Abstract
DNA polymerase δ (Polδ) is a high fidelity polymerase that plays a central role in replication from yeast to humans. We present here the crystal structure of the catalytic subunit of yeast Polδ in ternary complex with a template-primer and an incoming nucleotide. The structure, determined at 2.0Å resolution, catches the enzyme in the act of replication. The structure reveals how the polymerase and exonuclease domains are juxtaposed relative to each other and how a correct nucleotide is selected and incorporated. The structure also reveals the “sensing” interactions near the primer terminus that signal a switch from the polymerizing to the editing mode. Taken together, the structure provides a chemical basis for the bulk of DNA synthesis in eukaryotic cells and a framework for understanding the effects of mutations in Polδ̣ that cause cancers.
Accurate DNA replication is crucial for the maintenance of genomic stability and for the suppression of mutagenesis and carcinogenesis. Eukaryotic DNA polymerase δ (Polδ) is a high fidelity polymerase which plays an indispensable role in replication1–3. Deletion of the yeast Saccharomyces cerevisiae Pol3 gene, which encodes the catalytic subunit of Polδ, causes inviability, and mutational inactivation of its polymerase activity also confers lethality4–7. In addition to its high fidelity for DNA synthesis, Polδ achieves further enhancement of fidelity from its 3’→5’ proofreading exonuclease activity. A D400A mutation in murine Pol3 that inactivates its proofreading function causes a major increase in susceptibility to cancers, with nearly half of the homozygous D400A mice developing tumors after two months of age8, 9. Tumors also develop when Leu604 in the polymerase domain of murine Pol3 is changed to glycine or lysine10. Point mutations and single-nucleotide deletions have also been identified in human Pol3 in several cancer cell lines and sporadic colon cancers11, 12, as well as in Pol3 from the highly malignant Novikoff rat hepatoma cells13.
In addition to Polδ, eukaryotes possess three other B-family Pols, α, ε, and ζ1–3. Of these, Polα functions in the synthesis of lagging strand primers and its Pol function is essential for cell viability. Polα, however, synthesizes DNA with a lower fidelity than Polδ and it has no 3’→5’ proofreading exonuclease activity. Although the DNA polymerase domain of Polε can be deleted without any appreciable impairment of viability or other cellular functions in yeast cells1–3, this Pol has been suggested to have a role in the replication of the leading strand14. Polζ is a low fidelity Pol required for promoting synthesis through DNA lesions which block replication15. Despite decades of studies on Polδ, there is no structural information available for the catalytic subunit of this Pol (or for any other eukaryotic B-family Pol) that could yield detailed insights into its action mechanism. To date, most of the structural information on B-family Pols derives from distantly related viral and archaeal Pols16, including phage RB69 Pol17, which share < 15% sequence identity with the catalytic subunit of eukaryotic Pols α, δ, ε, and ζ.
We report here the structure of yeast Pol3 (residues 68 to 985), which exhibits the same DNA synthesis activity as the full-length protein, in ternary complex with a template-primer presenting G as the templating residue, and with dCTP as the incoming nucleotide. The structure, determined at 2.0Å resolution (Table 1), catches the enzyme in the act of replication, at a step just prior to the catalytic transfer of the dNTP onto the 3’ terminus of the primer. It provides a molecular basis for the high fidelity of DNA synthesis during replication in eukaryotic cells, and affords a context for understanding the effects of mutations that cause cancer.
Table 1.
Data collection, phasing and refinement statistics
| Native 1 | Au(CN)2 soak A |
Au(CN)2 soak B |
Native 2 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 83.7, 85.9, 87.3 | 83.6, 85.5, 87.2 | 84.2, 85.2, 86.9 | 81.1, 85.9, 86.9 |
| α, β, γ (°) | 90.0, 105.4, 90.0 | 90.0, 105.6, 90.0 | 90.0, 105.6, 90.0 | 90.0, 111.1, 90.0 |
| Resolution (Å) | 2.8 (2.9-2.8)* | 2.7 (2.8-2.7)* | 2.7 (2.8-2.7)* | 2.0 (2.07- 2.0)* |
| Rmerge | 8.3 (22.1) | 5.8 (14.6) | 8.5 (21.7) | 8.2 (45.6) |
| I / σI | 16.4 (3.2) | 21.6 (5.0) | 26.6 (6.1) | 26.3 (3.7) |
| Completeness (%) | 94.0 (68.3) | 97.1 (79.8) | 90.1 (63.2) | 98.3 (97.9) |
| Redundancy | 5.5(4.2) | 3.6(3.1) | 5.4(5.0) | 3.8 (3.8) |
| Refinement | ||||
| Resolution (Å) | 50.0-2.0 | |||
| No. reflections | 69856 | |||
| Rwork / Rfree | 19.3/23.4 | |||
| No. atoms | ||||
| Protein | 7029 | |||
| Ligand/ion | 602 | |||
| Water | 558 | |||
| B-factors | ||||
| Protein | 29.2 | |||
| Ligand/ion | 33.6 | |||
| Water | 35.9 | |||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.010 | |||
| Bond angles (°) | 1.23 |
Values in parentheses are for highest-resolution shell.
RESULTS
Overall architecture and arrangement
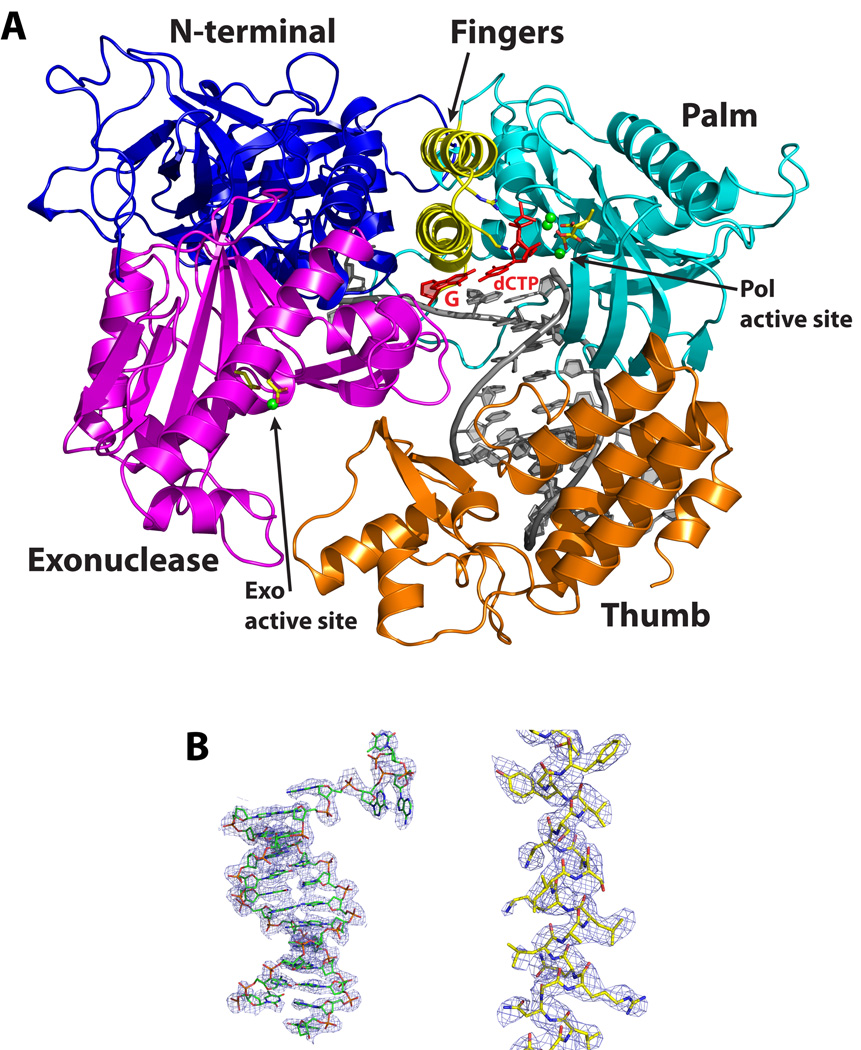
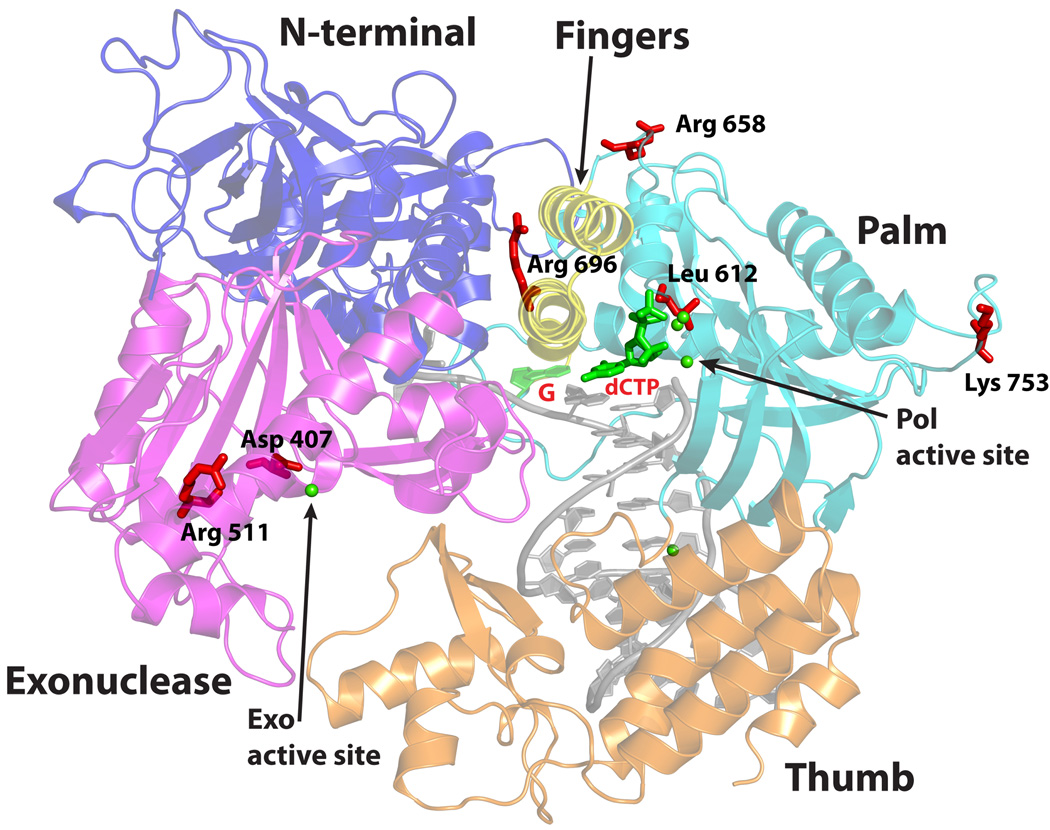
Pol3 embraces the template-primer with its palm, fingers, thumb, and exonuclease domains (Fig. 1). In addition, an N-terminal domain (NTD) caps the “top” of the polymerase and interacts with the last visible base on the unpaired segment of the template strand. The palm is composed of a mixed 6-stranded β-sheet (β23,β24,β25–β28) flanked by two long α helices (αQ and αR) from one side and a short helix (αM) from the other (Fig. 1 and Supplementary Fig. 1). The palm interacts with the replicative end of the DNA and carries the active site residues (Asp608 and Asp764) that catalyze the nucleotidyl transfer reaction. The fingers domain is dominated by two anti-parallel α-helices (αO and αP) that drape over the nascent G:dCTP base pair (Fig. 1). The thumb can be divided into two subdomains, the base (αS, αT, αW and αX) that connects to the palm, and the tip (β30, αU, β31 and αV) that packs against the exonuclease domain. The two subdomains cup the duplex portion of the template-primer and interact extensively with the sugar-phosphate backbones near the minor groove (Fig. 1). The exonuclease domain resembles the PAD (polymerase associated domain) in Y-family Pols15 in that it lies next to the fingers domain and over the DNA major groove. The exonuclease active site is defined by residues Asp321, Glu323 and Asp407 which cluster around a single Ca2+ ion. The polymerase and exonuclease active sites are separated by ~45Å in a direction roughly perpendicular to the DNA axis (Fig. 1). The unpaired segment of the template strand splays out of the polymerase active site at a sharp angle, such that the distance between the templating base and the 5’ unpaired base is > 10 Å. The unpaired nucleotides weave a contorted path between the base of the fingers and several segments of the exonuclease domain (Fig. 1). The duplex portion of the template-primer has a B-DNA–like conformation with average helical twist and rise values of ~33° and 3.3Å, respectively, and the phosphates are in standard BI configuration (ε = trans, ζ = gauche−). This is in contrast to the A-form DNA observed at the primer 3’ end in A-family Pols, including phage T7 Pol18, T. aquaticus (Taq) Pol I19, and the Bacillus Pol I fragment (BF)20.
Figure 1. Structure of the Pol3-DNA-dCTP ternary complex.
(A) The Pol3 palm, fingers, thumb, exonuclease, and the N-terminal domains are shown in cyan, yellow, orange, magenta and dark blue, respectively. DNA is in gray, template G and incoming dCTP are in red, and the Ca2+ ions are in green. The polymerase (Pol) and exonuclease (Exo) active sites are marked, and the residues that comprise the two active sites are highlighted. B) Experimental electron density at 2.7Ǻ after solvent flattening, around the 12/16 primer-template (left) and helix P (right; c.f. Supplementary Fig. 1). The density is contoured at 1.0σ.
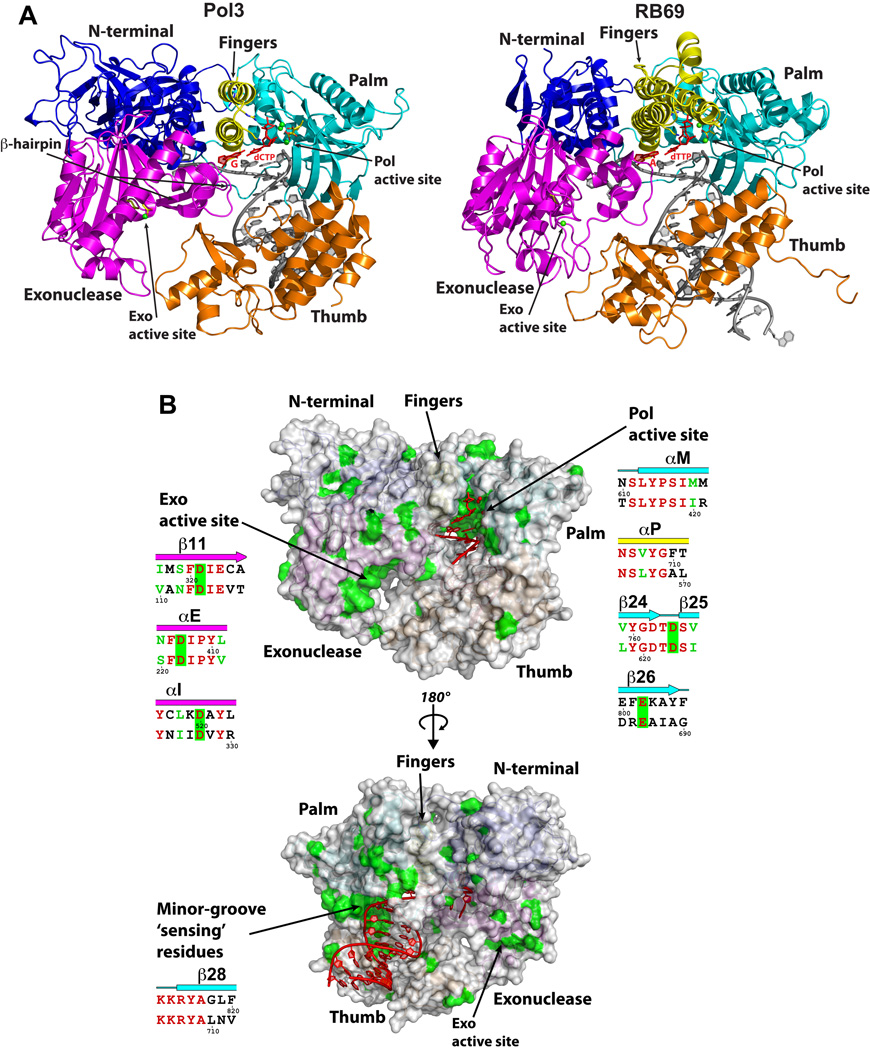
Despite the limited sequence identity, the Pol3 and RB69 Pol structures align with an rmsd of 2.50Å between the common structural elements (631 Cαs) (Fig. 2 and Supplementary Fig. 1). The palm is the most structurally conserved domain, aligning with an rmsd of 1.71Å (165 Cαs), while the NTD is the most structurally diverged region, aligning with an rmsd of 2.47Å (138 Cαs). The NTD in Pol3 is composed of three motifs (I, II and III) and is much more elaborate and extended than in RB69 Pol. It bears a strong resemblance to the NTD in the Herpes Simplex Virus (HSV) 1 Pol21 (Supplementary Fig. 2), and as we discuss below it may be involved in DNA and/or RNA binding. The Pol3 fingers domain, on the other hand, is simpler than in RB69 Pol (Fig. 2). Thus, whereas the RB69 Pol fingers domain consists of two exceptionally long and slightly curved anti-parallel α-helices capped at one end by a short α-helix, which protrude ~60Å from the base and pack against the distal portion of the palm domain, the fingers domain in Pol3 is shorter and straighter, composed of just two anti-parallel α-helices that extend ~40Å from the base (Fig. 2). Pol3 and RB69 Pol also differ in the configuration of a “β-hairpin” that extends from the exonuclease domain and has been implicated in the transition of the primer between the polymerase and exonuclease active sites22–24 (described below). Thus, whereas the β-hairpin in Pol3 is anchored in the major groove, it does not associate with DNA in the RB69 ternary complex17 (described below). Also, compared to RB69 Pol, the tip of the thumb in Pol3 is shifted “upwards” towards the 5’ end of the template by >9Å (Supplementary Fig. 3). Thus, the two regions of B-family polymerases that are most strongly implicated in active site switching, the β-hairpin22–24 and the hinge in the thumb24–26, show the most flexibility between Pol3 and RB69 structures.
Figure 2. Comparison between Pol3 and RB69 Pol ternary complexes.
(A) Pol3 and RB69 ternary complexes displayed in the same orientation, based on a superposition of their palm domains. The color scheme is the same as in Fig.1, where the palm, fingers, thumb, exonuclease, and the N-terminal domains are shown in cyan, yellow, orange, magenta and dark blue, respectively. DNA is in gray; template G (Pol3) and A (RB69), and incoming dCTP (Pol3) and dTTP (RB69) are in red; Ca2+ ions (Pol3 and Rb69) are in green. The polymerase (Pol) and exonuclease (Exo) active sites are marked, and the residues that comprise the two active sites are highlighted in stick form. (B) The Pol3 surface in two orientations. Highlighted in green are the sites of identical residues between Pol3 and Rb69 Pol (c.f. Supplementary Fig. 1), and highlighted in red is the DNA in the two orientations. Also highlighted, alongside the figures, are the “local” structure based sequence alignments between Pol3 (upper) and RB69 Pol (lower) (c.f. Supplementary Fig. 1) over the green “patches” of sequence identity. Note that the most prominent patches of sequence identity are centered over the Pol active site and the nascent base pair, the Exo active site, and the minor groove “sensing” residues.
Active site
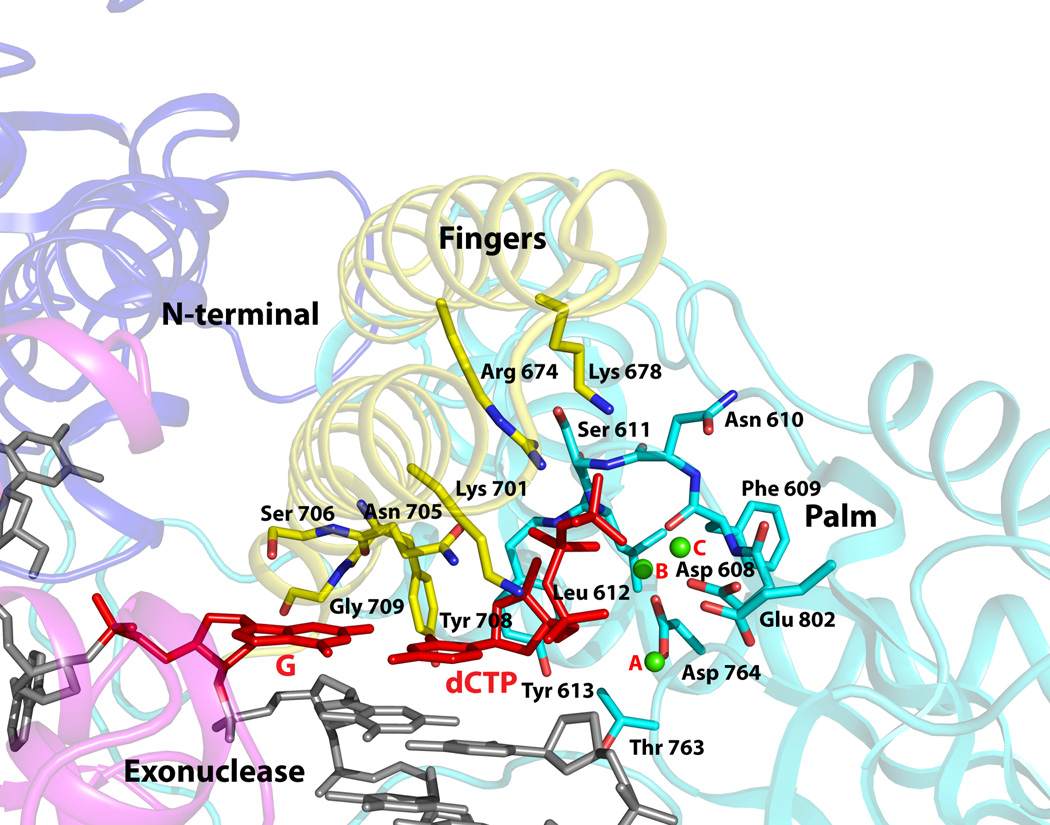
The incoming dCTP is bound to Pol3 with its triphosphate moiety interlaced between the fingers and palm domain (Fig. 3). The α-phosphate makes a hydrogen bond with Lys701 from the fingers domain, the β-phosphate accepts a hydrogen bond from the main chain amide of Leu612 from the palm domain, and the γ-phosphate makes direct hydrogen bonds with Arg674 and water-mediated hydrogen bonds with Lys678 from the fingers domain, and a hydrogen bond with the main chain amide of Ser611 from the palm domain (Fig. 3, Supplementary Fig. 4). In all, incoming dCTP is bound in a manner analogous to that seen in other Pol ternary complexes16, effectively drawing together the fingers and palm domains for catalysis and discrimination between a correct and an incorrect nascent base pair (see below). The dCTP sugar packs against Tyr613 (Fig. 3), buttressed by hydrogen bonds between its 3’OH and the main chain amide of the tyrosine as well as the β-phosphate. The sugar adopts a C3’-endo conformation, which casts its C2’ atom close (~3.5Å) to the aromatic plane of Tyr613 and, as in other DNA Pols (Tyr/Phe/Glu/His), provides a basis for the exclusion of ribonucleotides16. Arrayed between the dCTP triphosphate tail and the primer terminus are three calcium ions (A, B and C) and the acidic residues Asp608, Asp764, and Glu802 (Fig. 3). Ca2+ A and B are separated by ~3.7Å and are analogous to metals “A” and “B” in other DNA Pols, including RB69 and members of the A- and Y-families17–19, 27. Ca2+ C has not, to our knowledge, been observed previously in other DNA Pols, and is coordinated by the dCTP γ-phosphate and the Asp608 and Glu802 carboxylates and also possibly by Glu800 which is ~ 4Å away (Supplementary Fig. 4). Although calcium inhibits Polδ activity, the active site geometry is appropriate for catalysis, with the putative 3’OH located ~4Å from the dCTP α-phosphate and aligned with respect to the Pα-O3’ bond (angle of about 155°). Metals A and B are in a position to activate the primer 3’OH for its nucleophilic attack on the dNTP α-phosphate and to stabilize the pentacovlent transition state, and metal C is in a position to assist in the leaving of the β–γpyrophosphate (Supplementary Fig. 4).
Figure 3. Close-up view of the polymerase active site region.
The fingers, palm, exonuclease, and the N-terminal domains are colored in faded yellow, cyan, magenta, and blue. The DNA is colored gray, templating G and incoming dCTP are in red, and the Ca2+ ions (A, B and C) are in green. Highlighted and labeled are the acidic residues (Asp608, Asp764 and Glu802); residues that interact with the triphosphate moiety of incoming dCTP via side chain (Arg674 and Lys701), main chain (Ser611 and Leu612), and a water molecule (Lys678); residues that impinge on the dG-dCTP bases (Asn705, Ser706, Tyr708, and Glu709); and Tyr613 that stacks against the dCTP sugar. The residues are colored to match the domain they belong to.
To determine the possible significance of metal “C” for Pol 3 catalysis, we compared the DNA synthetic activity of wild type Pol3 with the mutant protein where the Glu800 and Glu802 have both been changed to alanines. To circumvent any confounding effects of nucleotide removal by the 3’ → 5’ exonuclease, we used Pol3 proteins which were devoid of exonuclease activity. As shown in Supplementary Table 1, compared to the wild type Pol3 protein, the Pol3 E800A E802A mutant protein exhibits an ~40 fold reduction in the efficiency of C incorporation opposite temple G and an ~ 25-fold reduction in the efficiency of A incorporation occurs opposite template T. We also examined the incorporation of incorrect nucleotides opposite templates G and T. Whereas the incorporation of G opposite template T by the mutant protein dropped ~ 45-fold (Supplementary Table 1), for the other wrong nucleotides with the mutant protein, we were unable to detect any misincorporation opposite either template even at very high dNTP concentrations at which the misincorporation by the wild type protein could be seen (data not shown). We conclude from these observations that Glu800 and Glu802 affect the efficiency of incorporation for both the correct and incorrect nucleotides, suggesting a possible role for the putative third metal binding site in modulating the catalytic efficiency of Pol3.
Interactions with the nascent base pair
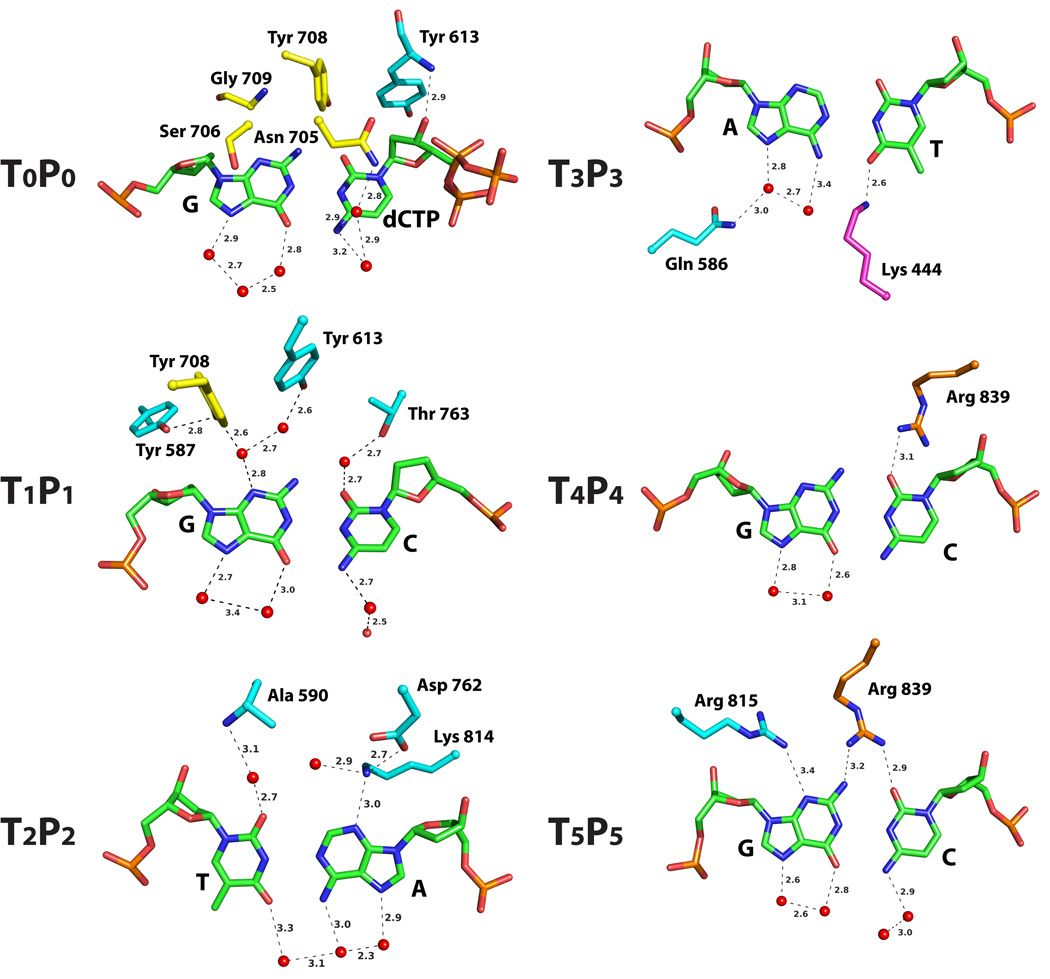
The high fidelity of Polδ is determined primarily by the shape of the pocket accommodating the nascent Watson-Crick (WC) base pair, between templating G and incoming dCTP. The pocket is shaped by residues Asn705, Ser706, Tyr708 and Gly709 from the fingers domain and Tyr613 from the palm domain (Figs. 3 and 4). Asn705 and Ser706 are perched over the C and G aromatic ring systems, while Tyr613, Tyr708 and Gly709 impinge on the minor groove. Specifically, Tyr613 packs against the sugar of C (see above), Tyr708 bisects the planes of G and C bases, and Gly709 abuts against N3 of G (Figs. 3 and 4). Although interactions with the nascent bases are primarily van der Waals in nature, the positioning of Asn705 and Ser706 polar atoms over the C and G π electrons and the positioning of Tyr708 σ charges next to the partial negative charges on the G and C acceptor atoms may provide an additional favorable electrostatic component. In contrast to the minor groove, the major groove is devoid of contacts, except for slight intrusion onto G by Ala 445 and Ile702.
Figure 4. Pol3-DNA base interactions.
The bases, at the replicative end of the template-primer, are labeled Tn-Pn (where T and P refer to template and primer strands, respectively, and the subscripts n (0, 1, 2 and 3) refer to the number of base pairs from the templating base position). The Pol3 residues are colored to match the domain they belong to: yellow for fingers, cyan for palm, and magenta for exonuclease. The water molecules are colored red. Dashed lines depict hydrogen bonds (with distances in Angstroms above the bonds).
“Sensing” interactions
Following the insertion of a nucleotide opposite the templating base, the base pair is translocated along the template-primer from the “insertion” T0-P0 to the “postinsertion” T1-P1 position (where T and P refer to template and primer strands, respectively, and the subscripts refer to the number of base pairs from the templating base position). The structure reveals, in the minor groove, a network of water mediated hydrogen bonds to G and C at T1 and P1 positions and a direct hydrogen bond to A at P2 position (Fig. 4). These hydrogen bonds to “universal” acceptor atom will be preserved as long as the base pairs have proper WC geometry. Specifically, Tyr613 and Tyr708 (buttressed by Tyr587) are bonded via two water molecules to N3 of G at T1, Thr763 is bonded via a single water molecule to O2 of C at P1, and Lys814 (buttressed by Asp762) makes a direct hydrogen bond with N3 of A at P2 position (Fig. 4). Tyr587, Asp762, Thr763 and Lys814 are conserved in B-family polymerases (Supplementary Fig. 1), and mutations of the analogous residues lead to reduction in DNA synthesis activity, probably because they disrupt DNA binding due to the loss of direct or water-mediated minor groove contacts28–32. Strikingly, Pol3 structure reveals interactions with bases as far away as T4-P4 and T5-P5 positions, wherein Arg839 in the linker between the palm and thumb domains makes direct hydrogen bonds in the minor groove to the P4 and P5 bases and Arg815 in the palm domain directly hydrogen bonds to the minor groove of T5 base (Fig. 4). Thus, in the event that a mismatch eludes detection near the primer terminus, it can be potentially sensed up to four base pairs away from the primer terminus by the direct readout of WC geometry.
β-hairpin
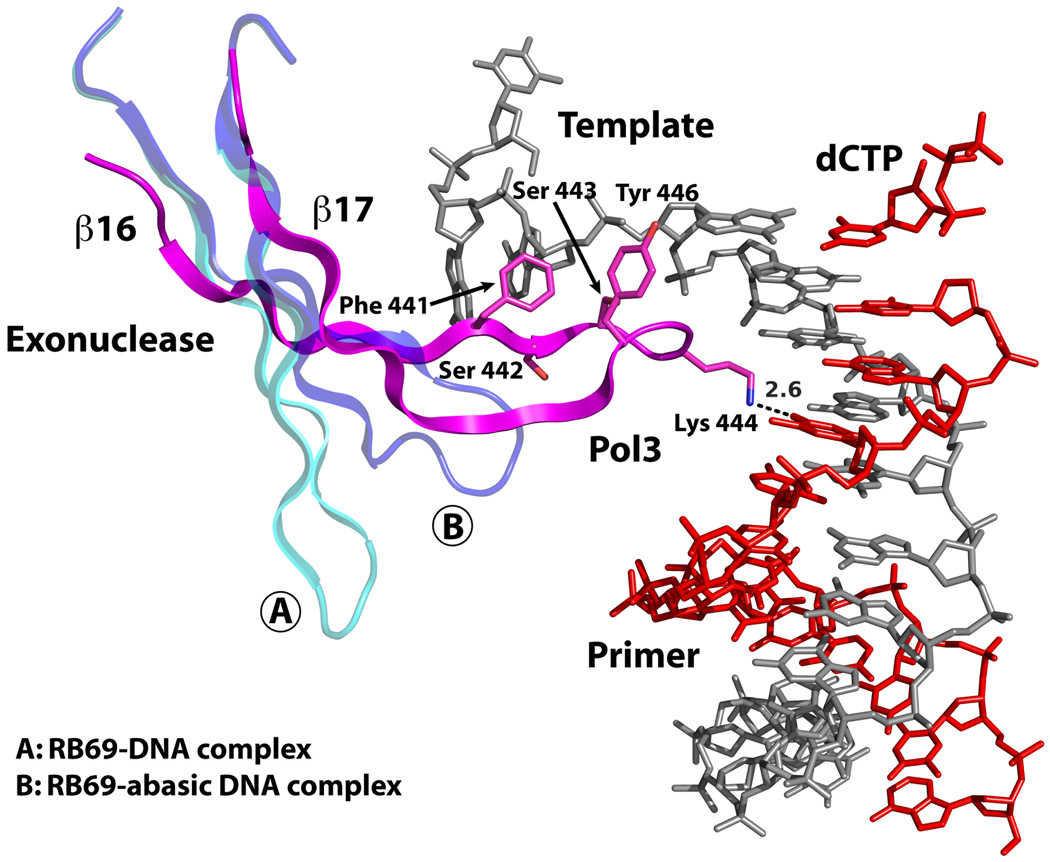
A long β-hairpin (β16 to β17) from the exonuclease domain inserts into the DNA major groove and Lys444 at its tip makes a direct hydrogen bond with O4 of T at position P3 (Figs. 4 and 5). However, the majority of interactions between the β-hairpin and the DNA occur with the unpaired segment at the 5’ end of the template strand (Fig. 5). This includes a water mediated hydrogen bond with N3 of A at position T−2 and an extensive set of van der Waals contacts with the sugar-phosphate backbone of nucleotides at positions T1 to T−1, with almost ~476Å2 of solvent accessible surface area buried at this interface. Based on mutational and deletion analysis, an analogous β-hairpin in RB69 and T4 Pols has been proposed to facilitate strand separation and the transition of the primer between the polymerase and exonuclease active sites22, 24. The idea is that the β-hairpin holds the template strand in place while the primer strand separates and migrates to the exonuclease active site22, 23. Curiously, the β-hairpin in RB69 Pol does not associate with DNA in the ternary complex with normal DNA17 (Fig. 5). And, in a binary complex with DNA containing an abasic residue at T0 position, it swings partly towards the DNA major groove and interacts with a portion of the single stranded template33 (Fig. 5). In contrast, the Pol3 β-hairpin is firmly entrenched in the DNA major groove and interacts extensively with the template; congruent with a role in active site switching (Fig. 5).
Figure 5. β-hairpin-DNA interactions.
In Pol3, an extended β-hairpin on the exonuclease domain inserts into the DNA major groove and interacts extensively with the unpaired portion of the template strand. Also shown for comparison are the β-hairpin positions in RB69 Pol complexed with normal DNA (A) or with DNA containing an abasic residue at the templating site (B).
Cancer mutations
A number of mutations in the mouse and human Pol3 gene have been shown to cause cancers. For example, D400A, L604G and L604K mutations in murine Pol3 that lower the fidelity of Polδ cause an increase in genomic instability and accelerate tumorigenesis8–10. The equivalent mutations in yeast Pol3, D407A, L612G and L612K, also cause an elevation in mutagenesis6, 34. From the structure, Asp407 is one of the three acidic residues (along with Asp321 and Glu323) that comprise the Pol3 exonuclease active site. Surprisingly, Leu612 does not contact incoming dNTP or the template-primer but rather packs against Tyr613 (Fig. 3). Thus, mutations at Leu612 may lower the fidelity of Polδ indirectly by aggravating interactions between Tyr613 and incoming dNTP and the base pair at T1P1 position (see above). In this context, one of the most puzzling mutation in yeast Pol3 is L612M, which causes not only a loss in Polδ fidelity but an error rate that is higher for certain mismatches (e.g. T.dGTP) over others (e.g A.dCTP)35. How this relatively conservative mutation can cause such varied error rates may only become clearer with the structure of the Pol3L612M mutant. Mutations in human Pol3 that cause cancers include R506H, R689W and S746I11, 12. An R648Q mutation has also been identified in the highly malignant Novikoff rat hepatoma cells13. The equivalent residues in yeast Pol3 are Arg511, Arg658, Arg696, and Lys753; wherein, Arg511 maps to the exonuclease domain and the others to the polymerase domain (Fig. 6). Intriguingly, Arg511, Arg658 and Lys753 are solvent exposed and far away from the exonuclease or the polymerase active site. As such, it is possible that these residues affect not so much the activity of Pol3 but rather its interactions with accessory subunits of the Polδ holoenzyme, which may accentuate the fidelity of Pol3. The cancerous effects of R689W mutation in human Pol3 may result from a difficulty in switching DNA between the polymerase and exonuclease active sites. The equivalent residue in yeast Pol3, Arg696, structurally links the two active sites (Fig. 6). It is sandwiched between Asp680 from the fingers domain and Glu539 from the exonuclease domain and a tryptophan at this position will break the salt-links that bind the two domains.
Figure 6. Mapping of cancer mutations.
Pol3 is the site of several mutations that accelerate tumorigenesis in mice and humans. The equivalent residues in yeast Pol3 (Asp407, Arg511, Leu612, Arg658, Arg696 and Lys753) are mapped onto the ternary complex to show their relative locations with respect to the polymerase (Pol) and exonuclease (Exo) active sites.
DISCUSSION
The replication of a large portion of the eukaryotic genome depends on Polδ, which synthesizes not only the lagging strand but also contributes to the synthesis of the leading strand. To keep the rate of mutations low, Polδ synthesizes DNA with high accuracy. A wrong nucleotide is incorporated only once in ~105 bases replicated, and the offending nucleotide can be further removed by the polymerase’s 3’→5’ exonuclease domain that provides an additional ~10–60-fold increase in the level of accuracy1–3.
High fidelity of Polδ
High fidelity of Polδ is determined primarily by shape of the Pol3 pocket accommodating the nascent WC base pair. Incoming dNTP binds in a manner analogous to that seen in other Pol ternary complexes16, effectively drawing together the fingers and palm domains for catalysis and discrimination between a correct and an incorrect nascent base pair. The fingers domain is in the “closed” conformation as in the other Pol ternary complexes. In A-family Pols the dNTP is believed to first bind a pre-insertion site on the fingers domain when it is in the “open” conformation before being ushered to the active site through the closure of the fingers domain16, while for B-family Pols kinetic evidence suggests that dNTP diffuses directly to the active site to provide a more direct check on correct versus incorrect base pairing36.
The binding pocket is shaped by residues Asn705, Ser706, Tyr708 and Gly709 from the fingers domain and Tyr613 from the palm domain. All of these amino acids are conserved in B-family Pols (Supplementary Fig. 1), and mutations of some of the analogous residues in phage RB69 Pol exhibit reduced base selectivity or reduction in DNA synthesis activity32, 37–39. Curiously, the Pol3 binding pocket, as in many other Pols, is largely devoid of contacts in the major groove. Accordingly, it is easier to see how Polδ rejects purine:pyrimidine mismatches than purine:purine or pyrimidine:pyrimidine mismatches. In purine:pyrimidine mismatches such as G:T and C:A the purine is displaced towards the minor groove and the pyrimidine towards the major groove40–42. Thus, given the shape of Pol3 binding pocket, the purine would sterically clash with Tyr708/Gly709 in the minor groove and the pyrimidine would lose some of the favorable van der Waals and electrostatic interactions. However, in purine:purine and pyrimidine: pyrimidine mismatches such as G.A and T.T the bases are displaced primarily towards the major groove, where there is little steric hindrance20, 42–44. In all, given the different steric properties of different mismatches it is surprising that Polδ discriminates them so uniformly, within ~2–3-fold of each other45. A deeper understanding of how Polδ occludes mismatches will require structures of Pol3 with different mismatches.
Proofreading by Polδ
How is a wrong nucleotide, once incorporated, sensed by Polδ for possible removal by the exonuclease domain? Strikingly, Pol3 interacts with bases as far away as the T5-P5 positions. Most of these interactions occur in the minor groove and involve direct or water mediated hydrogen bonds with “universal” acceptor atoms, which will only be preserved as long as the base pairs have proper WC geometry. Analogous hydrogen bonds in the minor groove occur near the T1-P1 and T2-P2 base positions in RB69 and A-family Pol ternary complexes17, 18, and the loss of these hydrogen bonds at a mismatch is believed to be the trigger that shifts the balance, for binding of the template-primer, from the polymerase to the exonuclease domain. DNA polymerization also greatly slows upon the incorporation of a wrong nucleotide, and is believed to provide the time window for the DNA to switch from the polymerase to the exonuclease active site23, 46, 47. Various mechanisms have been suggested for this stalling, including misalignment of the primer 3’OH for the in-line attack on incoming dNTP. In this regard, we note that Tyr613 and Tyr708 in Pol3 help to shape the binding pocket for nucleotide insertion as well as check for a mismatch at the T1-P1 position (Figs. 3 and 4). This raises the intriguing possibility that a mismatch at T1-P1 could contribute to the stalling of Pol3 via the repositioning of Tyr613 and Tyr708 and the reshaping of the pocket for nucleotide insertion.
DNA polymerases belonging to many different families undergo mismatch-induced stalling even when mismatches are up to four base pairs from the primer terminus20. The Pol3 structure reveals “direct” readout of bases as far away as the T4-P4 and T5-P5 positions (Fig. 4). Thus, in the event that a mismatch eludes detection near the primer terminus, it can be potentially sensed up to four base pairs away from the primer terminus by the direct readout of WC geometry. Interestingly, in A-family BF20 and C-family PolC48, the detection of remote replication errors has been attributed primarily to “indirect” readout mechanisms, wherein the mismatches propagate a distortion in the template strand to the pre-insertion site (BF), or disrupt interactions between the thumb domain and the phosphodiester backbones of the template and primer strands (PolC).
As noted previously, the exonuclease domain in A-family Pols lies on the opposite side of the palm domain to that in B-family Pols17, suggesting differences in the mechanisms by which the primer is transferred from the polymerase to the exonuclease active site. Also, in A-family Pols there appears to be no equivalent of a β-hairpin to facilitate strand separation or a “hinge” in the thumb domain to guide the primer from one active site to the other. (Mutations in the hinge region of the thumb have been shown to affect the ability of T4 Pol to switch the primer strand between the polymerase and exonuclease active sites24–26.) Together, these structural differences may reflect the much stronger exonuclease activity of B-family Pols when compared to A-family E.coli Pol I46, 49, and the different extent to which A- and B-family Pols use intramolecular (without dissociation) versus intermolecular (dissociation followed by reassociation) pathways for active site switching47, 50, 51.
Relationship to prokaryotic polymerases
Compared to RB69 Pol, the β-hairpin in Pol3 is anchored deeply in the major groove while the tip of the thumb is shifted towards the 5’ end of the template. Thus, the two regions of B-family polymerases that are most strongly implicated in active site switching, the β-hairpin and the hinge in the thumb, show the most flexibility between Pol3 and RB69 structures. The idea would be that the β-hairpin holds the template strand in place while the primer strand separates and is then guided to the exonuclease active site via interactions with the tip of the thumb22, 23. The observed flexibility in these two regions appears to be part of the mechanism for the transfer the primer from one active site to the other.
The Pol3 fingers domain is composed of just two anti-parallel α-helices (αO and αP) and is simpler than in RB69 Pol. Notably, helix P (or helix O in RB69 Pol) is analogous to helix O in the fingers domains of A-family Pols16. It is similarly draped over the nascent base pair and decorated with residues that ensure high fidelity. For example, residues Ly522, Tyr526, Gly527, and Tyr530 on helix O in T7 Pol18 structurally align with residues Lys701, Asn705, Ser706, and Tyr708 on helix P in Pol3 and make similar contacts, where, for example, Lys522/Lys701 makes a hydrogen bond with the triphosphate moiety of incoming dNTP, Tyr526/Asn705 and Gly527/Ser706 stack against the purine/pyrimidine aromatic rings, and Tyr530/Tyr708 impinge on the minor groove edge of the nascent base pair. One consequence of the more minimal fingers domain in Pol3 is that the gap through which the dNTP pyrophosphate is likely to exit following catalysis is significantly wider in Pol3, even when compared to RB6917. In contrast to the fingers domain, the NTD in Pol3 is much more elaborate and extended than in RB69 Pol. Strikingly, it bears a strong resemblance to the NTD in the Herpes Simplex Virus (HSV) 1 Pol21, and may play a role in DNA and RNA binding (discussed below).
Supplementary Figure 1 shows a structure based alignment of Pol3 and RB69 sequences, and Figure 2B shows how the limited sequence identity maps on the Pol3 surface. Although, much of the sequence between Pol3 and RB69 is variable there are three noteworthy “patches” of identity. First, the residues that interact with the template base and incoming dNTP and the acidic active site residues which help to coordinate the catalytic metals. Second, the exonuclease active site residues and, third, the minor groove “sensing” residues near the primer terminus. Thus, despite the evolutionary distance, the essential organization of the subdomains and the identity of residues involved in fidelity and proofreading are well conserved between viral and eukaryotic replicative DNA polymerases. Intriguingly, this conservation in structure between distant B family polymerases exceeds that for C family Gram-negative and Gram-positive bacterial replicative polymerases where, for example, the “OB domain” is located on opposite sides of the polymerase domain48.
Putative role of the NTD in DNA and RNA binding
The NTD in Pol3 is composed of three motifs (I, II and III), two of which may be involved in DNA and/or RNA binding (Supplementary Fig. 2). Motif I, for example, consists of two almost perpendicular β-sheets in the form of a β-barrel that has a similar structure and topology to the single stranded DNA binding domain (SSB) or the OB-fold in replication protein A (RPA)52 (Supplementary Fig. 5). Notably, the β-barrel lies ~20Å from the 5’ template base (at position T−4) and is part of a channel through which the template strand may pass ahead of the active site (with the β-hairpin acting as a “guide rail” along the channel) (Supplementary Fig. 6). Interestingly, a similar role has been proposed for the more classical OB-fold in the bacterial replicative polymerase PolC48, suggesting that sequestration of the template ~10–20 nt upstream of the active site may be a common feature amongst cellular replicative polymerases from different families.
Motif II strongly resembles the RNA binding motif (RNP or RRM) in ribonucleoproteins (Supplementary Fig. 5)53. In HSV Pol, motif II has been suggested to bind RNA or DNA and the NTD as a whole acting as a catalytic center for the excision of RNA primers during lagging strand DNA synthesis21. The NTDs in RB69 and T4 Pols have also been implicated in RNA binding. Both Pols are able to bind ribosome binding sites on their own mRNAs to repress their translation, and the NTD is thought to be involved in this process16. There is no evidence to our knowledge that Polδ can auto-regulate its synthesis by binding to its own mRNA, but as a lagging strand polymerase it can potentially run into the RNA primer of the “previous” Okazaki fragment. As such, the motif II of Pol3 NTD may play a role in the displacement and the binding of this initiator RNA for degradation by FEN1 and/or Dna2 nuclease(s) in a process of iterative “nick translation”54. Interestingly, in vitro studies have shown that the bacterial replisome can displace a collinear transcribing RNA polymerase, but the mRNA is retained and used by the leading strand polymerase to continue chain elongation55. It remains to be determined if a similar mechanism operates in a eukaryotic replisome but the finding of an RNP motif in Pol3 opens up the possibility (and provides a framework) for examining the broader role of RNA in Polδ function.
Supplementary Material
Acknowledgements
We thank the staff at BNL and APS for facilitating X-ray data collection. This work was supported by grant CA138546 from the U. S. National Institutes of Health.
Footnotes
Accession codes. Protein Data Bank. Coordinates and structure factors for the Pol3 ternary complex have been deposited with the accession code 3IAY.
Author contribution. M.K.S and R.E.J performed the experiments, and all of the authors contributed to the concepts and to the writing of the paper.
References
- 1.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 4.Boulet A, Simon M, Faye G, Bauer GA, Burgers PM. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. Embo J. 1989;8:1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell LH. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- 6.Simon M, Giot L, Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. Embo J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sitney KC, Budd ME, Campbell JL. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989;56:599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- 8.Goldsby RE, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc Natl Acad Sci U S A. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsby RE, et al. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 10.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flohr T, et al. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80:919–929. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 12.da Costa LT, et al. Polymerase delta variants in RER colorectal tumours. Nat Genet. 1995;9:10–11. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- 13.Popanda O, Flohr T, Fox G, Thielmann HW. A mutation detected in DNA polymerase delta cDNA from Novikoff hepatoma cells correlates with abnormal catalytic properties of the enzyme. J Cancer Res Clin Oncol. 1999;125:598–608. doi: 10.1007/s004320050322. [DOI] [PubMed] [Google Scholar]
- 14.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 16.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 17.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 18.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem. 2006;281:18193–18200. doi: 10.1074/jbc.M602414200. [DOI] [PubMed] [Google Scholar]
- 22.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 23.Reha-Krantz LJ, et al. The proofreading pathway of bacteriophage T4 DNA polymerase. J Biol Chem. 1998;273:22969–22976. doi: 10.1074/jbc.273.36.22969. [DOI] [PubMed] [Google Scholar]
- 24.Stocki SA, Nonay RL, Reha-Krantz LJ. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3' --> 5' exonuclease activities. J Mol Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- 25.Reha-Krantz LJ. Locations of amino acid substitutions in bacteriophage T4 tsL56 DNA polymerase predict an N-terminal exonuclease domain. J Virol. 1989;63:4762–4766. doi: 10.1128/jvi.63.11.4762-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu P, Nossal N, Benkovic SJ. Kinetic characterization of a bacteriophage T4 antimutator DNA polymerase. Biochemistry. 1998;37:14748–14755. doi: 10.1021/bi980835a. [DOI] [PubMed] [Google Scholar]
- 27.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 28.Saturno J, Lazaro JM, Blanco L, Salas M. Role of the first aspartate residue of the "YxDTDS" motif of phi29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J Mol Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 29.Blasco MA, Mendez J, Lazaro JM, Blanco L, Salas M. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J Biol Chem. 1995;270:2735–2740. doi: 10.1074/jbc.270.6.2735. [DOI] [PubMed] [Google Scholar]
- 30.Copeland WC, Wang TS. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J Biol Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- 31.Jacewicz A, Makiela K, Kierzek A, Drake JW, Bebenek A. The roles of Tyr391 and Tyr619 in RB69 DNA polymerase replication fidelity. J Mol Biol. 2007;368:18–29. doi: 10.1016/j.jmb.2007.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 33.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. Embo J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J Biol Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 35.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 37.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 38.Yang G, Lin T, Karam J, Konigsberg WH. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry. 1999;38:8094–8101. doi: 10.1021/bi990653w. [DOI] [PubMed] [Google Scholar]
- 39.Bebenek A, et al. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J Biol Chem. 2001;276:10387–10397. doi: 10.1074/jbc.M007707200. [DOI] [PubMed] [Google Scholar]
- 40.Hunter WN, Brown T, Anand NN, Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 41.Hunter WN, et al. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J Biol Chem. 1987;262:9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 42.Kennard O, Salisbury SA. Oligonucleotide X-ray structures in the study of conformation and interactions of nucleic acids. J Biol Chem. 1993;268:10701–10704. [PubMed] [Google Scholar]
- 43.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prive GG, et al. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 45.Fortune JM, et al. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 46.Capson TL, et al. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 47.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 48.Evans RJ, et al. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A. 2008;105:20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin TC, Karam G, Konigsberg WH. Isolation, characterization, and kinetic properties of truncated forms of T4 DNA polymerase that exhibit 3'–5' exonuclease activity. J Biol Chem. 1994;269:19286–19294. [PubMed] [Google Scholar]
- 50.Reddy MK, Weitzel SE, von Hippel PH. Processive proofreading is intrinsic to T4 DNA polymerase. J Biol Chem. 1992;267:14157–14166. [PubMed] [Google Scholar]
- 51.Joyce CM. How DNA travels between the separate polymerase and 3'–5'-exonuclease sites of DNA polymerase I (Klenow fragment) J Biol Chem. 1989;264:10858–10866. [PubMed] [Google Scholar]
- 52.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 53.Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat Struct Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- 54.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson RE, Prakash L, Prakash S. Yeast and human translesion DNA synthesis polymerases: expression, purification, and biochemical characterization. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- 57.Fortelle dL, Bricogne G. Maximum-likelihood heavy atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 58.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.