFM as well SEM and TEM allows verification that the sample preparation involved with these techniques has not substantially affected the surface topography. It was found that the basement membranes consist of a felt-like arrangement of fibers, bumps, and pores with feature sizes in the nanoscale and submicron range. Although the anterior basement membrane and Descemet's membrane have a similar topological appearance, features in the anterior basement membrane are larger and less densely packed. The profound impact of topography on corneal cell response has been reported. In particular, nanoscale to submicron topographic features have been shown to influence corneal epithelial cell alignment,130 adhesion,131 migration,132 and proliferation.133
Figure 5.
AFM tapping mode images of the human cornea. (A) AFM image of the surface of the anterior basement membrane. (B) AFM image of the surface of Descemet's membrane. These images reveal that the structural features of Descemet's membrane and smaller and more dense that those of the anterior basement membrane. Reprinted with permission from Abrams GA, Schaus SS, Goodman SL, Nealey PF, Murphy CJ. Nanoscale topography of the corneal epithelial basement membrane and Descemet's membrane of the human. Cornea. 2000;19:57–64. Copyright 2000 Lippincott Williams & Wilkins, Inc.
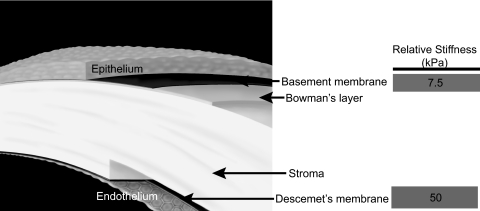
In addition to sensing substrate topography, cells sense and respond to the mechanical properties of the underlying substrate.33–35 Although SEM and TEM can be useful for obtaining nanoscale topographic data, the mechanical properties of the sample must be obtained without sample drying or fixing. A variety of techniques have been used to measure mechanical properties of the cornea, such as tensile testing and bulge testing. These methods, however, do not have the force sensitivity necessary to measure the mechanical properties of distinct spatially limited regions within the cornea such as the basement membranes or Bowman's layer. The AFM can uniquely provide the elastic modulus measurements of these layers. The value obtained for the modulus of the anterior basement membrane was 7.5 ± 4.2 kPa, whereas the modulus obtained on Descemet's membrane was 47.6 ± 16.5 kPa (Fig. 6).73 Although the topography of Descemet's membrane has characteristics similar to those of the anterior basement membrane, the topographic measurements reveal smaller pore sizes in Descemet's membrane, which creates a more compact structure. This important structural difference is consistent with the observed differences in elastic modulus. Mimicking the topographic and mechanical differences in these layers may be critical in the design of prosthetics. Results such as these have also been useful in directing the subsequent design and fabrication of synthetic substrates with biomimetic biophysical attributes for the study of cellular behaviors. Probing the mechanical properties of tissues can also provide insight into a disease state. In our laboratory, experiments are under way to characterize the elastic modulus of the human trabecular meshwork (HTM) (Russell P, et al. IOVS 2010;50:ARVO E-Abstract 3205). The HTM is responsible for regulating the aqueous humor outflow. Stiffening of the HTM may contribute to the increase in intraocular pressure and, therefore, the progression of glaucoma. The average value of the elastic modulus from six normal donor HTM samples was 3.5 kPa, whereas the average modulus from six glaucomatous HTMs was 108.7 kPa. This represents approximately a 30-fold increase in the stiffness of the HTM with glaucoma.
Figure 6.
Schematic of the human cornea identifying the epithelium, the anterior basement membrane, Bowman's layer, the stroma, Descemet's membrane and the endothelium. The elastic modulus of the anterior basement membrane and Descemet's membrane were determined by AFM to be 7.5 kPa and 50 kPa, respectively. Reprinted with permission from Last JA, Liliensiek SJ, Nealey PF, Murphy CJ. Determining the mechanical properties of human corneal basement membranes with atomic force microscopy. J Struct Biol. 2009;167:19–24. Copyright 2009 Elsevier.
Cornea and Photoablation.
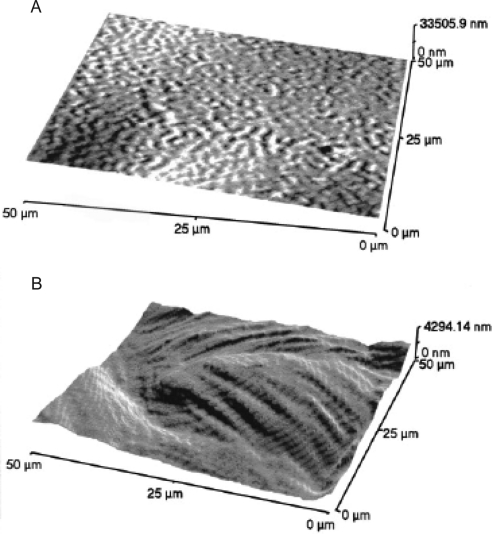
The epithelial surface has also been characterized before and after excimer laser photoablation in both human and porcine corneas.134–136 It was found that the photoablated surface was less regular and had increased surface roughness. In one study, both Bowman's layer and stroma were characterized after photoablation. Bowman's layer remained smooth, whereas the stromal surface increased in roughness (Fig. 7). In addition, AFM of the corneal surface after low-temperature laser treatment of the cornea found that though there was a loss of organization of the collagen bundles in the area of the treatment, there was no denaturation of the collagen fibers.137
Figure 7.
AFM images of the human cornea after laser ablation. (A) AFM image of Bowman's layer after 55 pulses. Bowman's layer is relatively smooth after the photoablation. (B) AFM image of the stroma after 320 pulses. The stromal surface becomes relatively rough after photoablation. The stromal layers can be identified. Reprinted with permission from Nógrádi A, Hopp B, Révész K, Szabó G, Bor Z, Kolozsvari L. Atomic force microscopic study of the human cornea after excimer laser keratectomy. Exp Eye Res. 2000;70:363–368. Copyright 2000 Elsevier.
Lens.
The elastic modulus of the intact primate lens has been measured by AFM to have a mean value of 1.720 kPa.74 The authors note that although the tip indented the lens capsule, the measured modulus also included a contribution from the epithelial cells and the softer, underlying cortex. These measurements were largely taken in the central region of the lens, and it would be of interest to determine whether the modulus varied in different regions such as the equatorial area compared with the central area. Investigating the mechanical properties of the lens at different ages may provide insight into the development of presbyopia.
The topography of lenses has been investigated with AFM. Imaging of rabbit lenses provided structural details, including fiber height and width, of fibers from both the nuclear and the cortical regions.138 This study was performed with fixed tissue, so it is unclear whether the topography of these regions is similar to the unfixed lens in situ. In a publication from the same group, the topographies of normal and cataractous canine lenses were compared, revealing a loss of structure in the epithelium during cataract progression.139
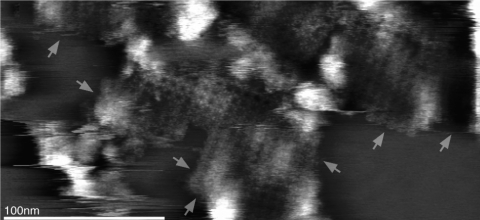
Several intriguing studies of the lens were conducted on native and cataractous lens fiber membranes.27,28,31 In these studies, the focus of the work centered on aquaporin 0. This protein represents approximately 50% of the membrane protein of the lens. Previously, this protein was named MIP or MP26. Aquaporin 0 is present in nonjunctional membranes in the lens, where it can form water pores, as well as at the thin junctions of the cells, where water conduction is not thought to occur. Proteolysis of the cytoplasmic amino and carboxyl terminals occurs during maturation of the fiber cells and results in the junctional aquaporin that is the closed form of this protein. Using the AFM to examine the topography of extracellular aquaporin, Buzhynskyy et al.28 imaged the extracellular loop of amino acids 35 to 38 and showed that there is a shift in the localization of this loop when aquaporin went from the junctional to the nonjunctional form. In normal lenses, they also showed that aquaporins formed square arrays surrounded by connexins. On examining cataractous lenses, they found that aquaporin tetramers had no associated connexins, suggesting the lack of transport between cells in the cataractous lens (Fig. 8).27
Figure 8.
High-resolution topograph of AQP0 junction arrays in the cataract lens membrane. No connexon was found at the AQP0 microdomain edges (arrows). Reprinted with permission from Buzhynskyy N, Girmens JF, Faigle W, Scheuring S. Human cataract lens membrane at subnanometer resolution. J Mol Biol. 2007;374:162–169. Copyright 2007 Elsevier.
Rhodopsin.
Rhodopsin is a G-protein–coupled receptor present in the rods of the retina. When a photon of light interacts with rhodopsin in the photoreceptors, the 11-cis retinal associated with opsin is altered. This initiates the phototransduction cascade, resulting in an electrical signal to the brain. In one study, the organization of rhodopsin in rod outer segment disc membranes from mouse retina was investigated with AFM in physiological conditions.29,30,140 High-resolution AFM images revealed that rhodopsin is arranged in dimers within paracrystalline domains. Although the dimer structure had been previously inferred, this report provided the first experimental evidence of the dimer structure. The AFM has also been used to access the stability of rhodopsin within the membrane with single-molecule force spectroscopy.141 These experiments involved bringing an AFM tip on top of a single rhodopsin molecule, resulting in an attachment of the molecule to the AFM tip. Pulling the AFM tip away from the membrane unfolded the molecule, revealing that rhodopsin could be characterized by structural segments, each within conserved sequence motifs of G-protein–coupled receptors. This result suggests that these segments play a role in the stabilization of rhodopsin within the membrane.
Bacteriorhodopsin, a proton pump, can be obtained as two-dimensional crystalline patches, or purple membrane, from Halobacterium salinarium. The structure of the bacteriorhodopsin within the membrane has been well characterized with AFM.142–147 These studies use high-resolution AFM imaging to obtain structural information at the molecular level in physiological conditions. It has been observed that conformational changes occurred within the membrane on illumination of the surface with white light.147,148 In one study an increase in the thickness of the membrane on illumination was monitored with an AFM tip, and a second study observed structural changes in the high-resolution images of the membrane after illumination. In both cases, the changes were reversible.
A second protein from H. salinarium can also be purified from the purple membrane and prepared on a substrate as a two-dimensional crystal. High-resolution AFM imaging was used to characterize surface structure, and it was found that the structure was dependent on the imaging force used, indicating differences in the stiffness over the surface of the membrane.
In addition to structural characterization, AFM force spectroscopy has been used to investigate the forces necessary to unfold and refold individual molecules in the membrane149–151 and to determine the surface charge and surface potential of the crystals.152–156 In addition, variations occurred in tip-sample interactions between the cytoplasmic side and the extracellular side of the membrane, with the cytoplasmic side having an apparent dome-like topography as a result of a strong tip-surface interaction with a long delay length.157 For a more in-depth look, Engel and Gaub have provided an excellent review on both the structural and the mechanical characterization of these membranes.142
Recoverin.
AFM force measurements were used in the study of recoverin. Recoverin is a Ca2+-binding protein found in the photoreceptors of the eye that plays a role in the adaptation of the eye after exposure to light. Binding of Ca2+ to the recoverin leads to a change in orientation of the myristoyl adduct and is, therefore, termed a Ca2+-myristoyl switch. The interactions between recoverin, which can be myristoylated at the N-terminal, and a lipid bilayer were recorded in the presence and absence of Ca2+.75 AFM showed that when Ca2+ was present, an adhesive force of 48 pN was measured between recoverin and the bilayers, but no binding was observed in the absence of this ion or with nonmyristoylated recoverin. The results from this study reveal that the binding of recoverin to the membrane is controlled by the myristoyl group.
Bruch's Membrane.
Both topographic imaging and force measurements have been used to characterize Bruch's membrane.158–160 In one study, a topographic image of the collagen fibers on the surface of Bruch's membrane was acquired. The adsorption of a collagen-binding peptide to this surface was also investigated with AFM, revealing that the peptide was bound to the groove of the collagen fiber.159 The ability to modify the surface of Bruch's membrane and thereby aid the attachment of retinal pigment epithelial cells is critical in the treatment of age-related macular degeneration.
AFM force spectroscopy with force mapping was also used to further characterize Bruch's membrane.160 Although both the structural and the chemical changes in the collagen have been investigated as they relate to the disease state, this study examined the adhesive properties of the collagen surface. AFM tips were modified with -COOH, -OH, -CH3, and -NH2 to map changes in adhesion across the membrane. With single-tip functionality the force maps reveal differences in adhesion across the surface that has been attributed to the arrangement of the collagen fibers. Although the statistical significance between measurements is questionable, the authors report that the highest mean adhesion was observed with the -COOH and -NH2 functionalized tips, attributed to the presence of hydrogen bonding and electrostatic interactions, respectively.160
Retinal Cells.
AFM force maps were also used to characterize the mechanical properties of human retinal pigment epithelium melanosomes from a 14-year-old donor and from a 76-year-old donor to evaluate potential age-related changes.72 These studies revealed that though the average Young's moduli of both sets of melanosomes were similar (12 and 16 MPa), the distribution of the modulus values was narrower for the 14-year-old samples. The modulus of the younger sample ranged from 5 MPa to 38 MPa, with the largest percentage of the data between 7 and 15 kPa. The modulus range from the older sample was similar, but with a more even distribution of the data between 5 MPa and 27 MPa. These modulus values are much larger than those typically observed for cells, suggesting a high packing density for the melanosomes. In addition, the older sample had a larger percentage of adhesive sites, as observed in the force curves. The adhesion is believed to be due to the presence of lipofuscin on the surface, with its presence confirmed with photoelectron emission spectroscopy. The increased presence of lipofuscin on the older sample may account for some of the age-related changes in the adsorption, fluorescence, and reactivity of the melanosomes.
As with the cornea, understanding the behavior of retinal cells on synthetic surfaces is critical for tissue engineering applications. Although cell attachment and proliferation on the synthetic surfaces can be monitored with optical microscopy, AFM can be used to quantitatively determine the nanoscale roughness of the surface. As previously mentioned, it is known that the biophysical properties of a substrate can have a profound effect on cell behavior.130–133 Changes in topography can be visualized with SEM, but AFM can easily provide quantitative roughness data. In particular, AFM, along with other complementary techniques, has been used to characterize the surfaces of polyhydroxymethylsiloxane and poly(hydroxybutyrate-co-hydroxyvalerate) substrates before and after surface treatments, including plasma treatments and ion irradiation, as substrates for bovine retinal cells (pericytes) and the retinal pigment epithelial cell line (D407), respectively.161,162
Inner Limiting Membrane.
The mechanical properties of the chick and mouse retinal inner limiting membrane (ILM) during development have been determined with AFM. An increase from 0.95 MPa to 3.30 MPa was reported for chick ILM between embryonic days 4 and 9.163 A similar increase was observed for the embryonic and adult mouse ILM, increasing from 3.81 MPa to 4.07 MPa. There was also an increase in the thickness of the chick ILM, from 137 nm on day 5 to 402 nm on day 9. The changes suggest that the basement membrane undergoes significant structural changes during development.
Conclusions
Although AFM is underutilized in vision research, many questions in vision science can be addressed by use of this technique. The ability of the AFM to provide high-resolution images and to sense small forces in the sample's native environment make this technique invaluable in the characterization of biological materials. Currently, the AFM has been used in vision research to determine the function of proteins within a cell membrane with high-resolution imaging, to explore and quantify ligand-receptor interactions, to characterize how nanoscale topography affects cellular behaviors, to determine the mechanical properties of tissues and cells in their native environment, and to help understand how a change in physical properties correlates with disease state. The AFM can be used alone or can be combined with other optical and spectroscopic techniques to provide complementary information, creating an even more powerful tool. The application of AFM will benefit evolving programs in cell, stem cell, and tissue engineering, with application to vision science.
Footnotes
Supported by National Eye Institute Grants R01EY016134 (CJM), R01EY017367 (PFN), and R01EY019475 (PR), and NEI-IPA (PR).
Disclosure: J.A. Last, None; P. Russell, None; P.F. Nealey, None; C.J. Murphy, None
References
- 1. Alessandrini A, Facci P. AFM: a versatile tool in biophysics. Measurement Sci Technol. 2005;16:R65–R92 [Google Scholar]
- 2. Franz CM, Puech PH. Atomic force microscopy: a versatile tool for studying cell morphology. Adhes Mech Cell Mol Bioeng. 2008;1:289–300 [Google Scholar]
- 3. Frederix P, Bosshart PD, Engel A. Atomic force microscopy of biological membranes. Biophys Jl. 2009;96:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris V, Kirby A, Gunning A. Atomic Force Microscopy for Biologists. London: Imperial College Press; 1999 [Google Scholar]
- 5. Muller DJ. AFM: a nanotool in membrane biology. Biochemistry. 2008;47:7986–7998 [DOI] [PubMed] [Google Scholar]
- 6. Muller DJ, Dufrene YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat Nanotechnol. 2008;3:261–269 [DOI] [PubMed] [Google Scholar]
- 7. Parot P, Dufrene YF, Hinterdorfer P, et al. Past, present and future of atomic force microscopy in life sciences and medicine. J Mol Recognition. 2007;20:418–431 [DOI] [PubMed] [Google Scholar]
- 8. Costa KD. Single-cell elastography: probing for disease with the atomic force microscope. Dis Markers. 2003;19:139–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasas S, Dietler G. Probing nanomechanical properties from biomolecules to living cells. Pflugers Arch Eur J Physiol. 2008;456:13–27 [DOI] [PubMed] [Google Scholar]
- 10. Radmacher M. Studying the mechanics of cellular processes by atomic force microscopy. Cell Mech. 2007:347–372 [DOI] [PubMed] [Google Scholar]
- 11. Santos NC, Castanho M. An overview of the biophysical applications of atomic force microscopy. Biophys Chem. 2004;107:133–149 [DOI] [PubMed] [Google Scholar]
- 12. Goksu EI, Vanegas JM, Blanchette CD, Lin WC, Longo ML. AFM for structure and dynamics of biomembranes. Biochim Biophys Acta Biomembr. 2009;1788:254–266 [DOI] [PubMed] [Google Scholar]
- 13. Wright CJ, Armstrong I. The application of atomic force microscopy force measurements to the characterisation of microbial surfaces. Surf Interface Anal. 2006;38:1419–1428 [Google Scholar]
- 14. Zhu J, Sun RG. Applications of atomic force microscopy to the structures and functions of single protein molecule. Chin J Anal Chem. 2006;34:735–740 [Google Scholar]
- 15. Gadegaard N. Atomic force microscopy in biology: technology and techniques. Biotechn Histochem. 2006;81:87–97 [DOI] [PubMed] [Google Scholar]
- 16. Noy A. Chemical force microscopy of chemical and biological interactions. Surf Interface Anal. 2006;38:1429–1441 [Google Scholar]
- 17. Ebner A, Wildling L, Zhu R, et al., eds. Functionalization of probe tips and supports for single-molecule recognition force microscopy. Top Curr Chem. 2008;285:29–76 [DOI] [PubMed] [Google Scholar]
- 18. Haupt BJ, Pelling AE, Horton MA. Integrated confocal and scanning probe microscopy for biomedical research. Sci World J. 2006;6:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikai A. Nanobiomechanics of proteins and biomembrane. Philos Trans R Soc Lond B Biol Sci. 2008;363:2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Withers JR, Aston DE. Nanomechanical measurements with AFM in the elastic limit. Adv Colloid Interf Sci. 2006;120:57–67 [DOI] [PubMed] [Google Scholar]
- 21. Seantier B, Giocondi MC, Le Grimellec C, Milhiet PE. Probing supported model and native membranes using AFM. Curr Opin Colloid Interface Sci. 2008;13:326–337 [Google Scholar]
- 22. Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: at the interface of biology, physics and chemistry. J R Soc Interface. 2008;5:15–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritort F. Single-molecule experiments in biological physics: methods and applications. J Phys Condensed Matter. 2006;18:R531–R583 [DOI] [PubMed] [Google Scholar]
- 24. Veerapandian M, Yun K. Study of atomic force microscopy in pharmaceutical and biopharmaceutical interactions: a mini review. Curr Pharmaceut Anal. 2009;5:256–268 [Google Scholar]
- 25. Cohen SR, Bitler A. Use of AFM in bio-related systems. Curr Opin Colloid Interface Sci. 2008;13:316–325 [Google Scholar]
- 26. Anderson MS, Gaimari SD. Raman-atomic force microscopy of the ommatidial surfaces of dipteran compound eyes. J Structural Biol. 2003;142:364–368 [DOI] [PubMed] [Google Scholar]
- 27. Buzhynskyy N, Girmens JF, Faigle W, Scheuring S. Human cataract lens membrane at subnanometer resolution. J Mol Biol. 2007;374:162–169 [DOI] [PubMed] [Google Scholar]
- 28. Buzhynskyy N, Hite RK, Walz T, Scheuring S. The supramolecular architecture of junctional microdomains in native lens membranes. EMBO Rep. 2007;8:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128 [DOI] [PubMed] [Google Scholar]
- 30. Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mangenot S, Buzhynskyy N, Girmens JF, Scheuring S. Malformation of junctional microdomains in cataract lens membranes from a type II diabetes patient. Pflugers Arch. 2009;457:1265–1274 [DOI] [PubMed] [Google Scholar]
- 32. Scheuring S, Buzhynskyy N, Jaroslawski S, Goncalves RP, Hite RK, Walz T. Structural models of the supramolecular organization of AQP0 and connexons in junctional microdomains. J Struct Biol. 2007;160:385–394 [DOI] [PubMed] [Google Scholar]
- 33. Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143 [DOI] [PubMed] [Google Scholar]
- 34. Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J Appl Physiol. 2005;98:1547–1553 [DOI] [PubMed] [Google Scholar]
- 36. Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys J. 2007;93:4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takai E, Costa KD, Shaheen A, Hung CT, Guo XE. Osteoblast elastic modulus measured by atomic force microscopy is substrate dependent. Ann Biomed Eng. 2005;33:963–971 [DOI] [PubMed] [Google Scholar]
- 38. Bergethon PR, Trinkaus-Randall V, Franzblau C. Modified hydroxyethylmethacrylate hydrogels as a modelling tool for the study of cell-substratum interactions. J Cell Sci. 1989;92(pt 1):111–121 [DOI] [PubMed] [Google Scholar]
- 39. Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gunn JW, Turner SD, Mann BK. Adhesive and mechanical properties of hydrogels influence neurite extension. J Biomed Mater Res A. 2005;72:91–97 [DOI] [PubMed] [Google Scholar]
- 41. Koh WG, Revzin A, Simonian A, Reeves T, Pishko M. Control of mammalian cell and bacteria adhesion on substrates micropatterned with poly(ethylene glycol) hydrogels. Biomed Microdevices. 2003;5:11–19 [Google Scholar]
- 42. Semler EJ, Lancin PA, Dasgupta A, Moghe PV. Engineering hepatocellular morphogenesis and function via ligand-presenting hydrogels with graded mechanical compliance. Biotechnol Bioeng. 2005;89:296–307 [DOI] [PubMed] [Google Scholar]
- 43. Shin H, Zygourakis K, Farach-Carson MC, Yaszemski MJ, Mikos AG. Modulation of differentiation and mineralization of marrow stromal cells cultured on biomimetic hydrogels modified with Arg-Gly-Asp containing peptides. J Biomed Mater Res A. 2004;69:535–543 [DOI] [PubMed] [Google Scholar]
- 44. Wallace C, Jacob JT, Stoltz A, Bi J, Bundy K. Corneal epithelial adhesion strength to tethered-protein/peptide modified hydrogel surfaces. J Biomed Mater Res A. 2005;72:19–24 [DOI] [PubMed] [Google Scholar]
- 45. Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia W, Liu W, Cui L, et al. Tissue engineering of cartilage with the use of chitosan-gelatin complex scaffolds. J Biomed Mater Res B Appl Biomater. 2004;71:373–380 [DOI] [PubMed] [Google Scholar]
- 47. Campbell GR, Chamley-Campbell JH. Smooth-muscle phenotypic modulation: role in atherogenesis. Med Hypotheses. 1981;7:729–735 [DOI] [PubMed] [Google Scholar]
- 48. Glukhova MA, Koteliansky VE. Integrins, cytoskeletal and extracellular matrix proteins in developing smooth muscle cells of human aorta. In: Schwartz SM, Mecham RP. eds. The Vascular Smooth Muscle Cell Molecular and Biological Responses to the Extracellular Matrix. New York: Academic Press; 1995:37–79 [Google Scholar]
- 49. Stedman HH, Sweeney HL, Shrager JB, et al. The Mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular-dystrophy. Nature. 1991;352:536–539 [DOI] [PubMed] [Google Scholar]
- 50. Stenmark KR, Mecham RP. Cellular and molecular mechanisms of pulmonary vascular remodeling. Ann Rev Physiol. 1997;59:89–144 [DOI] [PubMed] [Google Scholar]
- 51. Genes NG, Rowley JA, Mooney DJ, Bonassar LJ. Effect of substrate mechanics on chondrocyte adhesion to modified alginate surfaces. Arch Biochem Biophys. 2004;422:161–167 [DOI] [PubMed] [Google Scholar]
- 52. Georges PC, Hui JJ, Gombos Z, et al. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–G1154 [DOI] [PubMed] [Google Scholar]
- 53. Wells RG. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J Clin Gastroenterol. 2005;39:S158–S161 [DOI] [PubMed] [Google Scholar]
- 54. Zaman MH, Trapani LM, Siemeski A, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bowen WR, Lovitt RW, Wright CJ. Application of atomic force microscopy to the study of micromechanical properties of biological materials. BioTechnol Lett. 2000;22:893–903 [Google Scholar]
- 56. Canetta E, Adya AK. Atomic force microscopy: applications to nanobiotechnology. J Ind Chem Soc. 2005;82:1147–1172 [Google Scholar]
- 57. Docheva D, Padula D, Popov C, Mutschler W, Clausen-Schaumann H, Schieker M. Researching into the cellular shape, volume and elasticity of mesenchymal stem cells, osteoblasts and osteosarcoma cells by atomic force microscopy. J Cell Mol Med. 2008;12:537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hsieh CH, Lin YH, Lin S, et al. Surface ultrastructure and mechanical property of human chondrocyte revealed by atomic force microscopy. Osteoarthritis Cartilage. 2008;16:480–488 [DOI] [PubMed] [Google Scholar]
- 59. Jandt KD. Atomic force microscopy of biomaterials surfaces and interfaces. Surf Sci. 2001;491:303–332 [Google Scholar]
- 60. Li QS, Lee GY, Ong CN, Lim CT. AFM indentation study of breast cancer cells. Biochem Biophys Res Commun. 2008;374:609–613 [DOI] [PubMed] [Google Scholar]
- 61. Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J Biomech. 2001;34:1545–1553 [DOI] [PubMed] [Google Scholar]
- 62. Rabinovich Y, Esayanur M, Daosukho S, Byer K, El-Shall H, Khan S. Atomic force microscopy measurement of the elastic properties of the kidney epithelial cells. J Colloid Interface Sci. 2005;285:125–135 [DOI] [PubMed] [Google Scholar]
- 63. Rico F, Roca-Cusachs P, Gavara N, Farre R, Rotger M, Navajas D. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72:021914 [DOI] [PubMed] [Google Scholar]
- 64. Sirghi L, Ponti J, Broggi F, Rossi F. Probing elasticity and adhesion of live cells by atomic force microscopy indentation. Eur Biophys J. 2008;37:935–945 [DOI] [PubMed] [Google Scholar]
- 65. Wagh AA, Roan E, Chapman KE, et al. Localized elasticity measured in epithelial cells migrating at a wound edge using atomic force microscopy. Am J Physiol Lung Cell Mol Physiol. 2008;295:L54–L60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dufrene YF. Atomic force microscopy: a powerful molecular toolkit in nanoproteomics. Proteomics. 2009;9:5400–5405 Review [DOI] [PubMed] [Google Scholar]
- 67. Butt H-J. Electrostatic interaction in atomic force microscopy. Biophys J. 1991;60:777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gaboriaud F, Parcha BS, Gee ML, Holden JA, Strugnell RA. Spatially resolved force spectroscopy of bacterial surfaces using force-volume imaging. Colloids Surf B Biointerfaces. 2008;62:206–213 [DOI] [PubMed] [Google Scholar]
- 69. Zhang X, Chen A, De Leon D, et al. Atomic force microscopy measurement of leukocyte-endothelial interaction. Am J Physiol Heart Circ Physiol. 2004;286:H359–H367 [DOI] [PubMed] [Google Scholar]
- 70. Chandraprabha MN, Somasundaran P, Natarajan KA. Modeling and analysis of nanoscale interaction forces between Acidithiobacillus ferrooxidans and AFM tip. Colloids and Surfaces B: Biointerfaces. In press [DOI] [PubMed] [Google Scholar]
- 71. Berry M, McMaster TJ, Corfield AP, Miles MJ. Exploring the molecular adhesion of ocular mucins. Biomacromolecules. 2001;2:498–503 [DOI] [PubMed] [Google Scholar]
- 72. Guo S, Hong L, Akhremitchev BB, Simon JD. Surface elastic properties of human retinal pigment epithelium melanosomes. Photochem Photobiol. 2008;84:671–678 [DOI] [PubMed] [Google Scholar]
- 73. Last JA, Liliensiek SJ, Nealey PF, Murphy CJ. Determining the mechanical properties of human corneal basement membranes with atomic force microscopy. J Struct Biol. 2009;167:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ziebarth NM, Wojcikiewicz EP, Manns F, Moy VT, Parel JM. Atomic force microscopy measurements of lens elasticity in monkey eyes. Mol Vis. 2007;13:504–510 [PMC free article] [PubMed] [Google Scholar]
- 75. Desmeules P, Grandbois M, Bondarenko VA, Yamazaki A, Salesse C. Measurement of membrane binding between recoverin, a calcium-myristoyl switch protein, and lipid bilayers by AFM-based force spectroscopy. Biophys J. 2002;82:3343–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Binnig G, Quate CF, Geber C. Atomic force microscope. Phys Rev Let. 1986. 56:930. [DOI] [PubMed] [Google Scholar]
- 77. Jalili N, Laxminarayana K. A review of atomic force microscopy imaging systems: application to molecular metrology and biological sciences. Mechatronics. 2004;14:907–945 [Google Scholar]
- 78. West PE. Introduction to Atomic Force Microscopy: Theory, Practice, Applications. Pacific Nanotechnology; 2007 [Google Scholar]
- 79. Hansma PK, Cleveland JP, Radmacher M, et al. Tapping mode atomic-force microscopy in liquids. Appl Physics Lett. 1994;64:1738–1740 [Google Scholar]
- 80. Cappella B, Dietler G. Force-distance curves by atomic force microscopy. Surf Sci Rep. 1999;34:1–104 [Google Scholar]
- 81. Costa KD, Yin FCP. Analysis of indentation: implications for measuring mechanical properties with atomic force microscopy. J Biomech Eng Trans ASME. 1999;121:462–471 [DOI] [PubMed] [Google Scholar]
- 82. A-Hassan E, Heinz WF, Antonik MD, et al. Relative microelastic mapping of living cells by atomic force microscopy. Biophys J. 1998;74:1564–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hertz H. Uber die Beruhrung fester elastischer Korper. J Reine Angew Mathematik. 1882;92:156–171 [Google Scholar]
- 84. Sneddon IN. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int J Engng Sci. 1965;3:47–57 [Google Scholar]
- 85. Butt HJ, Jaschke M. Calculation of thermal noise in atomic-force microscopy. Nanotechnology. 1995;6:1–7 [Google Scholar]
- 86. Chen BY, Yeh MK, Tai NH. Accuracy of the spring constant of atomic force microscopy cantilevers by finite element method. Anal Chem. 2007;79:1333–1338 [DOI] [PubMed] [Google Scholar]
- 87. Clifford CA, Seah MP. The determination of atomic force microscope cantilever spring constants via dimensional methods for nanomechanical analysis. Nanotechnology. 2005;16:1666–1680 [Google Scholar]
- 88. Green CP, Lioe H, Cleveland JP, Proksch R, Mulvaney P, Sader JE. Normal and torsional spring constants of atomic force microscope cantilevers. Rev Sci Instrum. 2004;75:1988–1996 [Google Scholar]
- 89. Higgins MJ, Proksch R, Sader JE, et al. Noninvasive determination of optical lever sensitivity in atomic force microscopy. Rev Sci Instrum. 2006;77 [Google Scholar]
- 90. Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:3342–3342 [Google Scholar]
- 91. Neumeister JM, Ducker WA. Lateral, normal, and longitudinal spring constants of atomic-force microscopy cantilevers. Rev Sci Instrum. 1994;65:2527–2531 [Google Scholar]
- 92. Sader JE. Parallel beam approximation for v-shaped atomic-force microscope cantilevers. Rev Sci Instrum. 1995;66:4583–4587 [Google Scholar]
- 93. Sader JE, Chon JWM, Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev Sci Instrum. 1999;70:3967–3969 [Google Scholar]
- 94. Sader JE, Larson I, Mulvaney P, White LR. Method for the calibration of atomic-force microscope cantilevers. Rev Sci Instrum. 1995;66:3789–3798 [Google Scholar]
- 95. Senden TJ, Ducker WA. Experimental-determination of spring constants in atomic-force microscopy. Langmuir. 1994;10:1003–1004 [Google Scholar]
- 96. Torii A, Sasaki M, Hane K, Okuma S. A method for determining the spring constant of cantilevers for atomic force microscopy. Meas Sci Technol. 1996;7:179–184 [Google Scholar]
- 97. Clifford CA, Seah MP. Quantification issues in the identification of nanoscale regions of homopolymers using modulus measurement via AFM nanoindentation. Appl Surf Sci. 2005;252:1915–1933 [Google Scholar]
- 98. Muller DJ, Helenius J, Alsteens D, Dufrene YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol. 2009;5:383–390 [DOI] [PubMed] [Google Scholar]
- 99. Noy A, Vezenov DV, Lieber CM. Chemical force microscopy. Annu Rev Mater Sci. 1997;27:381–421 [Google Scholar]
- 100. Noy A. Strength in numbers: probing and understanding intermolecular bonding with chemical force microscopy. Scanning. 2008;30:96–105 [DOI] [PubMed] [Google Scholar]
- 101. Nagao E, Dvorak JA. An integrated approach to the study of living cells by atomic force microscopy. J Microsc. 1998;191:8–19 [DOI] [PubMed] [Google Scholar]
- 102. Francis LW, Lewis PD, Wright CJ, Conlan RS. Atomic force microscopy comes of age. Biol Cell. 2010;102:133–143 [DOI] [PubMed] [Google Scholar]
- 103. Lieberman K, BenAmi N, Lewis A. Fully integrated near-field optical, far-field optical, and normal-force scanned probe microscope. Rev Sci Instrum. 1996;67:3567–3572 [Google Scholar]
- 104. Noy A, Huser TR. Combined force and photonic probe microscope with single, molecule sensitivity. Rev Sci Instrum. 2003;74:1217–1221 [Google Scholar]
- 105. Park JW, Park AY, Lee S, Yu NK, Lee SH, Kaang BK. Detection of TrkB receptors distributed in cultured hippocampal neurons through bioconjugation between highly luminescent (quantum dot-neutravidin) and (biotinylated anti-TrkB antibody) on neurons by combined atomic force microscope and confocal laser scanning microscope. Bioconj Chem. 21:597–603 [DOI] [PubMed] [Google Scholar]
- 106. Hu DH, Micic M, Klymyshyn N, Suh YD, Lu HP. Correlated topographic and spectroscopic imaging by combined atomic force microscopy and optical microscopy. J Luminescence. 2004;107:4–12 [Google Scholar]
- 107. Callies C, Schon P, Liashkovich I, et al. Simultaneous mechanical stiffness and electrical potential measurements of living vascular endothelial cells using combined atomic force and epifluorescence microscopy. Nanotechnology. 2009;20:175104. [DOI] [PubMed] [Google Scholar]
- 108. Oreopoulos J, Yip CM. Combined scanning probe and total internal reflection fluorescence microscopy. Methods. 2008;46:2–10 [DOI] [PubMed] [Google Scholar]
- 109. Kassies R, Van der Werf KO, Lenferink A, et al. Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. J Microsc. 2005;217:109–116 [DOI] [PubMed] [Google Scholar]
- 110. Lal R, Proksch R. Multimodal atomic force microscopy: biological imaging using atomic force microscopy combined with light fluorescence and confocal microscopies and electrophysiologic recording. Int J Imaging Syst Technol. 1997;8:293–300 [Google Scholar]
- 111. Anderson MS. Locally enhanced Raman spectroscopy with an atomic force microscope. Appl Phys Lett. 2000;76:3130–3132 [Google Scholar]
- 112. Anderson MS, Pike WT. A Raman-atomic force microscope for apertureless-near-field spectroscopy and optical trapping. Rev Sci Instrum. 2002;73:1198–1203 [Google Scholar]
- 113. Moskalenko AV, Yarova PL, Gordeev SN, Smirnov SV. Single protein molecule mapping with magnetic atomic force microscopy. Biophys J. 2010;98:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Brayshaw DJ, Berry M, McMaster TJ. Optimisation of sample preparation methods for air imaging of ocular mucins by AFM. 2003:289–296 [DOI] [PubMed] [Google Scholar]
- 115. Round AN, Berry M, McMaster TJ, Corfield AP, Miles MJ. Glycopolymer charge density determines conformation in human ocular mucin gene products: an atomic force microscope study. J Struct Biol. 2004;145:246–253 [DOI] [PubMed] [Google Scholar]
- 116. Round AN, McMaster TJ, Miles MJ, Corfield AP, Berry M. The isolated MUC5AC gene product from human ocular mucin displays intramolecular conformational heterogeneity. Glycobiology. 2007;17:578–585 [DOI] [PubMed] [Google Scholar]
- 117. Miyagawa A, Kobayashi M, Fujita Y, et al. Surface ultrastructure of collagen fibrils and their association with proteoglycans in human cornea and sclera by atomic force microscopy and energy-filtering transmission electron microscopy. Cornea. 2001;20:651–656 [DOI] [PubMed] [Google Scholar]
- 118. Miyagawa A, Kobayashi M, Fujita Y, et al. Surface topology of collagen fibrils associated with proteoglycans in mouse cornea and sclera. Jpn J Ophthalmol. 2000;44:591–595 [DOI] [PubMed] [Google Scholar]
- 119. Yamamoto S, Hitomi J, Sawaguchi S, Abe H, Shigeno M, Ushiki T. Observation of human corneal and scleral collagen fibrils by atomic force microscopy. Jpn J Ophthalmol. 2002;46:496–501 [DOI] [PubMed] [Google Scholar]
- 120. Yamamoto S, Hitomi J, Shigeno M, Sawaguchi S, Abe H, Ushiki T. Atomic force microscopic studies of isolated collagen fibrils of the bovine cornea and sclera. Arch Histol Cytol. 1997;60:371–378 [DOI] [PubMed] [Google Scholar]
- 121. Meek KM, Fullwood NJ. Corneal and scleral collagens—a microscopist's perspective. Micron. 2001;32:261–272 [DOI] [PubMed] [Google Scholar]
- 122. Fullwood NJ, Hammiche A, Pollock HM, Hourston DJ, Song M. Atomic-force microscopy of the cornea and sclera. Curr Eye Res. 1995;14:529–535 [DOI] [PubMed] [Google Scholar]
- 123. Yamamoto S, Hashizume H, Hitomi J, et al. The subfibrillar arrangement of corneal and scleral collagen fibrils as revealed by scanning electron and atomic force microscopy. Arch Histol Cytol. 2000;63:127–135 [DOI] [PubMed] [Google Scholar]
- 124. Meller D, Peters K, Meller K. Human cornea and sclera studied by atomic force microscopy. Cell Tissue Res. 1997;288:111–118 [DOI] [PubMed] [Google Scholar]
- 125. Lydataki S, Lesniewska E, Tsilimbaris MK, et al. Observation of the posterior endothelial surface of the rabbit cornea using atomic force microscopy. Cornea. 2003;22:651–664 [DOI] [PubMed] [Google Scholar]
- 126. Lydataki S, Tsilimbaris MK, Lesniewska ES, Bron A, Pallikaris IG. Corneal tissue observed by atomic force microscopy. Methods Mol Biol. 2004;242:69–83 [DOI] [PubMed] [Google Scholar]
- 127. Sinniah K, Paauw J, Ubels J. Investigating live and fixed epithelial and fibroblast cells by atomic force microscopy. Curr Eye Res. 2002;25:61–68 [DOI] [PubMed] [Google Scholar]
- 128. Tsilimbaris MK, Lesniewska E, Lydataki S, Le Grimellec C, Goudonnet JP, Pallikaris IG. The use of atomic force microscopy for the observation of corneal epithelium surface. Invest Ophthalmol Vis Sci. 2000;41:680–686 [PubMed] [Google Scholar]
- 129. Abrams GA, Schaus SS, Goodman SL, Nealey PF, Murphy CJ. Nanoscale topography of the corneal epithelial basement membrane and Descemet's membrane of the human. Cornea. 2000;19:57–64 [DOI] [PubMed] [Google Scholar]
- 130. Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci. 2003;116:1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Karuri NW, Liliensiek S, Teixeira AI, et al. Biological length scale topography enhances cell-substratum adhesion of human corneal epithelial cells. J Cell Sci. 2004;117:3153–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Diehl KA, Foley JD, Nealey PF, Murphy CJ. Nanoscale topography modulates corneal epithelial cell migration. J Biomed Mater Res A. 2005;75:603–611 [DOI] [PubMed] [Google Scholar]
- 133. Liliensiek SJ, Campbell S, Nealey PF, Murphy CJ. The scale of substratum topographic features modulates proliferation of corneal epithelial cells and corneal fibroblasts. J Biomed Mater Res A. 2006;79:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Nogradi A, Hopp B, Revesz K, Szabo G, Bor Z, Kolozsvari L. Atomic force microscopic study of the human cornea following excimer laser keratectomy. Exp Eye Res. 2000;70:363–368 [DOI] [PubMed] [Google Scholar]
- 135. De Santo MP, Lombardo M, Serrao S, Lombardo G, Barberi R. Atomic force microscopy in ophthalmic surgery. 4th IEEE Conference on Nanotechnology Munich, Germany; 2004:562–564 [Google Scholar]
- 136. Lombardo M, De Santo MP, Lombardo G, Barberi R, Serrao S. Atomic force microscopy analysis of normal and photoablated porcine corneas. J Biomech. 2006;39:2719–2724 [DOI] [PubMed] [Google Scholar]
- 137. Matteini P, Sbrana F, Tiribilli B, Pini R. Atomic force microscopy and transmission electron microscopy analyses of low-temperature laser welding of the cornea. Lasers Med Sci. 2009;24:667–671 [DOI] [PubMed] [Google Scholar]
- 138. Antunes A, Gozzo FV, Nakamura M, et al. Analysis of the healthy rabbit lens surface using MAC mode atomic force microscopy. Micron. 2007;38:286–290 [DOI] [PubMed] [Google Scholar]
- 139. Antunes A, Gozzo FV, Nakamura M, Safatle AMV, Barros PSM, Toma HE. Atomic force imaging of ocular tissue: morphological study of healthy and cataract lenses. In: Mèndez-Vilas, Diaz J. eds. Modern Research and Educational Topics in Microscopy. Badajoz, Spain: Formatex; 2007:29–36 [Google Scholar]
- 140. Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259 [DOI] [PubMed] [Google Scholar]
- 142. Engel A, Gaub HE. Structure and mechanics of membrane proteins. Ann Rev Biochem. 2008;77:127–148 [DOI] [PubMed] [Google Scholar]
- 143. Heymann JB, Muller DJ, Landau EM, et al. Charting the surfaces of the purple membrane. J Struct Biol. 1999;128:243–249 [DOI] [PubMed] [Google Scholar]
- 144. Muller DJ, Heymann JB, Oesterhelt F, et al. Atomic force microscopy of native purple membrane. Biochim Biophys Acta. 2000;1460:27–38 [DOI] [PubMed] [Google Scholar]
- 145. Muller DJ, Sapra KT, Scheuring S, et al. Single-molecule studies of membrane proteins. Curr Opin Struct Biol. 2006;16:489–495 [DOI] [PubMed] [Google Scholar]
- 146. Muller DJ, Sass HJ, Muller SA, Buldt G, Engel A. Surface structures of native bacteriorhodopsin depend on the molecular packing arrangement in the membrane. J Mol Biol. 1999;285:1903–1909 [DOI] [PubMed] [Google Scholar]
- 147. Persike N, Pfeiffer M, Guckenberger R, Fritz M. Changes in the surface structure of purple membrane upon illumination measured by atomic force microscopy. Colloids Surfaces B Biointerfaces. 2000;19:325–332 [DOI] [PubMed] [Google Scholar]
- 148. Lewis A, Rousso I, Khachatryan E, Brodsky I, Lieberman K, Sheves M. Directly probing rapid membrane protein dynamics with an atomic force microscope: a study of light-induced conformational alterations in bacteriorhodopsin. Biophys J. 1996;70:2380–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kessler M, Gaub HE. Unfolding barriers in bacteriorhodopsin probed from the cytoplasmic and the extracellular side by AFM. Structure. 2006;14:521–527 [DOI] [PubMed] [Google Scholar]
- 150. Kessler M, Gottschalk KE, Janovjak H, Muller DJ, Gaub HE. Bacteriorhodopsin folds into the membrane against an external force. J Mol Biol. 2006;357:644–654 [DOI] [PubMed] [Google Scholar]
- 151. Cieplak M, Filipek S, Janovjak H, Krzysko KA. Pulling single bacteriorhodopsin out of a membrane: comparison of simulation and experiment. Biochim Biophys Acta Biomembr. 2006;1758:537–544 [DOI] [PubMed] [Google Scholar]
- 152. Hartley P, Matsumoto M, Mulvaney P. Determination of the surface potential of two-dimensional crystals of bacteriorhodopsin by AFM. Langmuir. 1998;14:5203–5209 [Google Scholar]
- 153. Knapp HF, Mesquida P, Stemmer A. Imaging the surface potential of active purple membrane. Surf Interface Anal. 2002;33:108–112 [Google Scholar]
- 154. Heinz WF, Hoh JH. Relative surface charge density mapping with the atomic force microscope. Biophys J. 1999;76:528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Butt HJ. Measuring local surface-charge densities in electrolyte-solutions with a scanning force microscope. Biophys J. 1992;63:578–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Muller DJ, Engel A. The height of biomolecules measured with the atomic force microscope depends on electrostatic interactions. Biophys J. 1997;73:1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zhong S, Li H, Chen XY, Cao EH, Jin G, Hu KS. Different interactions between the two sides of purple membrane with atomic force microscope tip. Langmuir. 2007;23:4486–4493 [DOI] [PubMed] [Google Scholar]
- 158. Mallick SB, Ivanisevic A. Study of the morphological and adhesion properties of collagen fibers in the Bruch's membrane. J Phys Chem B. 2005;109:19052–19055 [DOI] [PubMed] [Google Scholar]
- 159. Sistiabudi R, Ivanisevic A. Collagen-binding peptide interaction with retinal tissue surfaces. Langmuir. 2008;24:1591–1594 [DOI] [PubMed] [Google Scholar]
- 160. Mallick SB, Bhagwandin S, Ivanisevic A. Characterization of collagen fibers in Bruch's membrane using chemical force microscopy. Anal Bioanal Chem. 2006;386:652–657 [DOI] [PubMed] [Google Scholar]
- 161. Assero G, Satriano C, Lupo G, Anfuso CD, Marletta G, Alberghina M. Pericyte adhesion and growth onto polyhydroxymethylsiloxane surfaces nanostructured by plasma treatment and ion irradiation. Microvasc Res. 2004;68:209–220 [DOI] [PubMed] [Google Scholar]
- 162. Tezcaner A, Bugra K, Hasirci V. Retinal pigment epithelium cell culture on surface modified poly(hydroxybutyrate-co-hydroxyvalerate) thin films. Biomaterials. 2003;24:4573–4583 [DOI] [PubMed] [Google Scholar]
- 163. Candiello J, Balasubramani M, Schreiber EM, et al. Biomechanical properties of native basement membranes. FEBS J. 2007;274:2897–2908 [DOI] [PubMed] [Google Scholar]