Abstract
We have studied the role of core histone tails in the assembly of mitotic chromosomes using Xenopus egg extracts. Incubation of sperm nuclei in the extracts led to the formation of mitotic chromosomes, a process we found to be correlated with phosphorylation of the N–terminal tail of histone H3 at Ser10. When the extracts were supplemented with H1-depleted oligosomes, they were not able to assemble chromosomes. Selective elimination of oligosome histone tails by trypsin digestion resulted in a dramatic decrease in their ability to inhibit chromosome condensation. The chromosome assembly was also inhibited by each of the histone tails with differing efficiency. In addition, we found that nucleosomes were recruiting through the flexible histone tails some chromosome assembly factors, different from topoisomerase II and 13S condensin. These findings demonstrate that histone tails play an essential role in chromosome assembly. We also present evidence that the nucleosomes, through physical association, were able to deplete the extracts from the kinase phosphorylating histone H3 at Ser10, suggesting that this kinase could be important for chromosome condensation.
Keywords: assembly/chromosome/condensation/H3 phosphorylation/histone tails
Introduction
During mitosis, the relaxed interphase chromatin undergoes dramatic changes resulting in the formation of highly condensed mitotic chromosomes. Although the fascinating process of chromosome assembly was observed more than a century ago, it still remains poorly understood. An important contribution to the study of this process was due to the use of mitotic extracts isolated from Xenopus eggs: incubation of demembranated Xenopus sperm nuclei or somatic nuclei in such extracts results in the assembly of mitotic chromosomes (Lohka and Masui, 1983). Previously, we have shown that these in vitro assembled chromosomes exhibited identical physical properties to those of native chromosomes (Houchmandzadeh and Dimitrov, 1999). The use of mitotic extracts demonstrated that topoisomerase II is required for assembly and condensation, but not for structural maintenance of mitotic chromosomes (Hirano and Mitchison, 1993). The relatively easy biochemical procedure developed for the purification of these chromosomes made possible the identification of proteins specifically associated with them (Hirano and Mitchison, 1994; Hirano et al., 1997). These proteins, belonging to the SMC (structural maintenance of chromosomes) family, play an essential role in chromosome condensation (Hirano et al., 1997; Cubizollez et al., 1998). The SMC proteins exist in the form of two high molecular weight complexes (8S and 13S), termed condensins (Hirano et al., 1997). 13S condensin is phosphorylated in vitro by Cdc2, and this phosphorylation seems to be responsible for the triggering of chromosome condensation (Kimura et al., 1998)
Chromosome assembly is also accompanied by phosphorylation of histones H1 and H3 (Guo et al., 1995; Hendzel et al., 1997; Van Hooser et al., 1998). Linker histone H1 is highly phosphorylated at the beginning of mitosis in cells in culture and is rapidly dephosphorylated after anaphase, suggesting a role for this modification in chromosome condensation (Bradbury, 1992). However, recent experiments show that the absence of linker histones does not affect either chromosome condensation or nuclear assembly (Ohsumi et al., 1993; Dasso et al., 1994), thus arguing against a role for histone H1 hyperphosphorylation in these processes. On the contrary, histone H3 Ser10 phosphorylation is tightly correlated with chromosome condensation during both mitosis and meiosis (Hendzel et al., 1997). Additionally, this histone H3 modification seems to be required for the initiation but not the maintenance of the chromosome condensed state (Van Hooser et al., 1998). Recently it was also shown that H3 Ser10 phosphorylation is linked to chromosome condensation and segregation in vivo and is required for proper chromosome dynamics (Wei et al., 1999).
Both SMC proteins and topoisomerase II interact with DNA: 13S condensin induces an ATP-dependent positive supercoiling in closed circular DNA (Kimura and Hirano, 1997), and one of the topoisomerase II peptides, which is in close proximity to the DNA within the topoisomerase II–DNA complexes, was determined with high accuracy by using UV laser protein–DNA cross-linking (Hung et al., 1996). However, in vivo, DNA is packaged into nucleosomes and the effect of histones on the SMC protein and topoisomerase II interactions with nucleosomal DNA is not known.
The nucleosome is composed of two superhelical turns of DNA wrapped around an octamer of core histones consisting of two each of H2A, H2B, H3 and H4. The structure of the histone octamer has been determined by X–ray crystallography with high resolution: it represents a tripartite assembly with a centrally located (H3–H4)2 tetramer, flanked by two H2A–H2B dimmers (Arents et al., 1991). All four histones within the octamer consist of a rather elongated folded domain (the ‘histone-fold’) and externally located N–terminal tails with a large number of positively charged residues. The N–terminal tails are highly flexible and they contain the sites for different histone post-translational modifications (Arents et al., 1991; Luger et al., 1997). The tails are found to be active players in numerous vital functions in eukaryotic cells (Luger and Richmond, 1998). For example, acetylation of the tails seems to be involved in transcriptional regulation (Hebbes et al., 1989, 1994; Wolffe and Pruss, 1996; Wade et al., 1997). Strong support for this hypothesis comes from recent experiments demonstrating that basal transcription factors, transcription coactivators and transcription co-repressors possess histone acetyltransferase or histone deacetylase activities (for reviews, see Roth and Allis, 1996; Wolffe and Pruss, 1996; Wade et al., 1997). It was also shown that the N–terminal tails interact with repressor proteins and that they are essential for the assembly of specific protein complexes required for the establishment of chromatin silencing (Roth et al., 1992; Hecht et al., 1995). Additionally, acetylation at specific tail lysines is involved in chromatin assembly in vivo (Kaufman, 1996; Roth and Allis, 1996). The histone tails also seem to be implicated in nucleosome positioning in some yeast gene promoters (Roth et al., 1992).
In this work, we focused on the role of core histone tails in the in vitro assembly of mitotic chromosomes. Using Xenopus egg extract supplemented with native or tailless nucleosomes or GST–tail fusion proteins, we present evidence that the flexible histone tails are essential players in the chromosome condensation process. Our results also show that chromosome assembly is correlated with phosphorylation of histone H3 at Ser10 by a specific kinase and further reveal an involvement of this kinase in chromosome assembly.
Results
Inhibition of chromosome assembly in mitotic extracts by H1-depleted oligosomes
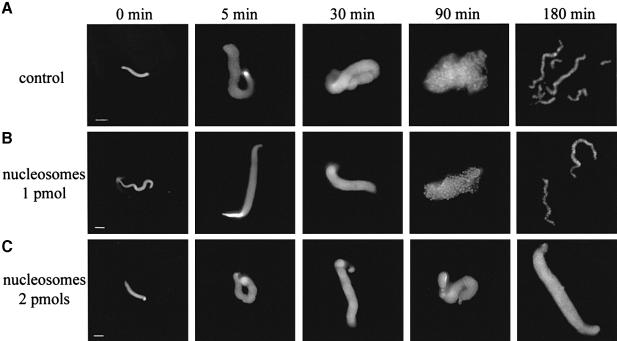
Incubation of demembranated sperm nuclei in a mitotic extract is accompanied by a series of well-defined sperm chromatin rearrangements, finally resulting in the formation of highly condensed mitotic chromosomes (Figure 1; Hirano and Mitchison, 1993). During the first 5 min of incubation, a dramatic decondensation of the snake-like shaped sperm nucleus is observed: the volume of the nucleus increases by about one order of magnitude. The sperm nucleus contains protamine-like proteins and a full complement of histones H3 and H4, and it is clearly deficient in histones H2A and H2B (Philpott et al., 1991; Philpott and Leno, 1992; Dimitrov et al., 1994). During the very rapid decondensation step, the protamine-like proteins are removed by the protein chaperone nucleoplasmin, and the core histones H2A and H2B, and B4 (the embryonic form of the linker histone) and HMG2 proteins are incorporated into the sperm chromatin (Dimitrov et al., 1994; S.Dimitrov and V.Gerson, unpublished observations). Incorporation of H2A and H2B histones in equimolar amounts to histones H3 and H4 allows the formation of proper nucleosome core particles, whilst the uptake of B4 and HMG2 proteins organizes the linker DNA (Dimitrov et al., 1994). Thus, during the decondensation step, the sperm nucleus is completely restructured and somatic-like type chromatin is assembled (Dimitrov et al., 1994). Since this decondensation step is very rapid compared with the whole chromosome condensation process (5 min versus 180 min, see Figure 1A), the protein factors responsible for the assembly process should ‘deal’ with the presence of nucleosomes. Indeed, it is known that at a higher sperm nuclei/extract ratio the chromosome assembly is inhibited. In order to check whether the nucleosomes are partners in this inhibition process, we prepared oligosomes, containing only DNA and core histones (for isolation of the oligosome material, see Materials and methods), and allowed chromosomes to assemble in their presence (Figure 1B and C). Oligosomes at a concentration of 1 pmol do not affect the assembly (Figure 1B), but increasing the concentration to 2 pmol leads to its complete inhibition: the process is stopped at the initial decondensation step (Figure 1C). It should be noted that this concentration of nucleosomes (2 pmol) is only twice that of nucleosomes (1 pmol) present in the sperm nuclei remodelled by the extract. Therefore, nucleosomes do not inhibit the function of nucleoplasmin, this protein being responsible for the decondensation step, but rather this is due to other factors acting on later stages of chromosome assembly.
Fig. 1. Inhibition of chromosome assembly by linker histone-depleted nucleosomes. A total of 60 000 demembranated sperm nuclei were incubated in 25 μl of mitotic extract for the times indicated in the absence (A) or presence of 1 (B) or 2 pmol (C) linker histone-depleted nucleosomes. The condensation intermediates were fixed and stained with Hoechst 33258. Well resolved individual chromosomes were observed after 180 min of incubation in the control (A) and in the experiment (B). Bars, 5 μm.
Naked DNA and tailless oligosomes do not affect chromosome formation
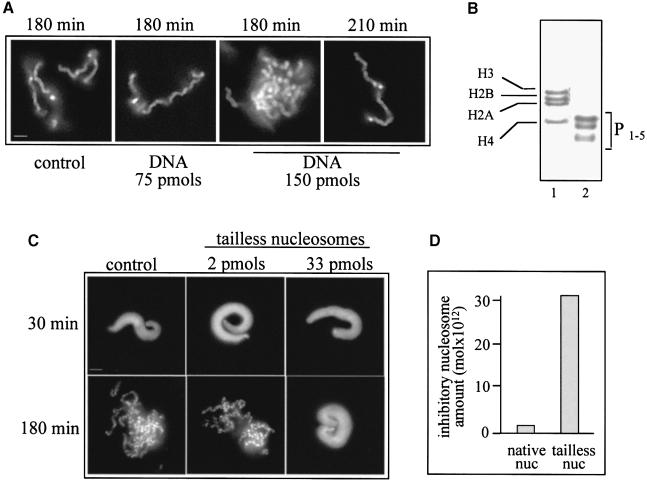
Since our oligosome preparation (Figure 2B) contains DNA and the histone octamer only, one of these two components could be the main target of the chromosome assembly factors. To determine whether DNA can titrate the chromosome assembly factors, we supplemented the extract with DNA and checked if the presence of DNA affects chromosome condensation. As shown in Figure 2A, even 150 pmol of nucleosomal DNA (75 times more than the nucleosome inhibitory concentration) shows no effect on chromosome formation. However, some slight effect on chromosome segration kinetics is observed in the presence of this large amount of DNA, since to obtain complete chromosome segregation 210 min of incubation are needed compared with 180 min in the presence of 75 pmol of DNA. These results clearly demonstrate that DNA cannot affect chromosome condensation. Therefore, this process must be inhibited by the histones within the nucleosome.
Fig. 2. Effect of DNA and of histone-tailless nucleosomes on chromosome condensation. (A) DNA does not affect chromosome condensation. Demembranated sperm nuclei (60 000) were incubated for the times indicated in 25 μl of extract in the absence (control) or presence of 75 or 150 pmol of nucleosomal DNA. The samples were fixed, stained with Hoechst 33258 and observed by fluorescence microscopy. Identical results were obtained when plasmid DNA was used instead of DNA isolated from nucleosomes. (B) An 18% SDS–PAGE of histones isolated from native (lane 1) and 3 min trypsin-digested (lane 2) nucleosomes. The positions of the core histones are indicated. P1–5 represent the trypsin-resistant peptides (the histone fold domains) designed as described by van Holde (1988). (C) A high concentration of tailless nucleosomes is necessary to inhibit chromosome condensation. Chromosome assembly was carried out as described in (A) in the absence and presence of 2 or 33 pmol of tailless nucleosomes. The samples fixed and stained with Hoechst 33258 after 30 and 180 min of incubation are shown. (D) Quantification of the experimental data presented in (C). Bars, 5 μm.
The histones within the octamer consist of folded domains and externally located flexible N–terminal tails (Arents et al., 1991). The histone tails are rather dynamic, and numerous reports in the literature have described their interactions with different protein factors (for reviews, see Wolffe and Pruss, 1996; Wade et al., 1997). We therefore hypothesized that the tails could interact with some chromosome assembly factors and thus could be essential players in the inhibition of chromosome condensation. In order to analyse the presumed interaction of the histone tails with chromosome assembly factors, we eliminated the histone tails of all four histones of the nucleosomes by trypsin digestion. As seen in Figure 2B, after 3 min incubation with trypsin, the tails are completely removed, while the structured histone fold domains are intact (it should be noted that elimination of the histone tails does not affect nucleosome structure, i.e. the histone fold domains are sufficient for proper folding of DNA around the tailless histone octamer; van Holde, 1988). Demembranated sperm nuclei and different amounts of tailless nucleosomes were next added to the mitotic extracts and the assembly was allowed to proceed. In parallel, experiments were carried out with extracts containing native oligosomes at the same concentrations. The data obtained (Figure 2C) show that tailless nucleosomes inhibit chromosome assembly at concentrations >15 times higher than that of the native nucleosomes (Figure 2D). These data demonstrated that the N–terminal histone tails of the histones within the nucleosome are essential for the inhibition of chromosome condensation.
Differential effect of individual histone tails on chromosome assembly
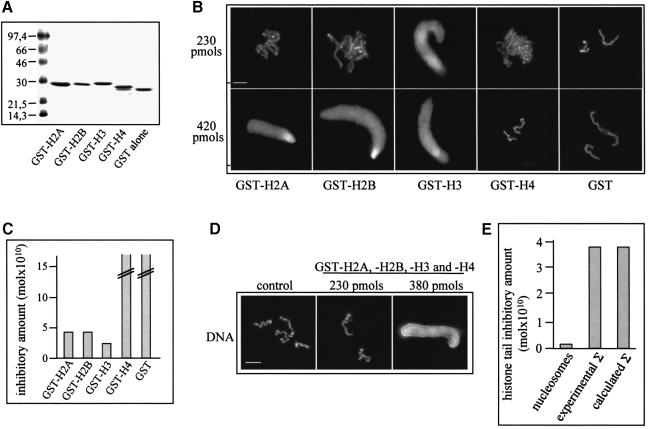
Is each individual histone tail involved in the inhibition of chromosome assembly? To answer this question, we prepared GST–histone tail fusion proteins (Figure 3A), titrated the mitotic extract with them and studied the chromosome assembly (Figure 3B). As a control, we used extracts supplemented with GST only. GST alone does not affect chromosome condensation even at the very high concentration of 10 nmol (Figure 3C). Three of the GST–histone tail fusion proteins (GST–H2A, GST–H2B and GST–H3) completely inhibit chromosome condensation, the best inhibitor being GST–H3 (Figure 3B and C). The absolute values of the inhibitory concentration of the different GST–tail fusions were found to vary in different extracts, but the ratio between them was preserved, i.e. the observed inhibitory concentration of GST–H3 was always 2–3 times lower than those of GST–H2A and GST–H2B. No inhibition of chromosome condensation was observed even at the highest concentration of GST–H4 used (1.8 nmol). We also carried out experiments with histone H3 and H4 chemically synthesized peptides corresponding to N–terminal amino acids 1–20 and we observed effects similar to their GST fusion counterparts (A.-E.de la Barre and S.Dimitrov, unpublished observations). Thus, we can conclude that the individual histone tails inhibit the process of chromosome assembly with different efficiencies.
Fig. 3. Individual histone tails differentially affect chromosome assembly. (A) A 12% SDS–PAGE of purified GST–histone tail fusion proteins. (B) The effect of each individual histone tail on chromosome condensation. The chromosome assembly reaction was performed under the conditions described in Figure 1 in the presence of 230 or 420 pmol of the different GST fusions and GST alone. (C) Quantification of the data presented in (B). The highest tested concentration of the GST–H4 fusion (1.8 nmol) and of GST alone (10 nmol) did not inhibit chromosome condensation. (D) Inhibitory effect on chromosome condensation induced by an equimolar mixture of the GST fusions at a total concentration of 230 or 380 pmol (the concentration of each individual tail was 25% of the total concentration). (E) Experimental Σ is a quantitative representation of the data shown in (D). Calculated Σ represents the inhibitory concentration of the equimolar mixture of GST fusions, calculated by using the experimental data for the inhibitory effect of each individual GST–histone tail. The quantity of nucleosomes necessary for complete inhibition of chromosome condensation is presented as the total amount of histone tails within the nucleosomes. Bars, 5 μm.
Histone tails act additively in inhibiting chromosome condensation
We have shown that nucleosomes at a concentration of 2 pmol (or 16 pmol histone tails within the nucleosome) completely inhibit chromosome assembly (Figure 1). To see whether the tails free in solution were as efficient inhibitors as those within the nucleosome, we supplemented the extract with an equimolar mixture of the individual tails and carried out the chromosome assembly. As seen from Figure 3D and E, the combination of the four GST fusions inhibits chomosome condensation at a total concentration (the sum of the concentrations of the individual tails) of 380 pmol (each individual tail was present at 95 pmol). It should be noted that this experimentally determined inhibitory concentration of total tails is essentially the same as that calculated with the assumption of independent and additive action of the tails during the inhibition of chromosome assembly (Figure 3E). Importantly, the observed inhibitory concentration of the tails free in solution is >20 times higher than that of the tails within the nucleosome (Figure 3E).
Chromosome assembly factors are found stably associated with nucleosomes
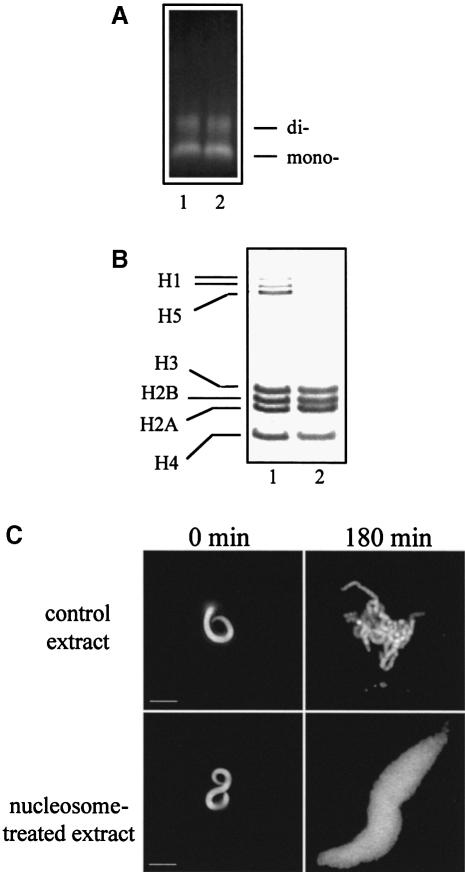
We have found that both nucleosomes and histone tails free in solution act as efficient inhibitors of chromosome condensation, suggesting that they are able to sequester some chromosome assembly factors. To check this hypothesis, we incubated the extract with an inhibitory concentration of oligosomes and removed them by centrifugation. Further, we studied whether the supernatant of the nucleosome-treated extract can still assemble chromosomes. As seen (Figure 4A and B), under the conditions of the centrifugation, the nucleosomes were pelleted quantitatively and they retained their full complement of core histones. However, the nucleosome-treated extract, in contrast to the nucleosome-non-treated (control) extract, lost its ability to assemble chromosomes: incubation of demembranated sperm nuclei in the nucleosome-treated extract results only in their decondensation (Figure 4C). Thus, some factors necessary for chromosome assembly remain physically associated with the nucleosomes present in the extract.
Fig. 4. Chromosome assembly factors are found associated with nucleosomes. Chromosome assembly was performed as described in Figure 1 in the presence of 2 pmol of exogenous nucleosomes per 25 μl of extract. The nucleosomes were pelleted by centrifugation in a bench-top centrifuge at 12 000 g for 15 min and the supernatant was removed and used in the chromosome assembly assay. (A) Nucleosomes were pelleted quantitatively under the conditions of the centrifugation. DNA was isolated from the nucleosome pellet and the calculated amount of this sample equivalent to 200 ng (lane 1) was loaded on a 1% agarose gel together with 200 ng of spectrophotometrically measured nucleosomal DNA (lane 2). The intensities of the signals of both samples are essentially identical, demonstrating a quantitative nucleosome pelleting. (B) Pelleted nucleosomes retained their full complement of core histones. Histones were isolated from 5 μg of pelleted nucleosomes and separated on an SDS–18% polyacrylamide gel (lane 2); lane 1, total histones isolated from hen erythrocyte nuclei. The positions of the core and the linker histones are given. (C) Nucleosome-treated extracts lose their ability to assemble chromosomes. Sperm nuclei were incubated for 180 min in the supernatants of control and nucleosome-treated extracts obtained after centrifugation, and the resulting structures were visualized with Hoechst 33258. Fixed sperm nuclei are shown for comparison. Bars, 5 μm.
Topoisomerase II and 13S condensin interact with similar efficiency with both native and tailless nucleosomes
We have shown that nucleosome-induced inhibition of chromosome condensation is determined by direct association of some chromosome assembly factors with the nucleosomes present in the extract. Which are these factors? Two known factors participating in chromosome assembly and interacting with DNA are topoisomerase II and SMC proteins (for reviews, see Poljak and Käs, 1995; Koshland and Strunnikov, 1996; Warburton and Earnshaw, 1997; Hirano, 1999). The SMC proteins form two high molecular weight complexes (13S and 8S), called condensins (Hirano et al., 1997). The 8S condensin is a heterodimer of two SMC proteins, XCAP-C and XCAP–E, while 13S condensin contains three additional non–SMC subunits, XCAP-H, XCAP-D2 and XCAP-G. 13S condensin is absolutely required for chromosome condensation, since 13S condensin-immunodepleted extracts are not able to condense chromosomes (Hirano et al., 1997; Hirano, 1998; Kimura et al., 1998).
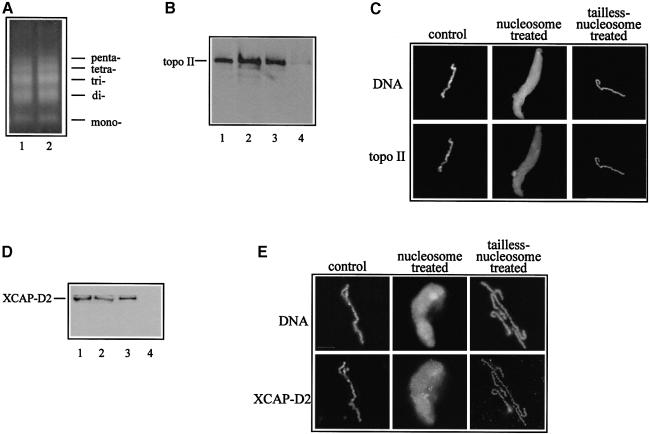
Do topoisomerase II and 13S condensin (relevant to chromosome condensation) interact with nucleosomes? Do histones, and N–terminal histone tails in particular, affect this interaction? To answer these questions, we incubated the extracts with both native and tailless nucleosomes, isolated the nucleosomes from the extract by centrifugation and checked the presence of topoisomerase II and 13S condensin in these two nucleosome samples by immunoblotting with highly specific antibodies against topoisomerase II and the XCAP-D2 protein (we used XCAP-D2 as a marker for 13S condensin since it is present only in this condensin form and not in the 8S form). High speed centrifugation quantitatively pelleted native and tailless nucleosomes (Figure 5A). The immunoblotting analysis shows that the same amount of topoisomerase II is present in precipitated native and trypsinized nucleosomes (Figure 5B). Importantly, the amount of topoisomerase II associated with the pelleted nucleosomes was found to be no more than 1% of that of the topoisomerase II present in the extract (Figure 5B). This is confirmed further by the immunofluorescent analysis of the presence of topoisomerase II on chromosome condensation intermediates assembled in the supernatants, isolated after centrifugation from both native and trypsinized nucleosome-treated extract (Figure 5C). The results obtained for XCAP-D2 are identical to those for topo– isomerase II (see Figure 5D and E). This demonstrates that: (i) topoisomerase II and 13S condensin interact similarly with both types of nucleosomes; and (ii) the amount of these proteins found associated with the nucleosomes is negligible compared with the amount present in the extract.
Fig. 5. Topoisomerase II and XCAP-D2 proteins interact with similar efficiency with both native and tailless nucleosomes. Native and tailless nucleosomes were incubated in mitotic extract (2 pmol of nucleosomes per 25 μl of extract) for 30 min at room temperature and the samples were loaded over 1.5 ml of a 5% sucrose cushion and centrifuged for 2 h in a TLS-55 (Beckman) swinging bucket rotor at 55 000 r.p.m. The upper fraction (extract) was removed and used for incubation with sperm nuclei, the sucrose cushion discarded and the pelleted oligosome fraction analysed. (A) The high speed centrifugation quantitatively pelleted native and tailless oligosomes. DNA was isolated from both types of pelleted oligosomes and dissolved in the same volume. The same aliquot from both samples was run on a 1% agarose gel and stained with ethidium bromide. Lane 1, tailless nucleosomal DNA; lane 2, native nucleosomal DNA. (B) Native and tailless nucleosomes were found associated with the same amount of topoisomerase II. Topoisomerase II associated with pelleted native and tailless nucleosomes detected by Western blotting with anti-topoisomerase II antibody. Lane 1, 1% of the extract used for incubation with nucleosomes; lanes 2 and 3, 5 μg of native and tailless pelleted oligosomes, respectively; lane 4, the pellet of the non-treated extract (control). (C) Immunofluorescence detection of topoisomerase II associated with sperm nuclei incubated for 180 min in control, nucleosome-treated and tailless nucleosome-treated extract, obtained after the high speed centrifugation. DNA was visualized with Hoechst 33258. (D) and (E), same as (B) and (C), but antiserum against XCAP-D2 was used for the immunodetection.
Phosphorylation of histone H3 at Ser10 correlates with the condensation of chromosomes in Xenopus egg extracts
Phosphorylation of histone H3 at Ser10 has been correlated with mitosis in different systems (Hendzel et al., 1997; Wei et al., 1998; Van Hooser et al., 1998). Evidence was presented recently that this specific H3 tail phosphorylation was required for proper chromosome condensation and segregation (Wei et al., 1999). If H3 phosphorylation is also required for chromosome assembly in Xenopus extract, this may explain, at least in part, the observed inhibitory properties of the histone H3 tail.
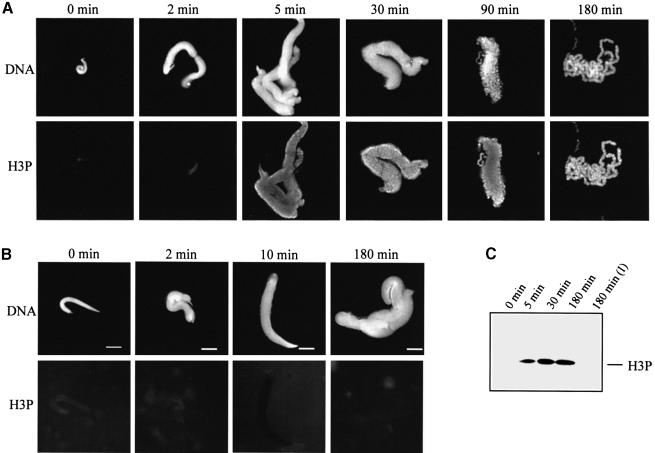
We first addressed this problem by studying the phosphorylation status of histone H3 during in vitro chromosome assembly. In these experiments, we used an antibody specific for Ser10-phosphorylated histone H3 (Hendzel et al., 1997). The immunofluorescence data presented in Figure 6A show that chromosome formation is accompanied by H3 phosphorylation. Histone H3 is not phosphorylated in sperm nuclei (Xenopus sperm nuclei contain a full complement of histones H3 and H4; see Philpott et al., 1991; Philpott and Leno, 1992; Dimitrov et al., 1994), but was phosphorylated in the decondensed sperm nuclei after only 5 min incubation in the extract. Phosphorylated H3 shows a specific pattern during chromosome assembly. First it appears on the contours of the decondensed sperm nuclei, but the next condensation intermediates, in addition to the contour labelling, also demonstrate a punctuated labelling, located ‘inside’. The fully assembled chromosomes are already uniformly labelled. The observed pattern of phosphorylated histone H3 is highly reproducible and is not related to accessibility, since anti-core histone antibodies stained each condensation intermediate uniformly (results not shown). Incubation of the sperm nuclei in the interphase extract did not result in H3 phosphorylation, thus demonstrating a mitotic-specific effect (Figure 6B). The immunoblotting analysis supports the above conclusion: phosphorylation of histone H3 is found only in the material incubated in the mitotic extracts (Figure 6C). The results of H3 phosphorylation kinetics demonstrate saturation after 30 min of incubation of sperm nuclei in the extract (Figure 6C).
Fig. 6. Mitosis-specific phosphorylation of histone H3 at Ser10. (A) Chromosomes were assembled under standard conditions in mitotic egg extract; aliquots were removed at the times indicated and fixed. DNA was visualized with Hoechst 33258 and phosphorylated histone H3 (H3P) was detected by indirect immunofluorescence using specific antibodies against histone H3 phosphorylated at Ser10. (B) Same as (A), but sperm nuclei were incubated in interphase extract isolated from Xenopus eggs. (C) Immunoblotting analysis of the time course of histone H3 phosphorylation. Sperm nuclei were incubated in either mitotic or interphase extracts for the times indicated, the samples were centrifuged on a bench-top centrifuge for 3 min at 10 000 g, and the proteins from the pellet were separated on an 18% SDS–polyacrylamide gel. After transfer, the phosphorylated H3 was visualized with anti-phosphorylated histone H3 antibody. The immunodetection for the sample incubated in the interphase extract for 180 min [180 min (I)] only is shown.
Effect of native and tailless nucleosomes on histone H3 phosphorylation
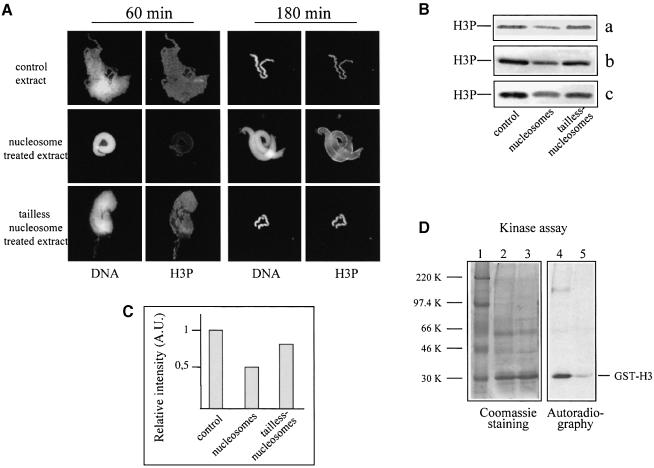
We next analysed the effect of native and tailless nucleosomes on histone H3 phosphorylation by studying chromosome assembly in the presence of both nucleosome samples. Phosphorylation of histone H3 was investigated again by using the above antibody specific for Ser10-phosphorylated histone H3. The structure observed in the presence of 2 pmol of native nucleosomes exhibited the contour pattern of phosphorylated histone H3, characteristic of the sperm nuclei decondensed upon 5 min incubation in the extract (compare Figures 7A and 6A). Tailless nucleosomes did not affect the distribution of phosphorylated histone H3 (Figure 7A). The immunoblotting analysis demonstrates that the degree of histone H3 phosphorylation of the sperm nucleus intermediates observed in the nucleosome-treated extract is lower than that of the chromosomes condensed in the control or tailless nucleosome-containing extract (Figure 7B and C). Again this result argues that the chromosome assembly reaction is arrested by the nucleosomes at a stage typical for the decondensed sperm nuclei, which exhibit lower H3 phosphorylation than fully condensed chromosomes (see the kinetics of H3 phosphorylation in Figure 6C). The pattern of H3 phosphorylation of sperm nuclei, observed after 3 h incubation in the extract from which the nucleosomes were removed by centrifugation, is also identical to that of decondensed nuclei (data not shown). Thus, nucleosomes inhibit chromosome assembly at a specific intermediate state with a characteristic phosphorylation pattern.
Fig. 7. A kinase activity phosphorylating histone H3 at Ser10 was found associated with nucleosomes. (A) Nucleosome-arrested chromosome condensation exhibits a specific pattern of histone H3 phosphorylation. Sperm nuclei were incubated for 60 or 180 min in mitotic extract, containing either native or tailless nucleosomes, and DNA and histone H3 phosphorylation were visualized by Hoechst 33258 and anti-phosphorylated histone H3 antibody. (B) Native nucleosomes present in the extract decrease the degree of histone H3 phosphorylation of the chromosome condensation intermediates. Extracts were incubated for 30 min with native or tailless nucleosomes, the nucleosomes were pelleted by centrifugation and the supernatants supplemented with sperm nuclei. The control and the two nucleosome-treated extracts were incubated for 180 min at room temperature and the remodelled sperm were pelleted by centrifugation. The proteins from the pellets were run on an SDS–18% polyacrylamide gel, and phosphorylated histone H3 was detected with a specific antibody; a, b and c represent the results of three independent experiments. (C) Quantification of the data presented in (B). (D) The pelleted native nucleosomes are associated with kinase activity phosphorylating histone H3. The pellets from the control and nucleosome-treated extracts (see above) were resuspended and supplemented with GST–H3 and [γ–32P]ATP. After 30 min incubation, the samples were precipitated with TCA and analysed on an SDS–12% polyacrylamide gel. Left, Coomassie staining of the gel; right, autoradiography. Lane 1, protein marker; lane 2, control pellet; lane 3, pellet from the nucleosome-treated extract; lanes 4 and 5, autoradiography of lanes 3 and 4, respectively. The positions of GST–H3 and the protein markers are indicated.
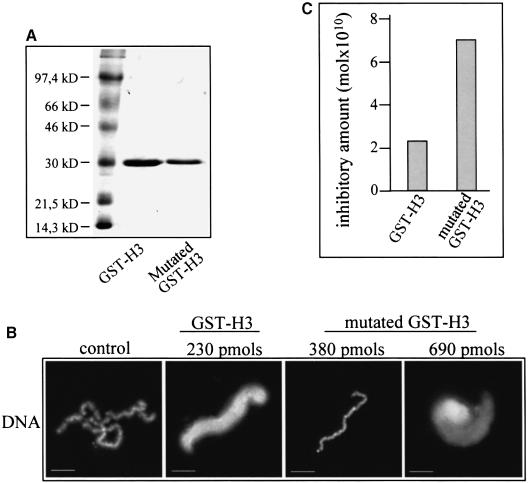
A simple explanation of the observed effect could be a partial sequestration by the nucleosomes of the kinase responsible for the phosphorylation of histone H3. To test this, we incubated the extract with native nucleosomes and, after pelleting, we checked the presence of histone H3 kinase activity within the pellet indirectly by a kinase assay. We used GST–H3 histone tail fusion protein as substrate for the kinase assay. As seen (Figure 7C), the pelleted nucleosomes exhibited histone H3 kinase activity. Therefore, the nucleosomes should interact with the kinase, presumably through the histone H3 tail. If this was really the case, one would expect a Ser10 GST–H3 mutant protein to exhibit a lower inhibitory effect on chromosome assembly. To check this, we prepared a GST–H3 mutant fusion with Ser10 substituted by alanine (Figure 8A). As demonstrated (Figure 8B), the mutant fusion, in contrast to GST–H3, does not arrest chromosome condensation at a concentration of 230 pmol. The inhibitory concentration for the GST–H3 mutant was found to be 700 pmol, i.e. ∼3 times higher than that of GST–H3 (Figure 8C). These data suggest that at least part of the inhibitory effect of the histone H3 tail is related to the sequestration of the kinase(s), phosphorylating histone H3 at Ser10.
Fig. 8. Effect of mutant (Ser10→Ala) GST–H3 fusion protein on chromosome assembly. (A) An 18% SDS–PAGE of GST–H3 and GST–H3 mutant proteins. The first lane contains a protein mass marker. (B) A higher concentration of GST–H3 mutant fusion is necessary for inhibition of chromosome condensation. Chromosomes were assembled for 180 min under standard conditions in the absence (control) and presence of GST–H3 and GST–H3 mutant fusions in the amounts indicated. Samples were fixed and stained with Hoecsht 33258. (C) Quantification of the data presented in (B).
Discussion
Incubation of demembranated sperm nuclei in mitotic extracts isolated from Xenopus eggs results in the formation of chromosomes with a good morphology and with physical properties essentially identical to those of chromosomes observed in vivo (Houchmandzadeh et al., 1997; Houchmandzadeh and Dimitrov, 1999). Thus, data obtained on the assembly of these chromosomes are relevant to the physiological mechanism. In addition, working with in vitro assembled chromosomes has a major advantage over studies using chromosomes from somatic cells: the in vitro condensed chromosomes can be manipulated in their natural assembly conditions. We used this advantage to study the role of core histone N–terminal tails in chromosome assembly.
Core histone tails are essential for the assembly of mitotic chromosomes
The conditions for in vitro assembly of chromosomes require an optimal sperm nuclei/extract ratio (Smythe and Newport, 1991; Hirano and Mitchison, 1993). Increasing the amount of sperm chromatin relative to that of the extract results in inhibition of the assembly: depending on the excess of sperm nuclei, the chromosome condensation is arrested at a specific intermediary stage (A.-E.de la Barre and S.Dimitrov, unpublished observations; this study). This suggests that some limiting chromosome assembly factors are sequestered by the sperm nuclei. Since during the very fast decondensation step (the decondensation is completed within the first 5–10 min of incubation in the extract) the sperm nuclei are completely remodelled and adopt a nucleosome-type structure (Philpott et al., 1991; Philpott and Leno, 1992; Dimitrov et al., 1994), we hypothesized that some of these chromosome assembly factors could be titrated by sperm nucleosomes. Indeed, our experiments clearly demonstrated that nucleosomes are strong inhibitors of chromosome assembly (Figure 1). Since tailless nucleosomes did not inhibit chromosome condensation (Figure 2), we concluded that the nucleosome inhibitory effect is effected through the histone non-structured N–terminal tails. This was confirmed by the experiments with GST–histone tail fusion proteins: individual histone tails inhibit the assembly process with differing efficiency within the range of 200–500 pmol (Figure 3). Interestingly, the combination of all tails affects chromosome condensation in an additive manner (Figure 3E), suggesting that they act independently in inhibiting this process. All these data allowed us to conclude that histone tails are essential players in chromosome assembly.
Unknown chromosome assembly factors are physically associated with nucleosomes
A simple way to explain the nucleosome-induced inhibition is to suggest a physical association between the nucleosomes incubated in the egg extract (essentially through their N–terminal tails) and some chromosome assembly factors. Our data demonstrated that this is indeed the case, since the supernatant obtained after centrifugation of nucleosome-treated extract is not able to assemble chromosomes (Figure 4C). Thus, some specific chromosome assembly proteins must be associated with the nucleosomes and could be pelleted together with them upon centrifugation.
Which are these proteins? Chromosome assembly is a very complex process and obviously many different factors are involved in it. However, participation in the assembly and maintenance of chromosomes has been clearly demonstrated for only two types of proteins: topoisomerase II and the SMC protein family, existing as higher molecular weight complexes called condensins, the form relevant for chromosome condensation being 13S condensin (Koshland and Strunnikov, 1996; Hirano et al., 1997; Warburton and Earnshaw, 1997; Hirano, 1998). Our Western blotting analysis and immunofluorescence results demonstrate that both topoisomerase II and 13S condensin interact with similar efficiency with both native and trypsinized nucleosomes (Figure 5B and C). Moreover, the amount of topoisomerase II and 13S condensin associated with each type of pelleted nucleosomes does not exceed 1% of the amount of these factors present in the extract. Therefore, the inability of nucleosome-treated extract to condense chromosomes is not due to nucleosome depletion of topoisomerase II and 13S condensin from the extract, but instead is due to other unknown chromosome assembly factors.
Nucleosomes are much better inhibitors than free tails in solution: >20 times more of an equimolar mixture of GST–histone tails is necessary to achieve a complete arrest of chromosome assembly (see Figure 3E). Thus, the histone tails within the nucleosome should be able to recruit chromosome assembly factors more efficiently than the free tails in solution. This presumed effect could be explained by a specific higher affinity chromosome assembly factor conformation of the histone tails within the nucleosomes. Indeed, in solution, the histone tails seem not to be structured, but when bound to nucleosomal DNA the tails of histones H3 and H4 adopt a highly structured conformation (Baneres et al., 1997; Luger et al., 1997; Luger and Richmond, 1998). These specific tail conformations as well as the close spatial localization of the different histone tails within the nucleosome could allow better interactions and consequently a greater association of histone tails with chromosome assembly factors.
A kinase, specifically phosphorylating histone H3 at Ser10, is a partner in chromosome condensation
Chromosome condensation is accompanied by specific phosphorylation of histone H3 Ser10 in different systems (Hendzel et al., 1997; Van Hooser et al., 1998; Wei et al., 1998), suggesting that both histone H3 mitotic-specific modification and a kinase activity may play an essential role in chromosome condensation. To check this hypothesis, we first asked whether histone H3 is phosphorylated during in vitro chromosome assembly by using a highly specific antibody, which recognizes Ser10-phosphorylated histone H3. The immunochemical data (Figure 6) clearly demonstrate that this is indeed the case. Next we showed that the nucleosome-arrested chromosome intermediates exhibit lower H3 phosphorylation as well as a specific intermediate H3 phosphorylation pattern (Figure 7A and B). This effect seemed to be determined by the partial depletion of the H3 kinase(s) due to its physical association with the nucleosomes upon their incubation in the extract (Figure 7C). We further confirmed this by demonstrating that GST–H3 fusion protein is a much better inhibitor of chromosome condensation than its mutant form, GST–H3 with Ser10 modified to alanine (Figure 8). Therefore, the histone H3 Ser10-specific kinase is an essential player in the chromosome condensation process. This conclusion is in agreement with the recent cell microinjection data, suggesting that competitive inhibition of H3 kinase prevents chromosome condensation and entry into mitosis (Van Hooser et al., 1998).
Histone H3 phosphorylation and chromosome condensation
Chromosome assembly exhibits condensation intermediates with a rather specific phosphorylation pattern (Figure 6). In addition, when the extract was supplemented with increasing amounts of nucleosomes, the chromosome condensation inhibition occurred at different condensation intermediary stages (depending on the amount of nucleosomes present), with a phosphorylation pattern very similar to some of the kinetic condensation intermediate patterns shown in Figure 6 (A.-E.de la Barre and S.Dimitrov, unpublished observations). In other words, when the assembly was arrested ‘artificially’ by the nucleosomes at an intermediary stage with a characteristic extent of condensation, this intermediate showed a histone H3 phosphorylation specific for the intermediate degree of assembly. Thus, the mitotic-specific histone H3 phosphorylation pattern could be viewed as a ‘signature’ of the extent of chromosome condensation. Obviously, this ‘signature’ is important for chromosome assembly, since phosphorylation of histone H3 is required for proper chromosome formation and segregation during mitosis and meiosis (Sauvé et al., 1999; Wei et al., 1999).
Recently it was suggested that histone H3 phosphorylation at Ser10 could act as a ‘chromatin receptor’ for 13S condensin and/or topoisomerase II (Hendzel et al., 1997; Wei et al., 1998, 1999; Hirano, 1999). However, histone H3 within the nucleosomes incubated in the extract is phosphorylated at Ser10 (A.-E.de la Barre and S.Dimitrov, unpublished observations), but nevertheless the amounts of 13S condensin and topoisomerase II associated with native and tailless nucleosomes were found to be the same. Thus, phosphorylation of histone H3 at Ser10 does not facilitate, as suggested, the binding of these chromosome condensation factors to chromatin.
In conclusion, the use of the in vitro system for chromosome assembly allowed us to demonstrate that chromatin is an active participant in the chromosome condensation process. This function of chromatin is carried out at least partially by the non-structured N–terminal histone tails through interactions with some chromosome assembly factors. One of these factors is found to be a kinase, phosphorylating histone H3 at Ser10. The cloning and identification of this kinase as well as other factors that interact with the core histone tails remain a challenge for future studies.
Materials and methods
Preparation of mitotic extracts
Mitotic extracts from Xenopus eggs were prepared essentially as described by Hirano and Mitchison (1993). Dejellied eggs were crushed in EB [80 mM β-glycerophosphate pH 7.3, 15 mM MgCl2, 20 mM EGTA and 1 mM dithiothreitol (DTT), supplemented with 10 μg/μl of leupeptin and apoprotin] by centrifugation for 20 min at 15 000 r.p.m. in an SW41 rotor (Beckman Instruments). The crude extracts (low speed supernatants) were transferred in 2 ml propylene tubes for the TLS-55 rotor (Beckman Instruments) and centrifuged at 52 000 r.p.m. for 2 h at 4°C. The top lipid layer was sucked up under vacuum, and the cytoplasmic fraction was collected carefully and recentrifuged at 4°C for 30 min at 52 000 r.p.m. (TLS-55 rotor) to remove residual membranes. The extracts (high speed supernatants) were aliquoted in 25 μl fractions, the aliquots were frozen immediately in liquid nitrogen and stored at –80°C until use. The extracts prepared in this way do not lose their ability to assemble chromosomes for at least 3 months.
Xenopus egg interphase extracts were isolated following strictly the protocol of Smythe and Newport (1991). The interphase extract was converted into a mitotic extract by adding cyclin BΔ90 (a non-degradable form of cyclin B). Recombinant cyclin BΔ90 was isolated as described (Holloway et al., 1993).
Preparation of sperm chromatin
The procedure used for preparation of sperm nuclei is based on the modification of the method of Smythe and Newport (1991). The testes from four frogs were placed in a Petri dish containing ice-cold buffer T (15 mM PIPES, 15 mM NaCl, 5 mM EDTA, 7 mM MgCl2, 80 mM KCl, 0.2 M sucrose, pH 7.4) and were rinsed a few times in this buffer. After cleaning the testes of blood and intestine contaminations, they were minced in 2 ml of buffer T using forceps, and centrifuged for 10 s at 800–1000 r.p.m. in a clinical centrifuge. The supernatant was centrifuged at 5000 r.p.m. for 5 min and the white upper layer of the pellet (the sperm) was carefully removed, trying to avoid red blood cells (the lower layer). This operation was repeated a few more times until all contamination from red blood cells was eliminated. The sperm were resuspended in 400 μl of buffer T, and 1.2 ml of buffer S (buffer T + 20 mM maltose + 0.05% lysolecithin) were added to it. The sperm suspension was incubated for 5–10 min at 23°C and placed on ice. The degree of demembranation was checked with a fluorescent microscope using Hoecsht 33258. If incomplete removal (non-uniform labelling of the sperm nuclei) of the membrane was achieved, the suspension was incubated at 23°C for an additional 5 min. The reaction was stopped by adding 4.8 ml of buffer R [buffer T + 3% bovine serum albumin (BSA)] and the material centrifuged at 2000 r.p.m. for 5 min. The pellet was resuspended in 5 ml of buffer R and recentrifuged under the same conditions. The demembranated sperm nuclei were resuspended in 300–400 μl of buffer T, their concentration measured, and the material was aliquoted in 5 μl fractions and stored at –80°C.
Isolation of nuclei and histone linker-depleted oligosomes
Chicken erythrocyte nuclei were isolated according to Mirzabekov et al. (1990). Oligosomes were prepared by digesting the nuclei with micrococcal nuclease. Linker histones H1 and H5 and non-histone proteins were removed from the oligosomes by fractionation in a 5–20% sucrose gradient containing 0.67 M NaCl as described (Mutskov et al., 1998). The concentration of the oligosomes was determined spectrophotometrically (1 OD260 = 50 μg of nucleosomes) and the nucleosome molar concentration calculated by assuming a nucleosome molecular mass of 200 kDa.
Preparation of tailless oligosomes
Tailless nucleosomes were prepared by digestion with trypsin (Mutskov et al., 1998). A 500 μl aliquot of linker histone-depleted oligosomes (0.5 mg/ml) in 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM EDTA was digested at 37°C for 3 min with trypsin (Sigma) at a trypsin:oligosome weight:weight ratio of 1:25. The reaction was terminated by addition of diisopropylfluorophosphate (Sigma) and the tailless histones were visualized on an 18% polyacrylamide gel containing SDS (Laemmli, 1970).
Expression and purification of GST–histone tail fusion proteins
GST–Drosophila histone tail constructs were a kind gift from Dr Carl Wu (Georgel et al., 1997). The mutation of Ser10 to alanine in the GST–H3 tail was made according to the standard oligonucleotide site-directed mutagenesis approach. The oligonucleotides used were two complementary oligonucleotides MC39-5′ (GCAAACTGCTCGCAAAGCGACTGGTG) and MC40-3′ (CACCAGTCGCTTTGCGAGCAGTTTGC) designed to contain the 5′ sequence of the H3 tail but with one base change of a T to a G, together with commercially available pGex internal sequencing primers. The fusion proteins were expressed in Escherichia coli strain BL21 and were purified using glutathione–Sepharose 4B (Pharmacia Biotech.) essentially as described by the manufacturer. The bound material was eluted from the resin by overnight incubation at 4°C in the elution buffer (10 mM glutathione in 50 mM Tris–HCl pH 8.0). The eluted fusion proteins were dialysed further overnight against EB and their integrity was checked by SDS–PAGE (Laemmli, 1970). The purity of the GST–histone tail fusion proteins was found to be >90%. The concentration of the fusions was determined both by Bradford assay and spectrophotometrically. The fusion proteins were concentrated using Centricon concentrators (Amicon). Their molar concentration was calculated assuming molecular masses of 33 kDa for GST–H2A, GST–H2B and GST–H4 and of 34 kDa for GST–H3.
Assembly of mitotic chromosomes
An aliquot (25 μl) of mitotic extract was thawed rapidly, diluted twice with EB and supplemented with the components of the ATP-regenerating system (20 mM phosphocreatine, 2 mM ATP pH 7.0 and 5 μg/ml creatine kinase, final concentration). A 1 μl aliquot of sperm nuclei (40 000–60 000 nuclei) was added to this solution and the material was incubated at 23°C. Aliquots of 5 μl were taken at different times after initial incubation and fixed immediately with 5 μl of the fix/stain buffer (Hoechst 33258 at 1 μg/ml in 200 mM sucrose, 10 mM HEPES pH 7.5, 7.4% formaldehyde, 0.23% DAPCO, 0.02% NaN3 and 70% glycerol). The assembly of chromosomes in interphase cyclin BΔ90- (the non-degradable form of cyclin B) activated extract was as described above, but 1 μl of cyclin BΔ90 was added to 25 μl of extract before dilution with buffer EB. The chromosome assembly properties of both extracts were very similar.
For chromosome assembly inhibition reactions, nucleosomes or GST–tail fusion proteins were first dialysed overnight against buffer EB and then used in the experiments. The inhibitory concentration Cinh was defined as the minimal concentration at which the assembly of chromosomes was arrested at the decondensation step. In the case of additivity of the action of GST–histone tails, the total inhibition effect totEinh (totEinh = 1/totCinh) was calculated as totEinh = H2AEinh + H2BEinh + H3Einh + H4Einh, where H2AEinh, H2BEinh, H3Einh and H4Einh were the values of the experimentally determined inhibitory effect for GST–H2A tail, GST–H2B tail, GST–H3 tail and GST–H4 tail, respectively.
Immunofluorescence
For the immunofluorescence analysis, 10 μl aliquots were removed at different times after the initiation of the assembly reaction and to each aliquot 40 μl of ice-cold 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; 125 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4 pH 7.5) were added, followed by incubation of the material for 45 min on coverslips in a moist chamber at 4°C. After additional incubation for 5 min in 50 ml of 4% PFA/PBS at 4°C, the slides were washed twice with PBS at 4°C for 5 min and covered with 200 μl of the specific antiserum at a suitable dilution (1:300 for anti-topoisomerase II antiserum and 1:100 for anti-XCAP-D2 antiserum) in antibody buffer [PBS containing 0.1% Triton X-100 and 5% lamb serum (mycoplasma screened, Gibco-BRL)]. The slides were washed three times for 10 min at 4°C with PBS-TB (PBS containing 0.1% Triton X-100, 0.5% BSA) and incubated for 30 min at 37°C with 200 μl of fluorescein-conjugated goat anti-rabbit IgG (20 μg/ml). This was followed by three washes (10 min each at 4°C) with PBS-TB. Finally, the chromosomes were counterstained with 8 μl of fix/stain buffer [Hoechst 33258 at 1 μg/ml in 200 mM sucrose, 10 mM HEPES pH 7.5, 7.4% formaldehyde, 0.23% DAPCO (Sigma), 0.02% NaN3 and 70% glycerol].
Immunoblotting
The immunoblotting procedure was carried out as described previously (Dimitrov and Wolffe, 1997). Briefly, after separation of the proteins in SDS–gels, they were transferred to nitrocellulose paper (Hybond-C, Amersham) and the filters were blocked for 1 h with PBS containing 5% non-fat dry milk, 0.2% Tween-20. They were then incubated for 1 h at room temperature with the appropriately diluted first antiserum (1:300 for anti-topoisomerase II antiserum and 1:100 for anti-XCAP-D2 antiserum) in PBS, 0.2% Tween-20, 10% calf serum. After three washes with PBS-T (PBS containing 0.5% Triton X-100), 0.4 M NaCl, the filters were treated for 1 h with peroxidase-conjugated secondary antibody in PBS, 0.2% Tween-20, 1% non-fat dry milk. Finally, after three washes with PBS-T, 0.4 M NaCl, the filters were developed using the enhanced chemiluminescence system (ECL, Amersham) as recommended by the manufacturer.
Kinase assay
The high speed centrifugation-precipitated oligosomes were resuspended in 50 μl of EB, 0.5 M NaCl, supplemented with 20 mM phosphocreatine pH 7.0 and 5 μg/ml creatine kinase. To this solution was added 1–3 μg of GST–H3 fusion protein and 20–30 μCi of [γ–32P]ATP (3000 Ci/mmol, Amersham), and the material was incubated for 30 min at room temperature. The reaction mixture was diluted with 100 μl of TE (10 mM Tris pH 8.0, 1 mM EDTA) and the proteins precipitated with 20% trichloroacetic acid (TCA; final concentration). After separation of the proteins on an SDS–12% polyacrylamide gel, the gel was dried and autoradiographed.
Acknowledgments
Acknowledgements
We thank Drs B.Houchmandzadeh, J.Bednar, S.Nonchev, T.Hirano, P.Adreassen and R.Margolis for critical comments on the manuscript, V.Legagneux and T.Hirano for antibodies against XCAP-D2 and topoisomerase II, S.Khochbin for useful discussions, and T.Lorca and M.Glotzer for the expression vector of cyclin B as well as for advice on cyclin B purification. We also appreciate the support of Dr Jean-Jacques Lawrence throughout the course of this work.
References
- Arents G., Burlingame, R.W., Wang, B.-C., Love, W.E. and Moudrianakis, E.N. (1991) The nucleosome core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left handed superhelix. Proc. Natl Acad Sci. USA, 88, 10148–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneres J.L., Martin,A. and Parello,J. (1997) The N-tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J. Mol. Biol., 273. [DOI] [PubMed] [Google Scholar]
- Bradbury E.M. (1992) Reversible histone modifications and the chromosome cell cycle. BioEssays, 14, 9–16. [DOI] [PubMed] [Google Scholar]
- Cubizollez F., Legagneux, V., Le Guellec, R., Isabelle, C., Uzbekov, R., Ford, C. and Le Guellec, K. (1998) pEg7, a new Xenopus protein required for mitotic chromosome condensation in egg extracts. J. Cell Biol., 143, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M., Dimitrov, S. and Wollfe, A.P. (1994) Nuclear assembly is independent of linker histones. Proc. Natl Acad. Sci. USA, 91, 12477–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S.I. and Wolffe, A.P. (1997) Fine resolution of histones by two-dimensional polyacrylamide gel-electrophoresis: developmental implications. Methods, 12, 57–61. [DOI] [PubMed] [Google Scholar]
- Dimitrov S., Dasso, M.C. and Wolffe, A.P. (1994) Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J. Cell Biol., 126, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P.T., Tsukiyama, T. and Wu, C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J., 16, 4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X.W., Th'ng, J.P.H., Swank, R.A., Anderson, H.J., Tudan, C., Bradbury, E.M. and Roberge, M. (1995) Chromosome condensation induced by fostreicin does not require p34cdc2 kinase activity and histone H1 hyperphosphorylation, but it is associated with enhanced histone H2A and H3 phosphorylation. EMBO J., 14, 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton, A.L., Thorne, A.W. and Crane-Robinson, C. (1994) Core histone acetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne, A.W. and Crane-Robinson, C. (1989) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Laroche, T., Strahl-Bolsinger, S., Gasser, S. and Grunstein, M. (1995) Histone H3 and H4 termini react with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Wei, Y., Mancini, M.A., Van Hooser, A., Ranalli, T., Brinkley, B.R., Bazett-Jones, D.P. and Allis, C.D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma, 106, 348–360. [DOI] [PubMed] [Google Scholar]
- Hirano T. (1998) SMC protein complexes and higher-order chromosome dynamics. Curr. Opin. Cell Biol., 10, 317–322. [DOI] [PubMed] [Google Scholar]
- Hirano T. (1999) SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev., 13, 11–19. [DOI] [PubMed] [Google Scholar]
- Hirano T. and Mitchison, T.J. (1993) Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol., 120, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. and Mitchison, T.J. (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell, 79, 449–458. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi, R. and Hirano, M. (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell, 89, 511–521. [DOI] [PubMed] [Google Scholar]
- Holloway S.L., Glotzer, M., King, R.W. and Murray, A.W. (1993) Anaphase is initiated by proteolysis rather than by the inactivation of maturation promoting factor. Cell, 73, 1393–1402. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B. and Dimitrov, S. (1999) Elasticity measurements show the existence of a thin rigid core inside mitotic chromosomes. J. Cell Biol., 145, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B., Marko,J.F., Chatenay,D. and Libchaber,A. (1997) Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J. Cell Biol., 1391–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung F., Luo, D., Sauve, D.M., Muller, M.T. and Roberge, M. (1996) Characterization of topoisomerase II–DNA interaction and identification of a DNA-binding domain by ultraviolet laser crosslinking. FEBS Lett., 380, 127–132. [DOI] [PubMed] [Google Scholar]
- Kaufman P.D. (1996) Nucleosome assembly: the CAF and the HAT. Curr. Opin. Cell Biol., 8, 369–373. [DOI] [PubMed] [Google Scholar]
- Kimura K. and Hirano, T. (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell, 90, 625–634. [DOI] [PubMed] [Google Scholar]
- Kimura K., Hirano, M., Kobayashi, R. and Hirano, T. (1998) Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science, 282, 487–490. [DOI] [PubMed] [Google Scholar]
- Koshland D. and Strunnikov, A. (1996) Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol., 12, 305–333. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lohka M.J. and Masui, M. (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science, 220, 719–221. [DOI] [PubMed] [Google Scholar]
- Luger K. and Richmond, T.J. (1998) The histone tails of the nucleosome. Curr. Opin. Genet. Dev., 8, 140–146. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader, A.W., Richmond, R.K., Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A.D., Pruss, D.V. and Ebralidze, K.K. (1990) Chromatin superstructure-dependent crosslinking with DNA of the histone H5 residues Thr1, Hist25 and His62. J. Mol. Biol., 211, 479–491. [DOI] [PubMed] [Google Scholar]
- Mutskov V., Gerber, D., Angelov, D., Ausio, J., Workman, J. and Dimitrov, S. (1998) Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol. Cell. Biol., 18, 6293–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi K., Katagiri, C. and Kishimoto, T. (1993) Chromosome condensation in Xenopus mitotic extracts without histone H1. Science, 262, 2033–2035. [DOI] [PubMed] [Google Scholar]
- Philpott A. and Leno, G.H. (1992) Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell, 69, 759–767. [DOI] [PubMed] [Google Scholar]
- Philpott A., Leno, G.H. and Laskey, R.A. (1991) Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell, 65, 569–578. [DOI] [PubMed] [Google Scholar]
- Poljak L. and Käs, E. (1995) Resolving the role of topoisomerase II in chromatin structure and function. Trends Cell Biol., 5, 348–354. [DOI] [PubMed] [Google Scholar]
- Roth S.Y. and Allis, C.D. (1996) Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell, 87, 5–8. [DOI] [PubMed] [Google Scholar]
- Roth S.I., Shimuzu, M., Johnson, L., Grunstein, M. and Simpson, T. (1992) Stable nucleosome positioning and complete repression by the yeast a2 repressors are disrupted by amino terminal mutations in histone H4. Genes Dev., 6, 411–425. [DOI] [PubMed] [Google Scholar]
- Sauvé D.M., Anderson, H.J., Ray, J.M., James, W.M. and Roberge, M. (1999) Phosphorylation-induced rearrangements of the histone H3 NH2-terminal domain during chromosome condensation. J. Cell Biol., 145, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C. and Newport, J.W. (1991) Systems for the study of nuclear assembly, DNA replication and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol., 35, 449–468. [DOI] [PubMed] [Google Scholar]
- van Holde K. (1988) Chromatin. Springer-Verlag KG, Berlin, Germany. [Google Scholar]
- Van Hooser A., Goodrich, D.W., Allis, D., Brinkley, B.R. and Mancini, M.A. (1998) Histone H3 phosphorylation is required for the initiation, but not for maintenance, of mammalian chromosome condensation. J. Cell Sci., 111, 3497–3506. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Pruss, D. and Wolffe, A. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. and Earnshaw, W.C. (1997) Untangling the role of topoisomerase II in mitotic chromosome structure and function. BioEssays, 19, 97–99. [DOI] [PubMed] [Google Scholar]
- Wei Y., Mizzen, C.A., Cook, R.G., Gorovsky, M.A. and Allis, C.D. (1998) Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl Acad. Sci. USA, 95, 7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Lanlan, Y., Bowen, J., Gorovsky, M.A. and Allis, C.D. (1999) Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell, 97, 99–109. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Pruss, D. (1996) Targeted chromatin disruption: transcription regulators that acetylate histones. Cell, 84, 817–819. [DOI] [PubMed] [Google Scholar]