Abstract
Aberrant expression of microRNAs (miRNAs) and the enzymes that control their processing have been reported in multiple biological processes including primary and metastatic tumours1–6, but the mechanisms governing this are not clearly understood. Here we show that TAp63, a p53 family member, suppresses tumorigenesis and metastasis, and coordinately regulates Dicer and miR-130b to suppress metastasis. Metastatic mouse and human tumours deficient in TAp63 express Dicer at very low levels, and we found that modulation of expression of Dicer and miR-130b markedly affected the metastatic potential of cells lacking TAp63. TAp63 binds to and transactivates the Dicer promoter, demonstrating direct transcriptional regulation of Dicer by TAp63. These data provide a novel understanding of the roles of TAp63 in tumour and metastasis suppression through the coordinate transcriptional regulation of Dicer and miR-130b and may have implications for the many processes regulated by miRNAs.
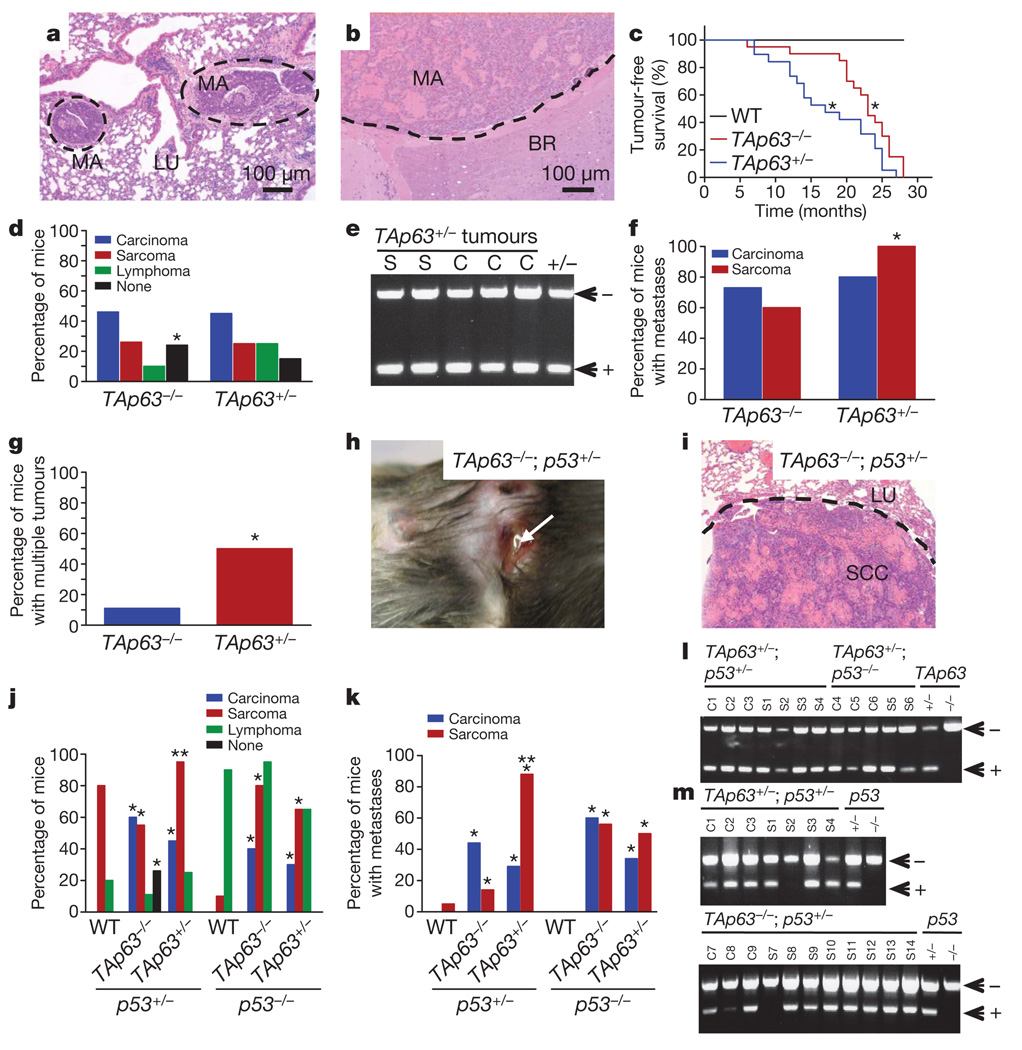
The precise roles of p63 in tumour suppression have been hotly debated7–10. Although some studies show p63 overexpression in human cancer7,8,11, some demonstrate a loss of p63 associated with tumour progression and metastasis8,12–14. Much of this controversy is due to the existence of multiple isoforms15. The full-length TA isoform of p63 bears structural and functional similarity to p53, whereas the ΔN isoforms of p63, which lack the transactivation (TA) domain, act primarily in dominant-negative fashion against p53, TAp63 and TAp73 (ref. 15). These activities suggest that TAp63 is a tumour suppressor gene and ΔNp63 is an oncogene; however, this has not been directly tested in vivo. To examine whether TAp63 is a tumour suppressor gene, we generated a cohort of 30 mice of each of the following genotypes: TAp63−/−, TAp63+/− and wild-type, and aged them for 2.5 years (Fig. 1 and Supplementary Fig. 1a, b). We found that both TAp63+/− mice and TAp63−/− mice developed spontaneous carcinomas and sarcomas (Fig. 1a–d, Supplementary Fig. 1a, b, and Supplementary Table 1) and had a significantly shorter lifespan than the wild-type cohort (Fig. 1c). Paradoxically, we noted that a larger proportion of the TAp63−/− mice (24%) were tumour free compared with TAp63+/− mice (15%) (Fig. 1c, d). These data suggest that TAp63 is a haploinsufficient tumour suppressor gene. Consistent with this finding, sarcomas (n = 10) and carcinomas (n = 10) from TAp63+/− mice retained the wild-type allele of TAp63 (Fig. 1e). TAp63+/− and TAp63−/− mice developed highly metastatic tumours (Fig. 1a, b, f, and Supplementary Fig. 1a, b), and 10% of these metastases were found in the brain (Fig. 1b), a rare finding in spontaneous mouse tumour models. Although equivalent numbers of carcinomas metastasized in the TAp63−/− and TAp63+/− mice, a greater number of sarcomas metastasized in TAp63+/− mice than in TAp63−/− mice (Fig. 1f, g). These data again indicate that heterozygosity for TAp63 results in a more severe phenotype in specific tissues.
Figure 1. TAp63−/− mice develop metastatic tumours.
a, b, Metastatic mammary adenocarcinoma in lung (LU; a) and brain (BR; b). c, Tumour-free survival curves; n = 30 and P ≤ 0.05. WT, wild type. d, Tumour spectra of the indicated mice genotypes; n = 30. e, LOH analysis of TAp63 in sarcomas (S) and carcinomas (C) from the indicated genotypes. f, g, Percentage of metastatic sarcomas and carcinomas (f) and multiple malignancies (g); n = 30. h, i, Metastatic squamous cell carcinoma (SCC) on skin (h) and in lung (LU; i). j, k, Tumour spectrum (j) and metastases (k) of the indicated mice genotypes; n = 30. Asterisk, statistical difference between TAp63/p53 mutant and p53+/− or p53−/− mice; two asterisks, statistical difference between two double-mutant genotypes; P ≤ 0.05. l, m, PCR analysis for LOH of TAp63 (l) or p53 (m) in carcinomas (C) and sarcomas (S) of the indicated genotypes.
To further understand the mechanisms employed by TAp63 as a tumour suppressor gene, we studied the effects of TAp63 on a p53+/− or p53−/− background8,16–18. Some human tumours show alterations in both p53 and p63 (refs 12, 13), and point mutant p53 binds and functionally inactivates p63 (refs 17, 18). To study these interactions, we generated six cohorts of 30 mice each: TAp63−/−;p53+/− and TAp63+/−;p53+/− for comparison with p53+/− mice, and TAp63−/−;p53−/− and TAp63+/−;p53−/− for comparison with p53−/− mice. Unlike p53+/− and p53−/− mice, TAp63/p53 compound mutant mice developed a remarkable number of metastatic tumours (Fig. 1h–k, Supplementary Fig. 1c–f and Supplementary Table 1)8,16. Whereas the TAp63+/−;p53+/− mice had a similar tumour spectrum to that of TAp63−/−;p53+/− mice (Supplementary Table 1), the sarcomas of the TAp63+/−;p53+/− mice were significantly more invasive and metastatic (88%) than those in the TAp63−/−;p53+/− cohort (14%) (Fig. 1k). In contrast, the carcinomas in the TAp63−/−;p53+/− cohort were more metastatic (44%) than those in the TAp63+/−;p53+/− group (29%) (Fig. 1k). We next examined sarcomas (n = 10) and carcinomas (n = 10) from TAp63+/−;p53+/− mice and found that the wild-type allele of TAp63 was retained (Fig. 1l). We also examined TAp63+/−;p53+/− tumours and TAp63−/−;p53+/− tumours for p53 loss of heterozygosity (LOH) and found that one out of ten sarcomas showed LOH of p53 in TAp63+/−;p53+/− tumours (Fig. 1m). A slightly higher frequency was noted in tumours from TAp63−/−;p53+/− sarcomas (one out of eight) and carcinomas (two out of eight), indicating that there is increased selective pressure to lose the wild-type p53 allele in the complete absence of TAp63 (Fig. 1m). However, the frequency of p53 LOH was lower than the previously reported frequency (ranging from 75–90%) of sarcomas from p53+/− mice8,16 and was more reminiscent of the frequency of p53 LOH in mouse models of Li–Fraumeni syndrome17, further supporting the concept that p53 and p63 interact in tumour suppression. We also analysed TAp63−/−;p53−/− and TAp63+/−;p53−/− mice and found that highly metastatic carcinomas and sarcomas developed in addition to the thymic lymphomas found in the p53−/− mice (Fig. 1j and Supplementary Table 1). Sarcomas (n = 5) and carcinomas (n = 5) from these mice retained the wild-type allele of TAp63 (Fig. 1l), providing further evidence that TAp63 is a haploinsufficient tumour suppressor gene.
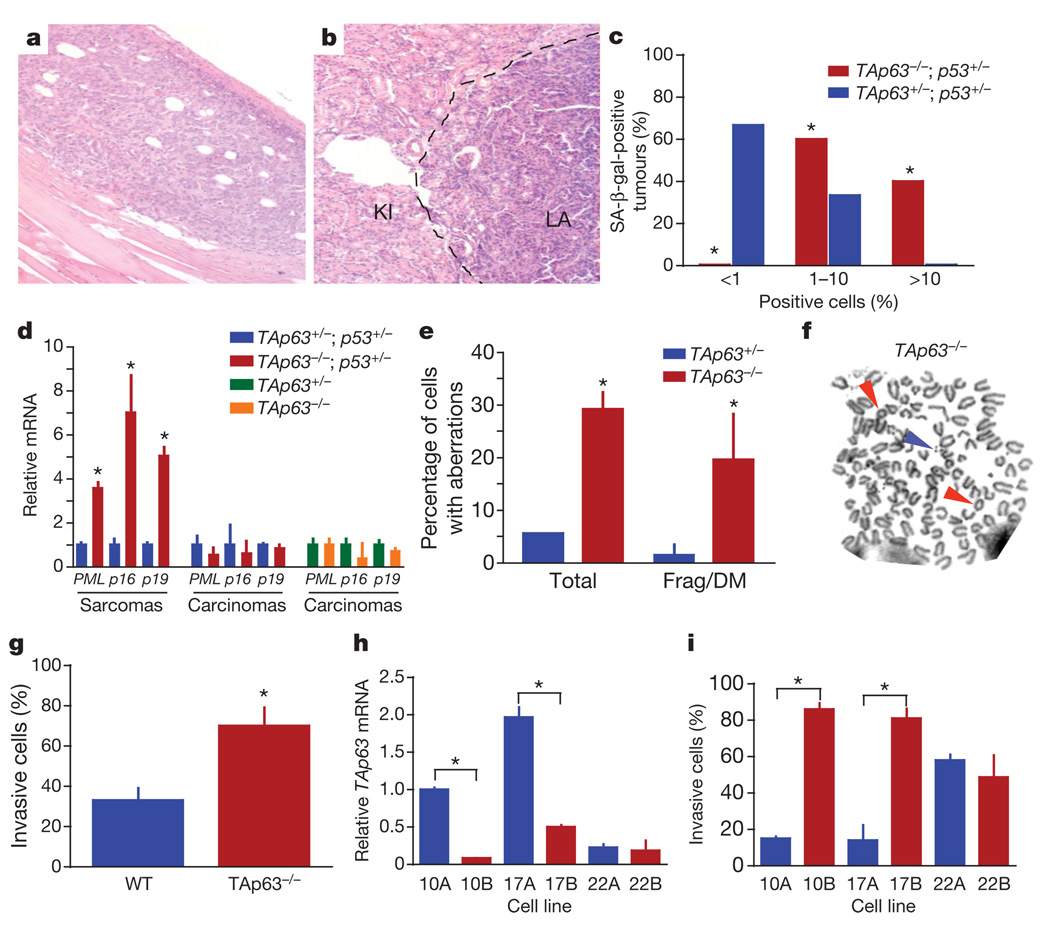
To understand how heterozygosity of TAp63 led to a more severe tumour phenotype than a complete loss of TAp63, we examined whether senescence in the TAp63−/− mice, which age prematurely19, inhibited tumorigenesis or metastasis. We found several examples of TAp63−/−;p53+/− mice that developed non-metastatic osteosarcomas together with metastatic carcinomas (Fig. 2a, b and Supplementary Fig. 1e, f). Tumours from these mice, along with sarcomas and carcinomas from TAp63+/−;p53+/− mice, were assayed for senescence-associated markers, namely SA-β-gal (Fig. 2c and Supplementary Fig. 2a, b) or PML (encoding promyelocytic leukemia protein), p16Ink4a and p19Arf (Fig. 2d). We found that osteosarcomas and rhabdomyosarcomas in the TAp63−/−;p53+/− mice expressed high levels of senescence markers, whereas the same tumour types in the TAp63+/−;p53+/− mice did not (Fig. 2c, d). None of the carcinomas from TAp63−/−;p53+/− or TAp63+/−;p53+/− mice was positive for these markers, indicating that TAp63 deficiency has a tissue-specific function in the induction of senescence. These results correlated with the level of aggressiveness and metastatic potential of these tumours.
Figure 2. TAp63-deficient tumours show high levels of senescence and genomic instability.
a, b, Non-metastatic osteosarcoma (a) and metastatic lung adenocarcinoma (LA) in kidney (KI) (b) from TAp63−/−;p53+/− mouse. c, Percentage of SA-β-gal-positive sarcomas; n = 6 of each genotype. d, Quantitative real-time polymerase chain reaction (qRT-PCR) for markers of senescence (PML, p16Ink4a and p19Arf) in the indicated tumours; n = 6 of each genotype. e, Percentage of TAp63−/− and TAp63+/− tumours with chromosomal aberrations determined by metaphase spreads; n = 6 of each genotype. Frag/DM, fragmented, double-minute chromosome. f, Representative metaphase spread from a TAp63−/− tumour. Coloured arrows indicate aberrations: rings (red arrows) and double minutes (blue arrow). g, Invasion assays in wild-type MEFs (n = 3) and TAp63−/− MEFs (n = 3). h, qRT-PCR for TAp63 in the indicated human squamous cell carcinoma (HNSCC) cell lines; n = 5. i, Invasion assays in HNSCC cell lines; n = 5. Error bars indicate s.e.m. Asterisk, P ≤ 0.05.
Genomic instability in tissues of TAp63−/− mice has been shown to be high19. To examine whether there is a similarly high level of genomic instability in tumours from TAp63−/−;p53+/− mice and whether this is correlated with tumour aggressiveness and metastasis, we performed γ-H2AX immunostaining and metaphase spreads on tumours from TAp63 mutant mice (Fig. 2e, f and Supplementary Fig. 2c–g). We did indeed find that TAp63−/− carcinomas showed high levels of γ-H2AX, polyploid cells and chromosomal aberrations, suggesting that carcinomas lacking TAp63 overcome senescence in epithelial tissues as a result of the acquisition of further genetic alterations. Although a decrease in TAp63 expression promotes metastasis in both tumour types, senescence is triggered in sarcomas, thus inhibiting progression, whereas in carcinomas, genomic instability ensues, permitting further genetic events favouring tumour progression.
Given the high level of metastasis in TAp63−/− mice, we examined whether TAp63−/− mouse embryonic fibroblasts (MEFs) had an enhanced invasive ability. TAp63−/− MEFs showed a 1.8-fold increase in invasion (Fig. 2g). To assess whether this finding could be generalized to human tumours, we examined primary human head and neck squamous-cell carcinomas (HNSCCs) (cell lines 10A, 17A and 22A) and matched metastatic lesions with markedly low levels of TAp63 (Fig. 2h) (cell lines 10B, 17B and 22B)20. Tumours with low levels of TAp63 showed an increased invasive ability (Fig. 2i), indicating that TAp63 is a critical regulator of cancer metastasis.
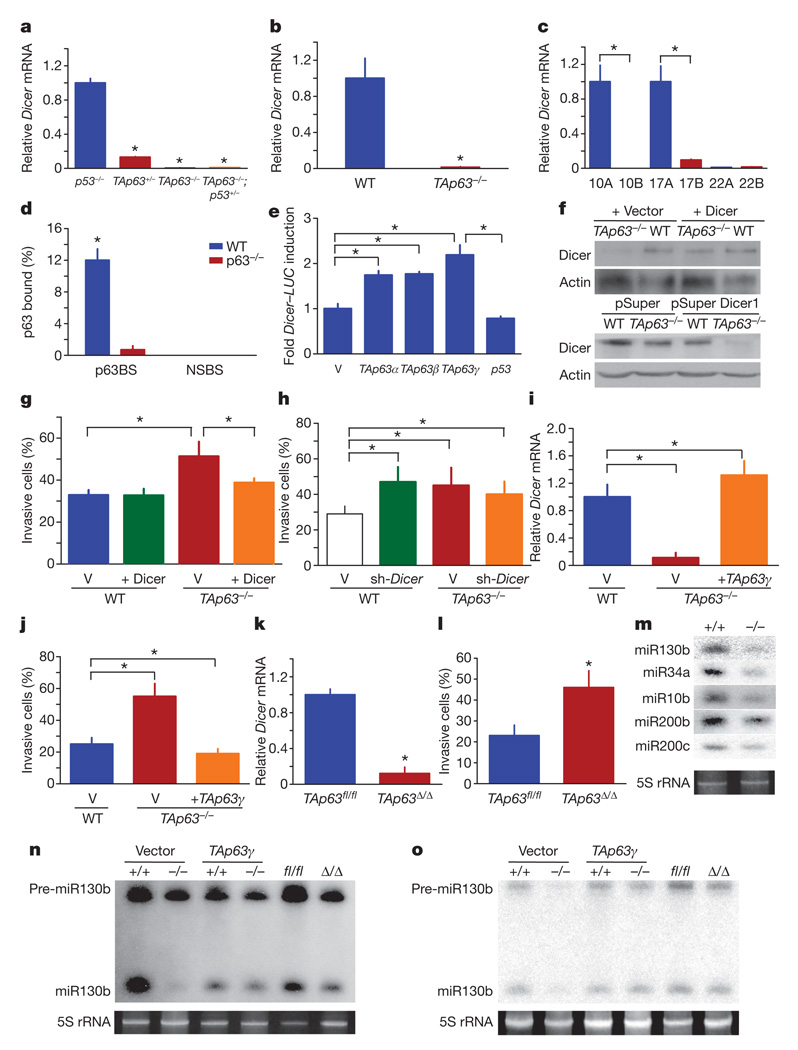
Our findings that TAp63+/− tumours are more aggressive than those from TAp63−/− mice are reminiscent of the phenotype of Dicer conditional knock out (Dicer1fl/+) mice intercrossed to the KrasLSL-G12D lung cancer model21, which indicate that Dicer is haploinsufficient for tumour suppression4, in a similar manner to our previous observations for TAp63. Additionally, Dicer1−/− mice show embryonic lethality (before embryonic day E8.5) similar to that in TAp63−/− mice on an enriched C57BL/6 background19; 35% of TAp63−/− embryos die between E6.5 and E8.5. Given these similarities, we examined whether messenger RNA levels of Dicer are dysregulated in tumours and cells lacking TAp63. TAp63−/− osteosarcomas, lung adenocarcinomas and mammary adenocarcinomas (n = 6) and MEFs expressed significantly lower levels of Dicer than p53−/− tumours and MEFs (Fig. 3a, b and Supplementary Fig. 3), suggesting that TAp63 is required for the transcription of Dicer. Similarly, we found that metastatic human HNSCCs (n = 46), lung adenocarcinomas and squamous cell carcinomas (n = 92) and mammary adenocarcinomas (n = 43) with low levels of TAp63 had low Dicer expression (Fig. 3c, Supplementary Fig. 4 and Supplementary Tables 2 and 3). These data indicate a critical role for TAp63 in the regulation of Dicer in metastasis in human cancer. Consistent with this is the observation that Dicer and Drosha levels have been shown to be low in human cancer20.
Figure 3. TAp63 regulates metastasis by transcriptional activation of Dicer.
a–c, qRT-PCR of Dicer mRNA in murine tumours (a), MEFs (b) and HNSCC cell lines (c); n = 5. d, qRT-PCR of ChIP assay using keratinocytes and indicating p63-binding site (p63BS) and nonspecific binding site (NSBS) on Dicer promoter; n = 3. e, Luciferase assay for Dicer; n = 5. V, pcDNA3 vector. f, Western blot analysis for Dicer expression in MEFs infected with the indicated vectors. g, h, Invasion assays of MEFs expressing Dicer (g) and MEFs expressing shRNA-Dicer (h); n = 3. i, qRT-PCR for Dicer in MEFs expressing TAp63γ; n = 3. j, Invasion assays of MEFs expressing TAp63γ; n = 3. k, qRTPCR for Dicer in wild-type (TAp63fl/fl) and TAp63-ablated (TAp63Δ/Δ) MEFs; n = 3. l, Invasion assays of TAp63Δ/Δ MEFs; n = 3. m, Northern blot analysis for the indicated mature miRNAs in wild-type and TAp63−/− MEFs. n, o, Northern blot analysis for miR-10b (n) or miR-130b (o) using MEFs of the indicated genotypes expressing vector (V) or TAp63γ. Error bars indicate s.e.m. Asterisk, P ≤ 0.05.
To determine whether Dicer is a transcriptional target of TAp63, we performed chromatin immunoprecipitation (ChIP) analysis for a site matching the p63 consensus binding site (Supplementary Table 4). We found that p63, and not p53, bound to the Dicer promoter (Fig. 3d and Supplementary Fig. 5a), suggesting that Dicer is a direct transcriptional target of p63. In addition, all isoforms of TAp63 (α, β and γ) were able to transactivate a Dicer–luciferase reporter gene with the p63-binding site (Fig. 3e), further indicating that TAp63 regulates Dicer transcriptionally.
To understand whether TAp63 controls metastasis through transcriptional regulation of Dicer, we re-expressed Dicer in TAp63−/− MEFs (Fig. 3f). These cells lost their ability to invade (Fig. 3g). Conversely, wild-type MEFs expressing a Dicer short hairpin RNA (shRNA)22 (Fig. 3f) showed increased invasiveness (Fig. 3h). These data indicate that low levels of Dicer lead to increased cell invasion, similar to that observed in TAp63-deficient cells.
To further explore the connection between TAp63 and Dicer in metastasis, we modulated levels of TAp63 in MEFs by expressing TAp63γ in TAp63−/− MEFs and human HNSCCs (Supplementary Figs 5b and 6a) and measured Dicer expression and invasion potential. Levels of Dicer increased to wild-type levels in TAp63−/− cells expressing TAp63γ (Fig. 3i and Supplementary Fig. 6b), which resulted in decreased invasion potential (Fig. 3j and Supplementary Fig. 6c). We also acutely deleted TAp63 after the introduction of adenovirus-Cre in TAp63fl/fl MEFs (Supplementary Fig. 5c). These TAp63Δ/Δ MEFs showed concomitant downregulation of Dicer (Fig. 3k) and a twofold increase in cellular invasion (Fig. 3l), indicating that regulation of Dicer by TAp63 is critical in the suppression of invasion.
Because Dicer expression is low in the absence of TAp63, we assessed the processing of miRNAs that have a recognized role in lung and mammary adenocarcinoma metastasis1–3,23, namely miR-10b, miR-200b, miR200c, miR34a and miR-130b, in TAp63−/− MEFs and found that their processing is defective (Fig. 3m). To examine whether modulating levels of TAp63 in cells would affect miRNA processing, we scored for processing of miR-10b in TAp63−/− MEFs reconstituted with TAp63γ and in TAp63-ablated (TAp63Δ/Δ) MEFs. When TAp63 was re-expressed, we observed a rescue of miRNA processing, and when TAp63 was acutely deleted, we observed a diminution of miRNA processing (Fig. 3n). The precursor of miR-130b (pre-miR-130b) was present at low levels in TAp63−/− MEFs (Fig. 3o), indicating that the primary transcript of miR-130b may be expressed at low levels in the absence of TAp63.
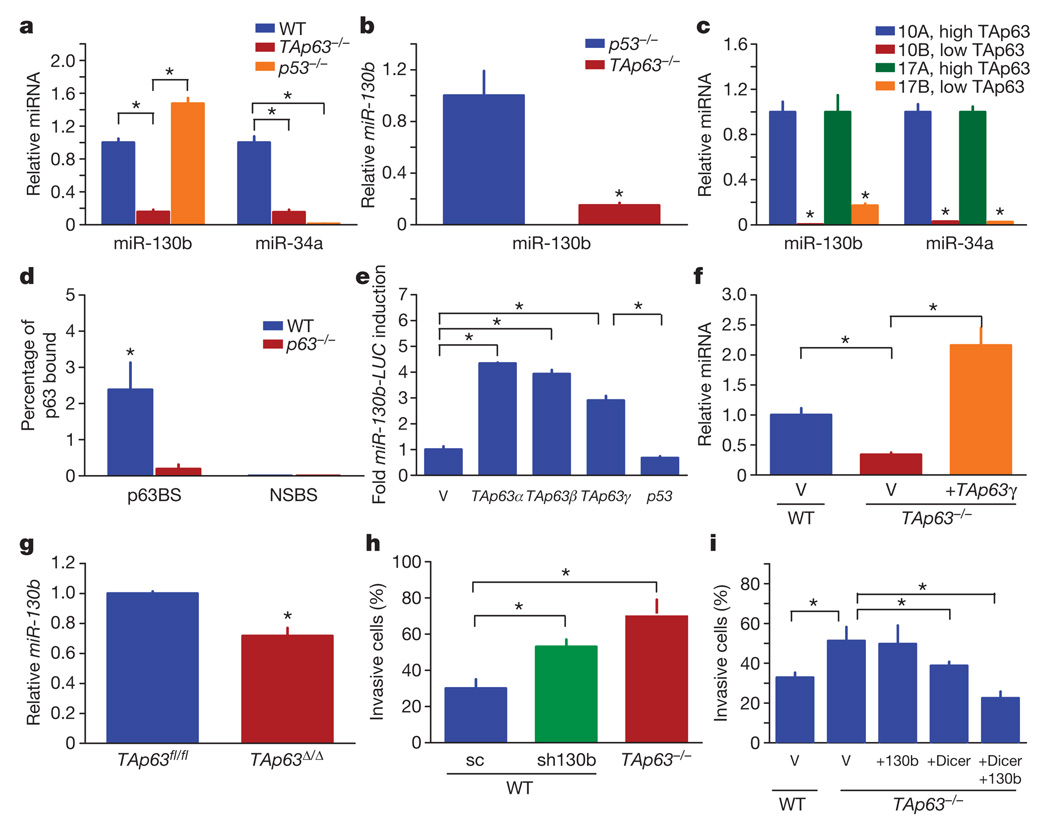
We found that levels of miR-130b were downregulated in TAp63−/− MEFs, suggesting a role for TAp63 in its regulation (Fig. 4a). We reasoned that other miRNAs, such as miR-34a, which have been shown to be a transcriptional target of p53 (ref. 23), may also be a target of TAp63. Indeed, miR-130b and miR-34a were downregulated in the absence of TAp63 (Fig. 4a–c and Supplementary Fig. 7a). miR130b was also low in high-grade lung adenocarcinomas and HNSCCs with low levels of TAp63 and Dicer (Supplementary Table 3). To assess whether p63 binds to the promoters of these miRNAs, we performed a ChIP analysis with identified putative p63-binding sites (Supplementary Table 4). We found a significant level of p63 binding at both the miR-130b and miR-34a promoters (Fig. 4d and Supplementary Fig. 7b–d). To determine whether p63 can directly transactivate miR-130b and miR-34a, we performed luciferase assays (Fig. 4e and Supplementary Fig. 7e) and found that TAp63 transactivated the miR130b reporter. We modulated the levels of TAp63 by overexpression or acute deletion in MEFs and found that levels of miR-130b correlated with levels of TAp63 (Fig. 4f, g). Taken together, these data indicate that miR-130b is a direct target of TAp63.
Figure 4. TAp63 regulates miR-130b in metastasis.
a–c, TaqMan reverse-transcriptase-mediated PCR (RT–PCR) for miR-130b and miR-34a in MEFs (a), murine tumours (b) and HNSCCs (c); n = 5. d, qRT-PCR of ChIP assay for miR-130b indicating p63-binding site (p63BS) and non-specific binding site (NSBS); n = 3. e, Luciferase assay for miR-130b; n = 3. f, g, TaqMan RT–PCR for miR-130b in wild-type and TAp63−/− MEFs expressing the indicated vectors (f) and wild-type (TAp63fl/fl) and TAp63-ablated (TAp63Δ/Δ) MEFs (g); n = 3. h, i, Invasion assays of MEFs infected with shRNA-miR-130b (sh-130b) (h) and wild-type and TAp63−/− MEFs expressing the indicated vectors (i); n = 3. Scrambled sequence (sc) shRNA was used as a negative control. Error bars indicate s.e.m. Asterisk, P ≤ 0.05.
To understand whether regulation of miR-130b by TAp63 has a function in the metastatic phenotype, we modulated levels of miR-130b in MEFs (Supplementary Fig. 7f–h) and assessed invasion (Fig. 4h, i). Wild-type MEFs with low levels of miR-130b showed increased invasion (Fig. 4h), and TAp63−/− MEFs expressing both miR-130b and Dicer showed decreased invasion (Fig. 4i) comparable to that of wild-type MEFs. This suggests that coordinate regulation of both Dicer and miR-130b by TAp63 is critical in suppressing metastasis.
We have shown that TAp63 is a suppressor of tumorigenesis and metastasis and that its complete inactivation results in tissue-specific antagonistic pleiotropy with respect to tumour progression including genomic instability and activation of senescence. Both the spectrum and the highly metastatic nature of tumours in TAp63-deficient mice are consistent with a role for TAp63 as a master regulator of metastasis. Furthermore, we have shown a role for transcriptional regulation by TAp63 of Dicer and miR-130b in metastasis. Given the diverse biological roles of both Dicer and TAp63, a complete understanding of the regulation of Dicer by TAp63 in other processes is a key question for the future.
METHODS SUMMARY
Generation of mice and tumour analysis
TAp63−/− mice19 were intercrossed with p53−/− mice16 to generate compound mutant TAp63/p53 mice on an enriched C57BL/6 background ((C57BL/6 = 95%) and (129/SvJae = 5%)). For analysis of tumour formation and metastatic disease, 30 mice of each genotype were aged for 2.5 years. Moribund mice were killed, following the approved guidelines of the IACUC at the University of Texas M. D. Anderson Cancer Center and analysed histologically by haematoxylin/eosin (H&E) staining as described previously8. Tumour-free survival of mice was plotted with PRISM5 software (GraphPad).
Migration and invasion assay
MEFs or mouse and human tumour cell lines were resuspended in 500 µl of DMEM with 10% FBS and 1 × penicillin/streptomycin (5 × 104 cells) and added to the top of each chamber containing BD BioCoat cell culture inserts (354578; BD Biosciences) or Matrigel Invasion Chamber (354480; BD Biosciences)19,24. Non-invasive cells were removed from the upper chamber. Cells remaining on the membrane were fixed, stained with Diff-Quik (Dade Behring, Inc.), and counted with a Zeiss AxioObserver A1 inverted microscope.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
We thank C.-g. Liu for technical assistance with miRNA analyses and A. Multani for metaphase analyses (funded by NCI CA16672); K. Y. Tsai for scientific discussions and comments on the manuscript; and D. Yu, J. Jackson and S. Rehman for reagents and advice. This work was supported by grants to E.R.F. from the American Cancer Society (RSG-07-082-01-MGO), the Susan G. Komen Foundation (BCTR600208) and the Hildegardo E. and Olga M. Flores Foundation. This work was supported in part by an NCI-Cancer Center Core Grant (CA-16672) (University of Texas M. D. Anderson Cancer Center), a Career Development Award from the Genitourinary Cancer SPORE (P50CA091846) to E.R.F., Lung Cancer SPORE (P50CA070907) to I.W. and U01DE019765 to A.E.-N. E.R.F. is a scholar of the Rita Allen Foundation and the V Foundation for Cancer Research.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions X.S., D.C. and E.R.F. designed the experiments and analysed the data. X.S., D.C., M.S.C., L.L., Y.J.G., Y.-L.L. and M.L.L. performed the experiments. A.E.-N. performed pathology analysis and provided human HNSCC samples. M.B.S. and I.W. provided human lung adenocarcinoma pathology information and RNA samples. C.J.C. performed statistical analysis on miRNA data. X.S., D.C. and E.R.F. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Baffa R, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 2.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449 doi: 10.1038/nature06174. 682-628. [DOI] [PubMed] [Google Scholar]
- 4.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores ER. The roles of p63 in cancer. Cell Cycle. 2007;6:300–304. doi: 10.4161/cc.6.3.3793. [DOI] [PubMed] [Google Scholar]
- 8.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Keyes WM, et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc. Natl Acad. Sci. USA. 2006;103:8435–8440. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 11.Hibi K, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl Acad. Sci. USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park BJ, et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60:3370–3374. [PubMed] [Google Scholar]
- 13.Urist MJ, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am. J. Pathol. 2002;161:1199–1206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park HR, et al. Low expression of p63 and p73 in osteosarcoma. Tumori. 2004;90:239–243. doi: 10.1177/030089160409000214. [DOI] [PubMed] [Google Scholar]
- 15.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 16.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 17.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Su X, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruya S, et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin. Cancer Res. 2004;10:3825–3830. doi: 10.1158/1078-0432.CCR-03-0370. [DOI] [PubMed] [Google Scholar]
- 21.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 23.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores ER, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.