Abstract
Mex67p and Mtr2p constitute an essential mRNA export complex that interacts with poly(A)+ RNA and nuclear pore proteins. We have identified Yra1p, an intranuclear protein with in vitro RNA–RNA annealing activity, which directly binds to Mex67p. The complex between Yra1p and Mex67p was reconstituted in vitro and shown by UV-crosslinking to bind directly to RNA. Mutants of YRA1 are impaired in nuclear poly(A)+ RNA export at restrictive growth conditions. ALY, the mouse homologue of Yra1p and a transcriptional co-activator, can bind in vitro to yeast and human Mex67p and partly complements the otherwise non-viable yra1 null mutant. Thus, Yra1p is the first RNA-binding protein characterized, which bridges the shuttling Mex67p/Mtr2p exporter to intranuclear mRNA transport cargoes.
Keywords: Mex67p/nuclear RNA export/nucleocytoplasmic transport/RNA chaperone/Yra1p
Introduction
Transport across the nuclear membrane occurs through the nuclear pore complexes (NPCs) and involves the coordinated interaction between structural components of the NPCs and shuttling transport receptors (Stoffler et al., 1999). Whereas a mechanistic picture of how proteins are imported into the nucleus or exported to the cytoplasm is emerging, much less is known about nuclear export of mRNA (Sträßer and Hurt, 1999). Although Ran has been implicated in exit of mRNA from the nucleus, a direct role has not yet been shown. Furthermore, a nuclear exportin that binds to pre-mRNA (either directly or indirectly via an hnRNP protein) in the presence of RanGTP has not been found. The role of Crm1p/Xpo1p, the export receptor for proteins containing the short leucine-rich type nuclear export signal (NES), in mRNA export is controversial. Data from yeast showed that Crm1/Xpo1p is involved in mRNA export (Stade et al., 1997), but no such evidence has been obtained from the vertebrate system (Fischer et al., 1995). In addition, it was reported recently that Crm1/Xpo1p does not participate directly in nuclear mRNA export in yeast (Neville and Rosbash, 1999).
Other factors considered to be involved in mRNA export are hnRNP proteins. It is widely accepted that naked pre-mRNAs are not export-competent, but require assembly into hnRNP particles for nuclear exit. Accordingly, mediators of nuclear mRNA export are thought to be shuttling hnRNP proteins, which could contribute in trans via their NES to the mRNA export process (Nakielny et al., 1997). To date, however, a cognate NES receptor for hnRNP proteins has not been found. Other components implicated in nuclear mRNA export have been identified predominantly in yeast. Among these were nucleoporins, components involved in RNA metabolism, importins and exportins (i.e. members of the importin/karyopherin β-family), components of the Ran system, and novel factors of unknown function (for review see Sträßer and Hurt, 1999). Of these latter factors, Gle1p (Murphy and Wente, 1996), Gle2p (Murphy et al., 1996; Bailer et al., 1998), Mex67p (Segref et al., 1997) and Mtr2p (Kadowaki et al., 1994; Santos-Rosa et al., 1998) are currently under investigation, in order to clarify their specific role in nuclear mRNA export. Furthermore, an interesting mRNA export mutant in yeast, Rat8p/Dbp5p, is an ATP-dependent DEAD-box RNA helicase, which is involved in poly(A)+ RNA export and localizes to the outer side of the nuclear envelope (Snay-Hodge et al., 1998; Tseng et al., 1998). Thus, the RNA-unwinding activity of Dbp5p may play a role in the disassembly of mRNP export complexes upon arrival in the cytoplasm.
Since Mex67p associates in vivo with nuclear pores, poly(A)+ RNA and Mtr2p, it may be an essential nuclear mRNA export factor that does not require importin β-like transport factors (Segref et al., 1997; Santos-Rosa et al., 1998). Moreover, TAP, the human homologue of Mex67p, was found to bind directly to the constitutive transport element (CTE) of viral RNA, thereby promoting nuclear RNA export (Grüter et al., 1998; Braun et al., 1999; Kang and Cullen, 1999). TAP was also shown to bind to poly(A)+ RNA in vivo, the FG-repeat domains of the nucleoporins CAN/Nup214 and human Rip, and to p15, which exhibits homology to NTF2. Most importantly, the expression of TAP and p15 in yeast can complement the otherwise lethal mex67/mtr2 double disruption mutant (Katahira et al., 1999). This showed that the Mex67p-mediated nuclear mRNA export pathway is conserved. Whereas the interaction of Mex67p with NPC components is established, its role in intranuclear ‘upstream’ events remains undetermined. We performed a synthetic lethal screen with a mex67 mutant, defective in the leucine rich repeat (LRR) domain. This led us to Yra1p, an essential nuclear protein with RNA–RNA annealing activity in vitro. We show that Yra1p is essential for nuclear export of mRNA. Since Yra1p binds directly to Mex67p, an RNA chaperone may be neccessary to target the newly transcribed pre-mRNAs to the Mex67p-mediated nuclear export pathway.
Results
A novel mex67 mutant allele is genetically linked to Yra1p, the major yeast protein with in vitro RNA–RNA annealing activity
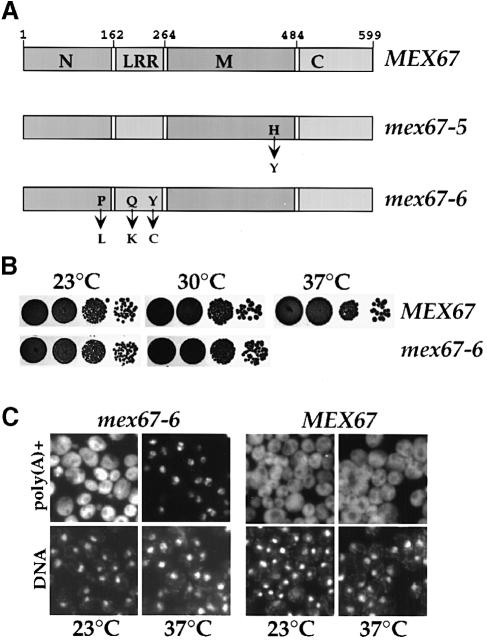
Mex67p exhibits several distinct domains, including an N-terminal (N), LRR, middle (M) and C-terminal (C) domain (Figure 1A; see also Segref et al., 1997; Santos–Rosa et al., 1998). Whereas the M domain binds to Mtr2p, the C–domain is important for nuclear envelope association (Segref et al., 1997; K.Sträßer, unpublished data). To study the role of the N+LRR domain of Mex67p, thermosensitive (ts) mutations in this region of Mex67p were generated. One derived allele, mex67-6, has mutations causing three amino acid changes P(117)→L, Q(199)→K, Y(232)→C (Figure 1A). mex67-6 cells, which grow well at 23 and 30°C but completely arrest at 37°C (Figure 1B), show a strong nuclear mRNA accumulation after being shifted to the non-permissive temperature (Figure 1C).
Fig. 1. A novel mex67-6 ts allele with mutations in the N+LRR domain is defective in nuclear mRNA export. (A) Domain organization of Mex67p and amino acid mutations within the mex67-5 (Segref et al., 1997) and mex67-6 allele. The Mex67p protein consists of an N–terminal (N), LRR, middle (M) and C–terminal (C) domain with the indicated amino acid boundaries. (B) Growth of the mex67-6 ts mutant. Equivalent numbers of cells were diluted in 10–1 steps, spotted onto YPD plates, and grown for 3 days at the temperatures indicated. (C) Nuclear accumulation of polyadenylated RNA in mex67-6 cells. Subcellular localization of poly(A)+ RNA was analyzed by in situ hybridization. MEX67 and mex67-6 cells were either grown at 23°C or shifted for 1 h to 37°C. Nuclear DNA was stained with 4′-6-diamidine-2-phenylindole (DAPI).
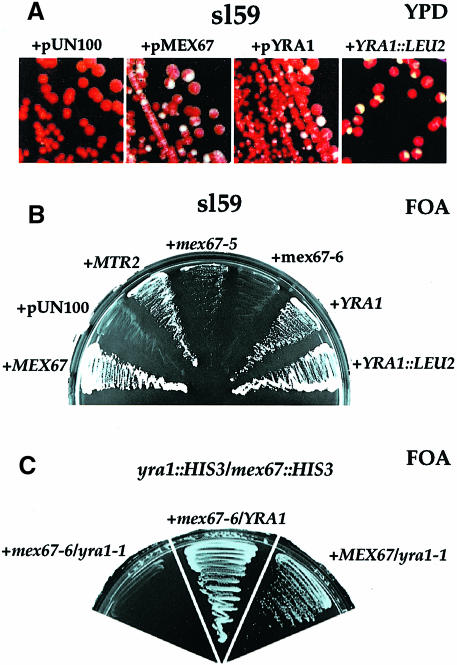
To identify components that are genetically linked to the N+LRR domain, the mex67-6 allele was exploited in a synthetic lethal (sl) screen, based on a red/white colony sectoring assay (Wimmer et al., 1992; see Materials and methods). One mutant, sl59, was obtained, which forms red colonies on yeast extract/peptone/dextrose (YPD) and cannot grow on 5′-fluoroorotic acid (5–FOA) plates (Figure 2A and B). The wild-type gene causing synthetic lethality with mex67-6 in sl59 was cloned by complementation and shown to be the essential yeast gene YRA1 described previously (Figure 2A and B). Yra1p was identified as the major yeast protein with RNA–RNA annealing activity (Portman et al., 1997). Its overexpression exhibits a G1 arrest with unbudded cells (Espinet et al., 1995). Yra1p is evolutionarily conserved and has homologues in Schizosaccharomyces pombe (L42551), Caenorhabditis elegans (CAB05180), mouse (U89876) and human (AF047002) (see later).
Fig. 2. Identification of YRA1, which functionally interacts with the N+LRR domain of MEX67. (A) The synthetically lethal mutant sl59 is complemented by YRA1. Red/white colony sectoring is restored in the mex67-6 synthetically lethal strain sl59 by transformation with plasmid-borne MEX67 (+pMEX67) or YRA1 (+pYRA1), but not with an empty plasmid (+pUN100). sl59 is also complemented by homologous integration of the cloned YRA1 gene (+YRA1::LEU2). (B) The sl59 mutant is also complemented by MTR2 and mex67-5. sl59 carrying plasmids pURA3-MEX67 and pTRP1-mex67-6 is unable to grow in the presence of 5–FOA, unless complemented either by transformation with wild-type MEX67, YRA1, or by integration of wild-type YRA1 into the yra1 locus of sl59. sl59 was also transformed with empty pUN100, pRS315-MTR2, pRS315-mex67-6 or pUN100-mex67-5. Transformants were streaked onto 5–FOA containing plates and grown for 3 days at 30°C. (C) Synthetic lethal interaction between mex67-6 and yra1-1 alleles. The yra1::HIS3/mex67::HIS3 shuffle strain (Table I) was transformed with plasmids giving the gene combinations mex67-6/yra1-1, mex67-6/YRA1 and MEX67/yra1-1. A synthetic lethal relationship was tested by streaking transformants on 5–FOA–containing plates.
To exclude the possibility that YRA1 was an extragenic low copy suppressor of sl59, as found for MTR2 (Figure 2B), the isolated YRA1 gene was used to replace the chromosomal YRA1 gene within sl59 by homologous recombination. Correctly integrated YRA1::LEU2 sl59 cells were no longer synthetically lethal (Figure 2A and B). This is genetic evidence that a mutated YRA1 gene locus in sl59 is responsible for the synthetic lethality in combination with the mex67-6 allele. To study how the different domains of Mex67p interact with Yra1p, synthetic lethal interactions were tested between yra1 and mex67 mutant alleles. One allele of MEX67, mex67-5, contains a single point mutation in the M domain, which leads to a weakened interaction with Mtr2p, a ts phenotype, and accumulation of poly(A)+ RNA at the restrictive temperature (Segref et al., 1997; Santos-Rosa et al., 1998). Interestingly, when transformed with this mex67-5 allele, sl59 was no longer synthetically lethal, although the mex67-5/yra1-59 combination causes a slow growing phenotype (Figure 2B). This points to an allele-specific synthetic lethal interaction between mutant yra1 and the mex67-6 allele. Taken together, these data suggest that the N+LRR domain, but not the M domain of Mex67p, is functionally linked to Yra1p (see below).
Yra1p is an intranuclear protein, which is not associated with nuclear pores
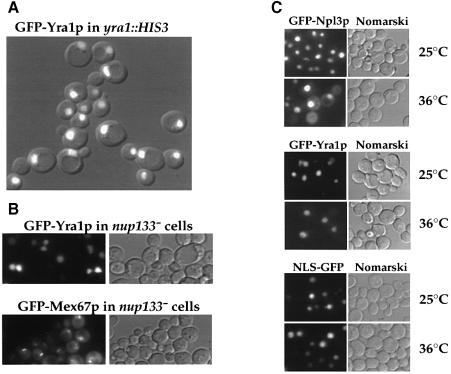
As shown by indirect immunofluorescence, Yra1p is a nuclear protein (Portman et al., 1997). In order to determine whether Yra1p is a nuclear resident or shuttling protein, the location of GFP-tagged Yra1p was analyzed in living cells. GFP–Yra1p, which is able to complement the otherwise non-viable yra1::HIS3 disruption mutant (data not shown), exhibits an exclusively intranuclear location (both in the nucleoplasm and nucleolus), with no indication of cytoplasmic or nuclear envelope staining (Figure 3A). To find out whether a minor pool of Yra1p associates with nuclear pores, the GFP–Yra1p location was determined in nup133– cells, in which nuclear pores and NPC-associated proteins cluster in one or several spots (Doye et al., 1994). Whereas GFP–Mex67p co-localized with clustered NPCs, GFP–Yra1p remained nuclear without the slightest indication of an NPC association in nup133– cells (Figure 3B). Thus, the major pools of Yra1p and Mex67p are found in different nuclear compartments (see Discussion).
Fig. 3. Yra1p is an intranuclear protein. (A) Fluorescence microscopy of yra1::HIS3 cells expressing GFP–Yra1p. Fluorescence microscopy and Nomarski pictures were merged. (B) Fluorescence microscopy of GFP–Yra1p and GFP–Mex67p, respectively, in nup133– cells. (C) GFP–Yra1p does not shuttle between the nucleus and the cytoplasm. The reporter constructs pGAL1::GFP–Npl3, pGAL::GFP–Yra1p, and NLS–GFP were expressed in the nup49-313 strain. Expression was stopped by shifting the cells to glucose-containing medium (not in the case of NLS–GFP, which has a constitutive promoter). Strains were further incubated at 23°C (permissive temperature) or shifted for 5 h to 37°C (restrictive temperature), before the intracellular location of GFP fusion proteins was analyzed by fluorescence microscopy. In all cases, the same exposure time was used.
Although Yra1p is a nuclear protein, it might shuttle between the nucleus and cytoplasm. To test this possibility, we utilized a recently described shuttling assay (Lee et al., 1996). This assay exploits a certain nup49 mutant (nup49-313), which is defective in protein import, but not in protein export at the restrictive temperature (Doye et al., 1994). The shuttling hnRNP-like protein Npl3p served as a positive control and a resident nuclear protein, NLS–GFP reporter protein, served as negative control. All three fusion proteins, GFP–Yra1p, GFP–Npl3p (both under the control of the GAL1 promotor) and NLS–GFP exhibit an exclusively intranuclear localization when expressed in the nup49-313 mutant under permissive conditions (Figure 3C). After repression of GFP–YRA1 and GFP–NPL3 expression and subsequent shift of nup49-313 cells to the restrictive temperature only GFP–Npl3p is exported to the cytoplasm, indicated by the cytoplasmic GFP signal (Figure 3C). In contrast, GFP–Yra1p remained nuclear with no significant GFP signal in the cytoplasm (Figure 3C). However, when we tested Yra1p shuttling in a heterokaryon assay in yeast (Flach et al., 1994), we could find preliminary evidence that Yra1p may shuttle from one nucleus to the other (K.Sträßer, unpublished data; also see Discussion).
YRA1 mutants accumulate poly(A)+ RNA inside the nucleus
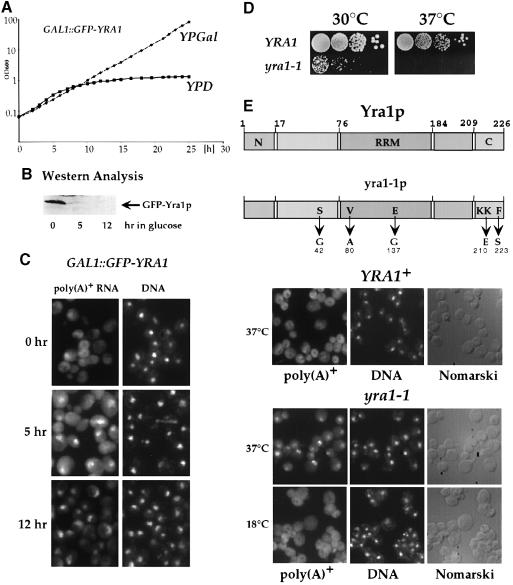
The genetic link between YRA1 and MEX67 suggests a role for Yra1p in nuclear mRNA export. To show this directly, both Yra1p depletion and ts mutants were analyzed for an impaired nuclear mRNA export. A deletion strain was constructed that harbors GAL1::GFP–YRA1. This strain is viable in galactose-containing medium, but stops growing 8–10 h after transfer to glucose-containing medium (Figure 4A), due to depletion of GFP–Yra1p (Figure 4B). Five hours after shifting to glucose-containing medium, many of the Yra1p-depleted cells accumulated poly(A)+ RNA inside the nucleus. After 12 h of depletion, almost all of the cells were blocked in mRNA export, with little residual poly(A)+ RNA signal in the cytoplasm (Figure 4C).
Fig. 4. YRA1 is essential for nuclear poly(A)+ RNA export. (A) Growth curve of the GAL1::GFP–YRA1 strain in galactose (YPGal) and glucose-containing medium (YPD) by measuring optical density at 600 nm. (B) Repression of GFP–Yra1p expression as determined by Western blot analysis using anti-GFP antibodies. The same amount of whole-cell extract derived from strain GAL1::GFP–YRA1 was analyzed after 0, 5 and 12 h in glucose-containing medium. (C) Subcellular localization of poly(A)+ RNA in GAL1::GFP–YRA1 cells was analyzed by in situ hybridization with a fluorescently labeled poly(dT) probe. The strain was grown in galactose-containing medium prior to transfer to glucose-containing medium for 5 and 12 h. Nuclear DNA was stained with DAPI. (D) Growth of the yra1-1 ts mutant. The same amount of yra1-1 and YRA1 cells, respectively, were diluted in 10–1 steps, spotted onto YPD plates, and grown for 3 days at 30 or 37°C. (E) Nuclear accumulation of poly(A)+ RNA in yra1-1 cells, as determined by in situ hybridization. yra1-1 cells were either grown at 18°C or shifted for 1 h to 37°C in YPD medium. Nuclear DNA was stained with DAPI, and Nomarski pictures were taken. A schematic representation of the yra1-1 allele with its five mutations in the context of the Yra1p domain organization (see also Figure 7) is also shown.
Since accumulation of poly(A)+RNA caused by depletion of Yra1p takes several hours, we sought to generate temperature-sensitive YRA1 mutants with a faster onset of the mRNA export defect. The YRA1 gene was mutagenized in vitro and three yra1 ts mutants were obtained. One of the three, yra1-1, was further analyzed (Figure 4D and E). The yra1-1 allele harbors five amino acid substitutions, where the two mutations in the C–terminal end mainly contribute to the ts phenotype (data not shown). Since the yra1-1 strain is already impaired in growth at 30°C (Figure 4D) and 23°C, we analyzed nuclear mRNA export at 18°C (permissive temperature) and 37°C (restrictive temperature). In yra1-1 cells grown at 18°C, a predominantly cytoplasmic poly(A)+ RNA signal is seen (Figure 4E). Strikingly, already 1 h after shifting to 37°C, all of the yra1-1 cells accumulate mRNA within the nucleus (Figure 4E). Accordingly, the short and conserved C–terminal domain of Yra1p fulfils a crucial function in nuclear mRNA export. These results are consistent with the finding that the yra1-1 allele when combined with the mex67-6 allele exhibits synthetic lethality (Figure 2C).
Yra1p directly associates with Mex67p
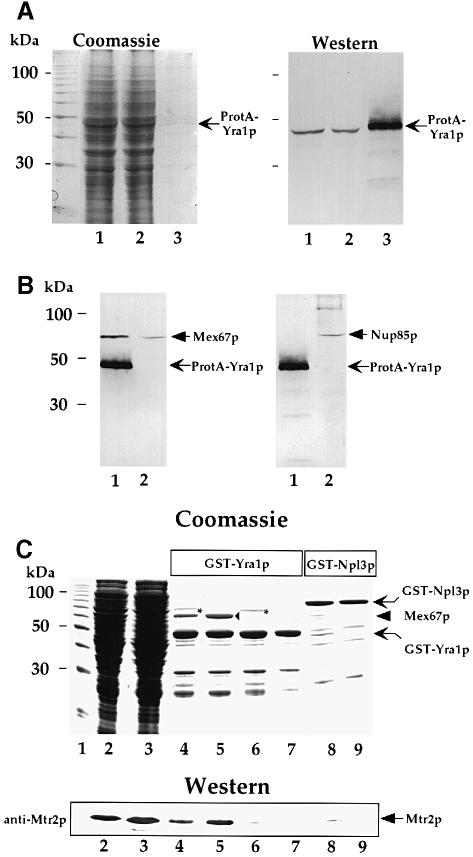
Although the bulk of Yra1p and Mex67p is located in different nuclear compartments (see above), a certain fraction of both proteins may come in direct contact. Therefore, we purified Yra1p from yeast and looked for co-purification of Mex67p. To facilitate isolation, Yra1p was tagged with protein A (ProtA) and expressed in the yra1::HIS3 null mutant. ProtA–Yra1p is functional and complements the non-viable yra1– strain (data not shown). Although ProtA–Yra1p is less efficiently affinity-purified than other ProtA fusion proteins, a faint Coomassie Blue-stainable band of the expected size can be seen in the SDS–polyacrylamide gel (Figure 5A, lane 3, Coomassie), which corresponds to the enriched ProtA–Yra1p fusion protein identified by immunoblotting (Figure 5A, lane 3, Western). In addition, Western blot analysis further indicates that Mex67p is present in the purified ProtA–Yra1p eluate (Figure 5B). When the same ProtA–Yra1p eluate was probed with anti-Nup85p antibodies, this nucleoporin was completely absent (Figure 5B). Thus, a pool of Mex67p can be affinity-purified with ProtA–Yra1p, but it remained uncertain whether this interaction is direct.
Fig. 5. Yra1p associates with Mex67p both in vivo and in vitro. (A) Affinity purification of ProtA–Yra1p. The whole-cell lysate (7.5 μl) (lane 1), the unbound fraction (lane 2) and the eluate from the IgG–Sepharose beads (lane 3) were analyzed by SDS–PAGE followed by Coomassie Blue staining, or Western blotting using anti-ProtA antibodies. (B) Western blot analysis of purified Yra1–ProtA (lane 1) and a whole cell lysate from strain RS453 (lane 2) using anti-Mex67p and anti-Nup85p antibodies. (C) In vitro binding of Mex67p to GST-tagged Yra1p. Bead-immobilized GST–Yra1p or GST–Npl3p purified from S.pombe was incubated with E.coli whole-cell lysates containing Mex67p/Mtr2p or Mtr2p or buffer. Bound proteins were analyzed by SDS–PAGE, followed by Coomassie Blue staining or Western analysis using anti-Mtr2p antibodies (lower panel). The positions of GST–Yra1p and GST–Npl3p, Mex67p (filled triangle), and an E.coli contaminating band (asterisk) are indicated. Lane 1, protein standard; lane 2, E.coli lysate with Mex67p/His6-Mtr2p complex; lane 3, E.coli lysate with His6-Mtr2p; lane 4, GST–Yra1p incubated with Mex67p/His6-Mtr2p lysate (–RNase); lane 5, GST–Yra1p incubated with Mex67p/His6-Mtr2p lysate (+RNase); lane 6, GST–Yra1p incubated with His6-Mtr2p lysate; lane 7, GST–Yra1p incubated with buffer; lane 8, GST–Npl3p incubated Mex67p/His6-Mtr2p lysate; lane 9, GST–Npl3p incubated with buffer.
To assess this experimentally, in vitro pull-down experiments were performed (Figure 5C). When recombinant glutathione S-transferase (GST)–Yra1p immobilized on glutathione (GSH) beads was incubated with a bacterial extract containing recombinant Mex67p (in the form of the Mex67p/His6-Mtr2p complex), nearly stoichiometric amounts of Mex67p were bound to GST–Yra1p, whereas most of the Escherichia coli proteins did not bind (Figure 5C, compare lanes 2, 4 and 7). Mtr2p could only be detected in the bound fraction by immunoblotting, since His6-Mtr2p comigrated at 25 kDa with one of the major GST–Yra1p breakdown products (Figure 5C, Western). An even greater extent of Mex67p binding (indicated by a closed triangle) to GST–Yra1p was seen when the E.coli lysate was treated with RNase prior to the binding assay, showing that this interaction is not dependent on RNA (Figure 5C, lane 5). On the other hand, RNase treatment caused the disappearance of the E.coli contaminant migrating above the Mex67p band (Figure 5C, compare lanes 4 and 5, asterisk). When an E.coli lysate that contains only His6-Mtr2p was incubated with GST–Yra1p beads, no 67 kDa band associated with GST–Yra1p appeared, proving that no protein other than Mex67p corresponds to the strong Coomassie Blue-stainable band at this position. Interestingly, His6-Mtr2p alone did not bind to GST–Yra1p above background levels (Figure 5C, lane 6; Coomassie and Western). Because Mex67p is largely insoluble in E.coli in the absence of Mtr2p (Santos-Rosa et al., 1998), we could not test whether Mex67p alone binds to GST–Yra1p. Npl3p, an hnRNP-like protein, was used as a negative control. Only background levels of binding were observed between Mex67p/Mtr2p to beads containing immobilized GST–Npl3p (Figure 5C, compare lanes 8 and 9). All this reveals that Yra1p specifically binds to Mex67p.
The reconstituted Yra1p–Mex67p complex can be UV-crosslinked to RNA
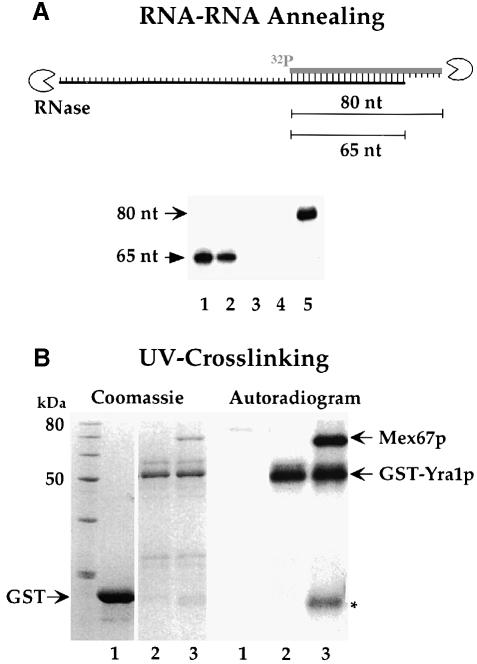
Recombinant GST–Yra1p purified from S.pombe was tested for RNA–RNA annealing activity as described previously (Portman et al., 1997). Two complementary RNA species, which are derived from the SSA1 gene and overlap for a length of 65 nucleotides, were in vitro transcribed. Only the short RNA was radiolabeled with 32P, so we could test Yra1p-dependent annealing of the two complementary RNAs through protection of double stranded RNA from RNAse T1 digestion (Figure 6A). When both RNAs were mixed and incubated at physiological temperatures in the absence of Yra1p, no significant annealing was found (Figure 6A, lane 4). Similarly, when cold RNA was omitted and only radiolabeled RNA was incubated with Yra1p, no RNAse T1 protection was seen (Figure 6A, lane 3). However, when both RNAs were incubated with increasing amounts of Yra1p, the expected 65 nucleotide long RNA–RNA hybrid was protected from the RNAse T1 attack (Figure 6A, lanes 1 and 2). This shows that recombinant Yra1p expressed in S.pombe has RNA–RNA annealing activity, consistent with earlier findings using Yra1p expressed in E.coli (Portman et al., 1997). Although Yra1p was shown to be the major yeast RNA annealing protein, it did not UV-crosslink to pre-mRNA in vivo (Portman et al., 1997). Therefore, we tested the property of purified Yra1p to associate with RNA in vitro using radiolabeled SSA1 mRNA (nucleotides 818–1152). We recently showed that SSA1 mRNA exits the nucleus via the Mex67p-dependent nuclear export pathway (Hurt et al., 2000). When the purified GST–Yra1p immobilized on GSH-beads was incubated with 32P-labeled SSA1 RNA and UV-irradiated, GST–Yra1p was crosslinked to RNA (Figure 6B, lane 2). There was no radiolabeled RNA associated with GST–Yra1p, when UV irradiation was omitted (data not shown). Furthermore, GST alone was not crosslinked to RNA to any extent (Figure 6B, lane 1). This shows that Yra1p can bind to RNA. Since Mex67p interacts with RNA both in vivo and in vitro (Segref et al., 1997; Santos-Rosa et al., 1998), we wanted to test whether complex formation between Yra1p and Mex67p influences the ability of Mex67p to bind to RNA. Accordingly, the GST–Yra1p beads were pre-incubated with the recombinant purified Mex67p/Mtr2p complex to reconstitute the ternary complex (see above) before radiolabeled RNA was added and UV-crosslinking performed. Under these conditions, both GST–Yra1p and Mex67p were efficiently crosslinked to radiolabeled RNA (Figure 6B, lane 3). A weak signal can be seen in the autoradiogram at the approximate size of Mtr2p. We showed previously that Mtr2p can be crosslinked to RNA, although weakly compared to Mex67p (Santos-Rosa et al., 1998). Since this signal was sometimes also observed for GST–Yra1p alone (data not shown), it could also be a crosslinked GST–Yra1p breakdown product. In conclusion, Yra1p and Mex67p retain RNA-binding activity when organized in a complex and can be efficiently UV-crosslinked to RNA.
Fig. 6. Yra1p has RNA–RNA annealing activity and can be UV-crosslinked together with Mex67p to RNA. (A) RNA–RNA annealing assay. In vitro transcribed SSA1 sense (32P-labeled) and anti-sense (non-labeled) RNAs were incubated with or without recombinant, purified Yra1p. RNA–RNA hybrid formation was analyzed by digestion of single-stranded RNA with RNase T1. Double-stranded and thus protected RNA fragments were separated on a 10% polyacrylamide/8 M urea gel, before autoradiography was performed. Lane 1, ∼40 ng Yra1p with both 32P-labeled and unlabeled RNA; lane 2, ∼20 ng Yra1p with both 32P-labeled and unlabeled RNA; lane 3, ∼40 ng Yra1p with only 32P-labeled RNA; lane 4, no Yra1p, but both 32P-labeled and unlabeled RNA; lane 5, untreated sense 32P–labeled RNA. (B) UV-crosslinking of RNA to Yra1p and Mex67p. 32P-labeled SSA1 RNA (bp 818–1152) was incubated with GST, GST–Yra1p on beads, and GST–Yra1p bound to Mex67p/Mtr2p on beads. After UV-irradiation, the non-crosslinked RNA was digested with RNase A and RNase T1, and proteins were analyzed by SDS–PAGE, followed by Coomassie Blue staining and autoradiography. Lane 1, GST; lane 2, GST–Yra1p bound to beads; lane 3, GST–Yra1p with bound recombinant Mex67p/Mtr2p complex on beads.
Mouse ALY is homologous to yeast Yra1p
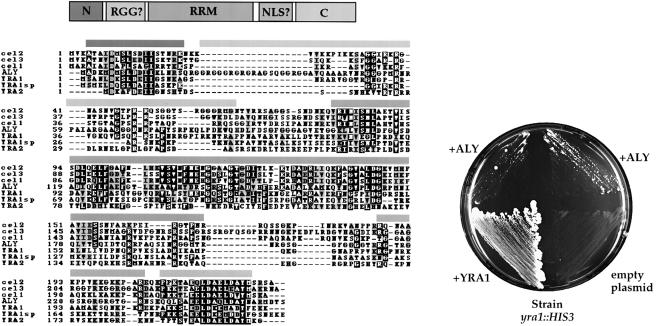
Yra1p is evolutionarily conserved and has homologues in different organisms including mouse (Figure 7, upper panel). Common to all of the Yra1p homologues is their relatively small size (∼25 kDa), strongly conserved N– and C–terminal ends, and a central RNA-binding domain with RNPI and RNPII consensus motifs (Figure 7, upper panel). Between the conserved N–peptide and the RNA-binding domain is a less conserved sequence of variable length, which tends to be rich in arginines and glycines, and in the case of ALY exhibits a RGG box pattern. Between the RNA-binding domain and the conserved C–domain lies a sequence that resembles a bipartite NLS (e.g. in Yra1p).
Fig. 7. Mouse ALY complements the yra1– mutant. Upper panel: Schematic representation of the Yra1p/ALY domain organization, which includes a short N-terminal domain (N), a RGG box (only in ALY, but not Yra1p), and RNA-binding domain (RRM), a putative NLS and a C-terminal (C) domain; the different YRA1 homologues (including ALY) are aligned. Lower panel: restoration of growth of the yra1::HIS3 strain by expression of mouse ALY. The YRA1 shuffle strain (Table I) was transformed with 2μ plasmids pNOPA2A-ALY (ALY), pNOPA2A empty (empty plasmid), or ARS–CEN plasmid pNOPGFPA-YRA1 (YRA1). In the case of ALY, two individual transformants were first grown on selective SDC –ade plates, and then restreaked on 5-FOA. On this plate, only cells that have lost the pURA3-YRA1 plasmid, and thus are yra1::HIS3 can grow. When ALY is expressed, small colonies became visible after 3 days incubation at 30°C. Plates were incubated for 13 days, prior to being photographed.
The mouse homologue of Yra1p, ALY (Figure 7, left panel), is a ubiquitously expressed nuclear protein, which acts as a transcriptional coactivator. It binds to the transcription factors LEF–1 and AML–1, which are required for T–cell receptor α enhancer regulation (Bruhn et al., 1997). To test whether mouse ALY can functionally replace Yra1p in yeast, the ALY cDNA was placed under the control of the yeast NOP1 promoter, inserted into a 2μ ADE2 plasmid, and transformed into the YRA1 shuffle strain (yra1::HIS3, pURA3-YRA1). When transformants were incubated on 5–FOA-containing plates (to select for colonies that lost the pURA3-YRA1 plasmid), slow growing colonies formed after several days (Figure 7, lower panel). In contrast, the shuffle strain carrying an empty ADE2 plasmid did not grow on 5–FOA-containing plates (Figure 7, lower panel). This shows that ALY can restore growth of the otherwise non-viable yra1– strain. However, this complementation is inefficient, since the colonies growing on 5–FOA plates remained small compared to YRA1+ colonies.
Finally, we tested whether GST–ALY can interact with Mex67p and its human homologue TAP in vitro. Similar to GST–Yra1p, GST–ALY was also able to bind to recombinant Mex67p–Mt2p complex and human TAP (data not shown).
These data show that mouse ALY and yeast Yra1p exhibit functional similarities (see Discussion).
Discussion
The mechanism of Mex67p-mediated mRNA export from the nucleus to the cytoplasm is still unknown. To date, it is uncertain whether in vivo Mex67p binds to mRNA with the help of co-factors. Because we could not find an interaction between Mex67p and the major yeast hnRNP-like protein Npl3p, which is also implicated in nuclear mRNA export, we searched genetically for putative ‘upstream’ factors of Mex67p. For this, we exploited the mex67-6 ts allele, which maps to the N+LRR domain. Our rationale was that the leucine-rich region (LRR) of Mex67p might mediate such an interaction. Indeed, mex67-6 led us to YRA1, which encodes the major yeast RNA–RNA annealing protein (Portman et al., 1997).
One clue as to how Mex67p is coupled to Yra1p came from biochemical analysis. Both in vivo and in vitro data show that Mex67p physically associates with Yra1p. In vivo, only a pool of Mex67p is bound to Yra1p, which is consistent with the finding that the bulk of Yra1p is intranuclear, whereas Mex67p is associated with the nuclear envelope. However, these are steady-state distributions, which do not permit conclusions about the in vivo dynamics. Mex67p very likely shuttles, since Tap, the functional human homologue of Mex67p, shuttles between the nucleus and cytoplasm (Kang and Cullen, 1999; Katahira et al., 1999). In addition, Mex67p was detected by immuno-electron microscopy inside the nucleus as well as in the cytoplasm (Santos-Rosa et al., 1998). The final proof that Yrap1 can directly associate with Mex67p came from in vitro reconstitution experiments, which revealed that Yra1p is a Mex67p-binding protein. GST–Yra1p specifically ‘fishes out’ recombinant Mex67p present in an E.coli lysate. Thus, Mex67p and Yra1p can form a stoichiometric complex. The subnuclear compartments in which this complex assembles and dissociates, respectively, remains to be shown (see below).
Since Yra1p most likely binds to intranuclear mRNA transcripts, it is possible that Mex67p, which was shown to bind to poly(A)+ RNA (Segref et al., 1997), gains contact to pre-mRNA through physical interaction with Yra1p. Thus, targeting of Mex67p to PolII transcripts could be mediated by Yra1p. Interestingly, a mouse homologue of Yra1p, ALY, associates with the activation domains of transcription factors LEF–1 and AML–1, which are required for T–cell receptor α enhancer regulation (Bruhn et al., 1997). Assuming that ALY not only plays a role in transcription initiation, but also in subsequent pre-mRNA assembly, ALY could in principle recruit mRNA export factors such as Tap/Mex67p to transcriptionally active genes. Interestingly, the mouse ALY when expressed in yeast partly restores growth of the otherwise non-viable yra1::HIS3 null mutant, showing that at least part of the Yra1p/ALY function is conserved. Future work will address the question of whether transcription initiation is coupled to nuclear export of pre-mRNA. Interestingly, recombinant GST-tagged ALY can also bind to human TAP and yeast Mex67p (K.Sträßer and E.Hurt, unpublished results).
Mex67p can bind to RNA in vitro (Santos-Rosa et al., 1998). Tap, the human homologue of Mex67p, binds directly and with high affinity to a viral RNA CTE, but less efficiently to mRNA (Grüter et al., 1998). The CTE consists of two double-stranded stems and single-stranded loops, but already a single loop and an adjacent bulge are sufficient for the nuclear export of CTE–RNA and interaction with TAP (Grüter et al., 1998; Braun et al., 1999). Taking these findings into account, it is possible that the RNA–RNA annealing activity of Yra1p found in vitro generates CTE-like RNA secondary structures within transcripts, thus facilitating binding of Mex67p–Tap to pre-mRNA. However, the in vitro RNA–RNA annealing activity, typically found for many hnRNP proteins (Portman and Dreyfuss, 1994), may be fortuitous and not related to the in vivo function of these RNA binding proteins. It is therefore also conceivable that Yra1p and Tap/Mex67p may bind cooperatively to pre-mRNA to form export-competent mRNP particles. The stage at which Yra1p detaches from nuclear pre-mRNA during nuclear export is not known. Since we cannot say with certainty whether or not Yra1p shuttles between the nucleus and cytoplasm, Yra1p may either dissociate from Mex67p and mRNA at an intranuclear stage or after export to the cytoplasm.
In conclusion, we have identified Yra1p as a nuclear RNA-binding protein, which is essential for nuclear mRNA export. Strikingly, this RNA-binding protein physically contacts the mRNA export factor Mex67p. This points to a direct link between the intranuclear pre-mRNA assembly and nuclear export machinery.
Materials and methods
Yeast strains, DNA recombinant work and microbiological techniques
The strains used in this study are listed in Table I. Microbiological techniques, plasmid transformation, plasmid recovery, gene disruption, mating, sporulation of diploids and tetrad analysis were done essentially as described (Santos-Rosa et al., 1998). DNA recombinant work such as restriction analysis, end-filling, ligations and PCR amplifications was performed according to Maniatis et al. (1982).
Table I. Yeast strains.
| RS453 | MATa/α,ade2/ade2,his3/his3,leu2/leu2,trp1/trp1,ura3/ura3 |
| MEX67 shuffle | MATa,ade2,his3,leu2,trp1,ura3,mex67::HIS3 (pRS316-MEX67) |
| mex67-6 | MATa,ade2,his3,leu2,trp1,ura3,mex67::HIS3 (pRS314-mex67-6) |
| MEX67/mex67-6 | MATα,ade2,ade3,his3,leu2,trp1,ura3,mex67::HIS3 (pHT4467-MEX67, pRS314-mex67-6) |
| sl59 | MATα,ade2,ade3,his3,leu2,trp1,ura3,mex67::HIS3,yra1-59 (pHT4467-MEX67,pRS314-mex67-6) |
| GFP–MEX67 | MATa,ade2,his3,leu2,trp1,ura3,mex67::HIS3 (pRS315-NOP1::GFP–MEX67) |
| YRA1 shuffle | MATa,ade2,his3,leu2,trp1,ura3,yra1::HIS3 (pRS316-YRA1) |
| yra1-1 | MATa,ade2,his3,leu2,trp1,ura3,yra1::HIS3 (pRS314-yra1-1) |
| GFP–YRA1 | MATa,ade2,his3,leu2,trp1,ura3,yra1::HIS3 (pRS315-NOP1::GFP–YRA1) |
| GAL1::YRA1 | MATa,ade2,his3,leu2,trp1,ura3,yra1::HIS3 (pGAL1::GFP–YRA1) |
| ProtA–TEV-YRA1 | MATa,ade2,his3,leu2,trp1,ura3,yra1::HIS3 (pUN100-NOP1::ProtA–TEV-YRA1) |
| MEX67/YRA1 shuffle | MATa,ade2,his3,leu2,trp1,ura3,mex67::HIS3,yra1::HIS3 (pRS316-YRA1, pRS316-MEX67) |
| nup133– | MATa,ade2,his3,leu2,trp1,ura3,nup133::TRP1 |
| nup49-313 | MATa,ade2,his3,leu2,trp1,ura3,nup49::TRP1 (pUN100-nup49-313) |
Plasmids
Plasmids pRS314-MEX67, pUN100-MEX67, pUN100-mex67-5, pRS315-MTR2, and pHT4467-MEX67 were described previously (Segref et al., 1997; Santos-Rosa et al., 1998). An NheI restriction site was generated by site-directed PCR mutagenesis before the start codon of MEX67 (pRS314-NheI-MEX67). This site was convenient for generation of ts mutants within the N+LLR domain (see below). pRS315-mex67-6 was derived from pRS314-mex67-6 (see below) by subcloning the mex67-6 open reading frame (ORF) containing a SacI fragment into SacI-cut pRS315. For the UV-crosslinking experiments, pBluescriptIIKS(+)-SSA1 (bp 818–1152) was generated by amplifying the corresponding SSA1 ORF DNA by PCR, creating a 5′ SacI and 3′ XbaI site. This DNA was inserted into pBluescriptIIKS(+), cut with the same enzymes. pBluescriptIIKS(+)-SSA1 (bp 1131–1461), which was used for the RNA–RNA annealing assay, was constructed in a similar manner.
For expression of ALY in yeast under the control of the NOP1 promoter, the coding region of ALY was cloned into the high copy yeast expression vector pNOPA2A (Katahira et al., 1999), yielding pNOPA2A-ALY. The ORF of ALY was amplified by PCR creating a 5′ BamHI and 3′ XmaI site. The PCR product was cloned into pNOPA2A, cut with the same restriction enzymes. The YRA1 shuffle strain was transformed with pNOPA2A-ALY, using the ADE2 selectable marker, and transformants were streaked onto 5–FOA-containing plates.
Isolation of the mex67-6 ts allele
Ts mutants of MEX67 were generated by mutagenic PCR and gap repair as described (Santos-Rosa et al., 1998). In detail, a DNA fragment of MEX67 encompassing the sequence between 247 bp 5′ of the start codon and 200 bp 3′ of the BglII site in the LRR domain was amplified by PCR under suboptimal conditions [5 mM MgCl2, 0.5 mM MnCl2, dGTP, dCTP, dTTP (1 mM each), 0.2 mM dATP, 1 μg template DNA, 5 U Taq DNA polymerase]. Vector pRS314-NheI-MEX67 was cut with NheI and BglII, releasing the sequence coding for the N and LRR domain. The mutagenized PCR product and the cut vector were co-transformed into the MEX67 shuffle strain. Approximately 1000 TRP+ transformants were restreaked on 5–FOA plates to shuffle out pRS316-MEX67. Ura– colonies were re-streaked onto YPD and incubated at 23 and 37°C. pRS314-mex67* plasmids were recovered from ts strains, retransformed into the shuffle strain, in order to confirm the plasmid-dependent ts phenotype, and sequenced. A total of seven temperature-sensitive mex67 alleles was isolated.
Synthetic lethal screen with the mex67-6 allele
The synthetic lethal screen was performed as described earlier (Wimmer et al., 1992; Segref et al., 1997). The used screening strain (see Santos–Rosa et al., 1998), however, harboured plasmid pRS314-mex67-6, instead of pRS314-mex67-5. Approximately 15 000 colonies, which survived the UV-mutagenesis, were screened for a red, non-sectoring phenotype and death on 5–FOA. Candidate sl mutants were transformed with pUN100-MEX67, pRS315-mex67-6 or pRS315-MTR2, and checked for restoration of red/white colony sectoring and growth on 5–FOA. One candidate, sl59, fulfilled the requirements to be sl with mex67-6. sl59 was transformed with a pUN100-LEU2 based yeast genomic library, and a complementing pUN100 plasmid with a 10 kb genomic insert containing the YRA1 gene as well as four other ORFs was recovered. Deletion of the sequences coding for the other four ORFs gave pUN100-YRA1, which complemented sl59.
YRA1 gene constructions
The yra1::HIS3 null construct was made by PCR amplification of ∼400 bp 5′ and 3′ of the YRA1 gene, which were attached via BamHI sites to the 5′ and 3′ sites, respectively, of the HIS3 gene (obtained from YDp-H) and inserted into pBluescriptIIKS(+). pRS316-YRA1 was constructed by subcloning a YRA1 containing SacI–HindIII fragment from pUN100-YRA1 (see above) into pRS316. pRS314-YRA1 was subcloned from pRS316-YRA1 by inserting the YRA1 containing SacI–XhoI fragment of pRS316-YRA1 into pRS314.
For integration of the authentic YRA1 at the presumptive mutant yra1 gene locus in sl59, the LEU2 gene, isolated as a BamHI fragment from plasmid YDp-L, was inserted into a SnaBI site, which is ∼300 bp upstream of the YRA1 ORF. From this construct, a SacI–HindIII fragment was isolated, which contains another 1.2 kb upstream of the YRA1 gene and 1.2 kb downstream of the YRA1 gene. This 5.7 kb DNA construct was cloned into pBluescriptIIKS(+). The YRA1::LEU2-containing fragment was released by digestion with SacI and HindIII and transformed into sl59. LEU+ transformants, which carried YRA1::LEU2 at the YRA1 locus (as verified by PCR Southern), were analyzed for a red/white sectoring phenotype and growth on 5–FOA.
In order to generate ts alleles of YRA1, the sequence between the start codon and ∼400 bp downstream of YRA1 was amplified by PCR under suboptimal conditions (see above). This PCR product was co-transformed into the MEX67 shuffle strain together with linearized plasmid pRS314-YRA1, which was cut with EcoRI and BglII releasing approximately the C–terminal half of the YRA1 ORF. The screen for ts mutants was performed as the screen for me-x67 ts mutants (see above).
ProtA-tagged YRA1 was constructed by PCR amplification of the YRA1 coding sequence creating a NcoI 5′ and a BamHI site 3′ of YRA1 and insertion of this PCR product into the pNOPPATA vector (Hellmuth et al., 1998). A GFP-tagged version of YRA1 (pNOPGFPA-YRA1) was cloned by subcloning the YRA1 containing PstI fragment of pNOPPATA-YRA1 into the pNOPGFPA vector (Hellmuth et al., 1998). pGAL1::GFP–YRA1 was constructed by subcloning the GFP–YRA1 containing SphI–BamHI fragment of pNOPGFPA-YRA1 into the same sites of pGALPATG1L (M.Künzler, unpublished data).
L20GST-YRA1 (used for expression of recombinant Yra1p in S.pombe) was constructed by PCR amplification of the YRA1 ORF without the intron by ‘sew’ PCR with primers creating NcoI and BamHI sites at the 5′ and 3′ of the YRA1 ORF, respectively. This PCR product was cloned into pBluescript-eGFP. From this construct a BamHI–SalI fragment containing the ORF of YRA1 was subcloned into L20GST (Braspenning et al., 1997), previously cut with the same enzymes.
Gene disruptions and strain constructions
The YRA1 gene was disrupted in the diploid RS453 strain by transformation with yra1::HIS3 (see above) and selection of HIS+ transformants. Heterozygous transformants with the correctly integrated yra1::HIS3 null allele were sporulated and tetrads dissected. A 2:2 segregation for viability was found, confirming that YRA1 is an essential gene (Portman et al., 1997). Haploid yra1::HIS3 cells could be recovered, when the diploids carried pRS316-YRA1. This strain died on 5–FOA plates. A mex67::HIS3/yra1::HIS3 (+pRS316-MEX67/pRS316-YRA1) double shuffle strain was generated by mating of the corresponding shuffle strains and tetrad analysis. Analysis of synthetic lethality between me-x67 and yra1 mutant alleles was done by transformation of the MEX67–YRA1 double shuffle strain with plasmids containing mex67-6, yra1-1, or the corresponding wild-type alleles and testing for growth on 5–FOA plates.
Shuttling assay using nup49-313 cells
The shuttling assay was performed essentially as described (Lee et al., 1996). Briefly, strain nup49-313 was transformed with pGAL1::GFG-YRA1, pADH::NLS–GFP–lacZ, and pGAL1::GFP–NPL3, respectively. Cells also carried the pASZ11 plasmid harbouring the ADE2 gene. Cells were grown in selective medium containing raffinose at 25°C to an OD600 of ∼0.25. Galactose was added to a final concentration of 2% and cells further grown for 2 h. Cells were washed once in YPD and resuspended in an equal volume YPD. After 2 h incubation, the cultures were split and incubated at 25 and 36°C for 5 h, respectively. The localization of the GFP reporter proteins was analyzed in the fluorescein channel of a Zeiss Axioskop fluorescence microscope. Photographs were taken with a Xillix Microimager charge-coupled device camera, ensuring that the same exposure time was used in all cases.
Affinity-purification of ProtA–Yra1p
Affinity purification of ProtA–Yra1p was done as described earlier (Siniossoglou et al., 1996) with the following modifications. Yeast spheroplasts (2 g) were lysed in 20 ml lysis buffer (150 mM KCl, 20 mM Tris–HCl pH 8.0, 5 mM MgCl2, 1% Triton-100, protease inhibitors). The soluble supernatant was incubated with 200 μl IgG–Sepharose beads (Amersham Pharmacia Biotech AB, Sweden), previously treated with TST (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.05% Tween-20) and 0.5 M acetic acid (pH 3.5) for 1 h at 4°C. Beads were washed with 10 ml TST. Since ProtA–Yra1p was eluted by pH 3.5 treatment very inefficiently (ProtA–Yra1p may aggregate under these conditions), it was eluted with SDS sample buffer lacking dithiothreitol (DTT). Supernatants and eluates were analyzed by SDS–PAGE, followed by Coomassie Blue staining or Western blotting, using antibodies against Mex67p or Nup85p.
Purification of recombinant Yra1p expressed in S.pombe and GST pull-down experiments
Schizosaccharomyces pombe was transformed with L20GST-YRA1. Transformants were grown in EMM medium (Stratagene) containing 5 mM thiamine as described (Braspenning et al., 1997). Cells were induced by washing with EMM medium without thiamine, resuspended in EMM medium without thiamine at an OD595 of 0.02, and grown for 21 h. Cells were lysed with glass beads in TNN buffer (100 mM Tris–HCl pH 8.0, 100 mM NaCl, 1% Nonidet P-40, protease inhibitors). After centrifugation, the pellet was resuspended in TNN buffer containing 500 mM NaCl, followed by sonication. After the second centrifugation, the derived supernatant was incubated with TNN–washed glutathione Sepharose beads (Amersham Pharmacia Biotech AB, Sweden) for 1 h at 4°C. For thrombine cleavage, the beads were washed with Universal buffer [20 mM HEPES pH 7.0, 100 mM KOAc, 2 mM Mg(OAc)2, 0.1% Tween-20, 10% glycerol, 5 mM β–mercaptoethanol, protease inhibitors] (Künzler and Hurt, 1998), followed by incubation with 1 μl thrombine at room temperature for 30 min. The cleaved Yra1p was eluted with 100 μl elution buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 2.5 mM CaCl2, 0.1% β–mercaptoethanol, 0.1% Tween-20).
For in vitro binding of GST–Yra1p or GST–ALY (Bruhn et al., 1997) to the recombinant Mex67p/Mtr2p complex or human TAP, beads with bound GST–Yra1p or GST–ALY were prepared. Beads bound to GST, or GST–Npl3p (kindly provided by Dr J.Braspenning) served as negative controls; these were expressed in S.pombe with the help of plasmid L20GST. The beads containing GST or GST fusion proteins were washed with Universal buffer. Then, the supernatant of lysed E.coli BL21 cells, containing recombinant Mex67p/Mtr2p complex, Mtr2p alone or human TAP, was incubated with these beads for 1 h at 4°C. For RNase treatment, the E.coli supernatant containing Mex67p/Mtr2p was incubated with RNase A (0.5 mg/ml final concentration) for 1 h at 37°C. The beads were washed with Universal buffer, and bound proteins eluted with SDS sample buffer containing 100 mM DTT, before analysis by SDS–PAGE and Coomassie Blue staining or Western blotting.
RNA–RNA annealing assay
The RNA–RNA annealing assay was similar to the one described in Portman and Dreyfuss (1994), but in vitro transcribed RNAs derived from SSA1 were used. Plasmid pBluescriptIIKS(+)-SSA1 (bp 1131–1461) was linearized with StyI (for radiolabeled RNA) or with SacI and blunt ended with T4 DNA polymerase (for cold RNA) and used as template in the in vitro transcription reaction (Maniatis et al., 1982). Recombinant Yra1p, purified from S.pombe, was pre-incubated in a total volume of 5 μl in 2× annealing buffer [40 mM HEPES pH 7.5, 200 mM KCl, 1 mM Mg(OAc)2, 1 mM DTT, 0.1 mg/ml BSA, 8 U/100 μl RNasin] for 10 min at 30°C. The RNA solution was prepared freshly with 2 nM cold and 1 nM 32P-α-CTP-labeled RNA final concentration, which were incubated separately at 65°C for 5 min, chilled on ice, and mixed. An aliquot (5 μl) of the mixed RNAs was added to the Yra1p-containing solution and incubated at 30°C for 10 min. For digestion of single-stranded RNA, RNase T1 (1 U/μl final concentration) was added and the solution incubated at 37°C for 15 min. Ten microlitres of stop solution (0.2% SDS 1 mg/ml proteinase K, 2.5 mg/ml E.coli tRNA, 600 mM NaOAc pH 5.2, 0.2 U/μl RNasin) were added and again incubated at 37°C for 15 min. The solution was extracted with 20 μl phenol:cholorform:isoamyl alcohol (25:24:1) and precipitated with 2.5 volumes of EtOH. Pellets were washed with 80% EtOH and resuspended in 10 μl formamide loading buffer. RNA fragments were resolved on a 10% polyacrylamide/8 M urea gel. The gel was fixed in 10% acetic acid/30% EtOH, dried, and exposed to a X-OMAT AR film (Kodak).
UV-crosslinking of RNA to recombinant Yra1p
UV-crosslinking was performed according to Kessler et al. (1997). As RNA served a radiolabeled transcript of pBluescriptIIKS(+)-SSA1 (bp 818–1152), which was linearized with XbaI and used as a template in the in vitro translation reaction (Maniatis et al., 1982). GST–Yra1p was purified on GSH beads as described above. The beads were then incubated with FPLC–purified Mex67p–Mtr2p complex [purified as described in Santos-Rosa et al. (1998)], in order to form a ternary complex, or incubated with buffer. Fifteen microlitre samples containing either GST, GST–Yra1p bound to GSH-beads, or GST–Yra1p with prebound Mex67p–Mtr2p complex on GSH beads, 1.5 nM labeled RNA (corresponding to 140 000 c.p.m.) and crosslinking buffer [1 mM MgCl2, 75 mM K(OAc)2, 1.5 mM tRNA, 0.5 U/μl RNasin] were incubated at 30°C for 5 min. The probes were transferred to parafilm and irradiated at room temperature with UV light for 10 cycles with the autocrosslink function of the UV Stratalinker 1800. RNase A was added to a final concentration of 0.9 mg/ml and RNase T1 to a final concentration of 10 U/15 μl. The RNA was digested for 30 min at 37°C. SDS-sample buffer was added to the probes, which were separated on a 12% SDS–PAGE gel. The gel was stained with Coomassie Blue, dried, and exposed to a Kodak X-OMAT AR film.
Miscellaneous
SDS–PAGE and Western blot analysis were performed according to Siniossoglou et al. (1996). In situ hybridization of poly(A)+ RNA and fluorescence microscopy of GFP fusion protein-expressing yeast cells, including expression in nup133::HIS3 cells, were performed as described previously (Doye et al., 1994; Santos-Rosa et al., 1998).
Acknowledgments
Acknowledgements
The excellent technical assistance of Birgit Schmelzl in the initial phase of this project and help by Dr Joris Braspenning for S.pombe expression is acknowledged. We are grateful to Dr R.Grossschedl (Genzentrum München, Germany) for providing a plasmid containing the ALY and GST–ALY cDNA. We thank Drs George Simos, Christian Ungermann and Jun Katahira for critical reading of the manuscript. E.C.H. was recipient of grants from the Deutsche Forschungsgemeinschaft (SFB352) and the Human Frontiers Science Program (HFSP).
References
- Bailer S.M., Siniossoglou, S., Podtelejnikov, A.V., Hellwig, A., Mann, M. and Hurt, E.C. (1998) Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J., 17, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braspenning J., Manetti, R., Zumbach, K., Meschede, W., Gissmann, L. and Tommasino, M. (1997) A general purification protocol for e7 proteins from ‘high- and low-risk’ human papillomavirus types expressed in the yeast Schizosaccharomyces pombe. Protein Expr. Purif., 10, 192–201. [DOI] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach, E., Schmitt, C. and Izaurralde, E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn, A. and Grosschedl, R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed] [Google Scholar]
- Doye V., Wepf, R. and Hurt, E.C. (1994) A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J., 13, 6062–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinet C., de la Torre, M.A., Aldea, M. and Herrero, E. (1995) An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast, 11, 25–32. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber, J., Boelens, W.C., Mattaj, I.W. and Lührmann, R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Flach J., Bossie, M., Vogel, J., Corbett, A., Jinks, T., Aker Willins, D. and Silver, P.A. (1994) A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol. Cell. Biol., 14, 8399–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K. and Izaurralde, E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Hellmuth K., Lau, D.M., Bischoff, F.R., Künzler, M., Hurt, E.C. and Simos, G. (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol., 18, 6364–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Sträßer,K., Segref,A., Bailer,S., Schlaich,N., Presutti,C., Tollervey,D. and Jansen,R. (2000) Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi, M., Chen, S. and Tartakoff, A.M. (1994) Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell, 5, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.B. and Cullen, B.R. (1999) The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev., 13, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Sträßer, K., Podtelejnikov, A., Mann, M., Jung, J.J. and Hurt, E.C. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M., Henry, M.F., Shen, E., Zhao, J., Gross, S., Silver, P.A. and Moore, C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′–end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler M. and Hurt, E.C. (1998) Cse1p functions as the nuclear export receptor for importin α in yeast. FEBS Lett., 433, 185–190. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Henry, M. and Silver, P.A. (1996) A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev., 10, 1233–1246. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.T. and Sambrook,J. (1982). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. [Google Scholar]
- Murphy R., Watkins, J.L. and Wente, S.R. (1996) GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell, 7, 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. and Wente, S.R. (1996) An RNA-export mediator with an essential nuclear export signal. Nature, 383, 357–360. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Fischer, U., Michael, W.M. and Dreyfuss, G. (1997) RNA transport. Annu. Rev. Neurosci., 20, 269–301. [DOI] [PubMed] [Google Scholar]
- Neville M. and Rosbash, M. (1999) The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J., 18, 3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D.S. and Dreyfuss, G. (1994) RNA annealing activities in HeLa nuclei. EMBO J., 13, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman D.S., O'Connor, J.P. and Dreyfuss, G. (1997) Yra1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA, 3, 527–537. [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Panté, N. and Hurt, E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R. and Hurt, E.C. (1997) Mex67p which is an essential factor for nuclear mRNA export binds to both Poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer, C., Rieger, M., Doye, V., Tekotte, H., Weise, C., Emig, S., Segref, A. and Hurt, E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot, H.V., Goldstein, A.L. and Cole, C.N. (1998) Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J., 17, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford, C.S., Guthrie, C. and Weis, K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Fahrenkrog, B. and Aebi, U. (1999) The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol., 11, 391–401. [DOI] [PubMed] [Google Scholar]
- Sträßer K. and Hurt, E. (1999) Nuclear RNA export in yeast. FEBS Lett., 452, 77–81. [DOI] [PubMed] [Google Scholar]
- Tseng S.S.L., Weaver, P.L., Liu, Y., Hitomi, M., Tartakoff, A.M. and Chang, T.H. (1998) Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J., 17, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer C., Doye, V., Grandi, P., Nehrbass, U. and Hurt, E. (1992) A new subclass of nucleoporins that functionally interacts with nuclear pore protein NSP1. EMBO J., 11, 5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]