Abstract
Aminoacyl-tRNA synthetases of higher eukaryotes possess polypeptide extensions in contrast to their prokaryotic counterparts. These extra domains of poorly understood function are believed to be involved in protein–protein or protein–RNA interactions. Here we showed by gel retardation and filter binding experiments that the repeated units that build the linker region of the bifunctional glutamyl-prolyl-tRNA synthetase had a general RNA-binding capacity. The solution structure of one of these repeated motifs was also solved by NMR spectroscopy. One repeat is built around an antiparallel coiled-coil. Strikingly, the conserved lysine and arginine residues form a basic patch on one side of the structure, presenting a suitable docking surface for nucleic acids. Therefore, this repeated motif may represent a novel type of general RNA-binding domain appended to eukaryotic aminoacyl-tRNA synthetases to serve as a cis-acting tRNA-binding cofactor.
Keywords: NMR spectroscopy/protein–RNA interactions/RNA-binding domain/tRNA synthetase
Introduction
Aminoacyl-tRNA synthetases (aaRSs) catalyze the aminoacylation of tRNA molecules, the main decoding step in the transfer of genetic information (Schimmel and Söll, 1979). Protein–RNA interactions play a central role in this essential process (Arnez and Cavarelli, 1997). aaRSs from eukaryotes possess specific protein modules, absent in their prokaryotic counterparts (Mirande, 1991). These supplementary domains are generally found as N- or C–terminal polypeptide extensions, with sizes ranging from ∼40 to 300 amino acid residues for the polypeptide extensions of human seryl- and valyl-tRNA synthetases, respectively. The function of these extensions is poorly understood, but they provide eukaryotic synthetases with polyanion-binding properties (Alzhanova et al., 1980; Mirande, 1991).
From fly to human, the multifunctional glutamyl-prolyl-tRNA synthetase (EPRS) displays a eukaryotic-specific domain made of several units of a conserved motif of 50 amino acid residues (Cerini et al., 1991; Fett and Knippers, 1991). This motif is repeated in tandem in three or six units that build the linker region connecting the two synthetase domains. A homologous sequence pattern also exists as singular or plural elements in six monofunctional aaRSs where it constitutes N- or C–terminal extensions (Figure 1). Glutamyl- or prolyl-tRNA synthetases lacking this repeated segment are still active, suggesting that it is not essential for catalysis (Cerini et al., 1991; Kerjan et al., 1992; Ting et al., 1992; Stehlin et al., 1998). However, human histidyl-tRNA synthetase or Bombyx mori glycyl-tRNA synthetase with a deletion of this repeat at their N–terminus were found to have lost about one order of magnitude of catalytic activity (Raben et al., 1994; Wu et al., 1995). The in vivo physiological relevance of this conserved motif has been supported by the observation that overexpression of the six tandemly repeated motifs of EPRS in transgenic flies led to a sterility phenotype (Cerini et al., 1997).
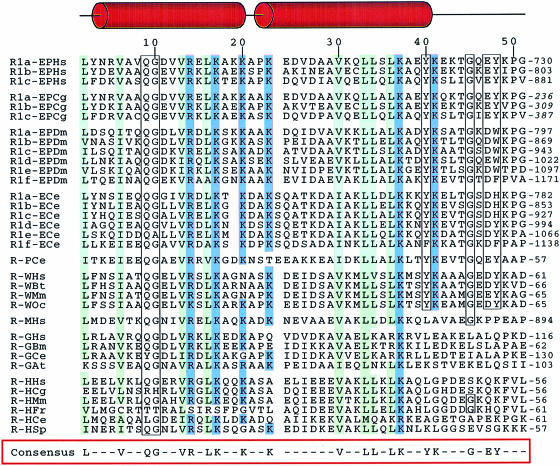
Fig. 1. Alignment of the conserved RNA-binding motifs appended to eukaryotic aaRS. The first capital letters of the protein represent the amino acid substrate, while the final two letters are for the species [Hs (Homo sapiens) stands for human, Cg (Cricetulus griseus) for hamster, Dm (Drosophila melanogaster) for fly, Ce (Caenorhabditis elegans) for nematode, Bt (Bos taurus) for cow, Mm (Mus musculus) for mouse, Oc (Oryctolagus cuniculus) for rabbit, Bm (Bombyx mori) for silkworm, At (Arabidopsis thaliana) for cress, Fr (Fugu rubripes) for pufferfish and Sp (Schizosaccharomyces pombe) for yeast]. Residues that match the consensus sequence (defined as residues conserved in 80% of the repeated sequences) are boxed. Conserved hydrophobic residues are in green, basic residues are in blue. The sequence numbers given on the top line relate to the sequence of the R1b motif from hamster used to determine its solution structure. The regions that form helices in R1b are indicated above the sequence alignment.
The native form of the B.mori glycyl-tRNA synthetase displays general RNA-binding properties, whereas an N–terminally truncated derivative does not (Wu et al., 1995). The repeats of EPRS from human (Rho et al., 1998) have also been characterized as RNA ligands. By affinity co-electrophoresis, several nucleic acids were shown to form stable complexes. However, this domain shows no significant sequence homology with any other known RNA-binding protein. On the other hand, these repeated units have also been involved in generating protein–protein interaction sites. The repeats of EPRS were shown to bind isoleucyl-tRNA synthetase with an apparent dissociation constant of 3 μM (Rho et al., 1998).
In this work, we tested and established that the linker domain of EPRS, made of three repeated units, referred to as the R3 domain, had general RNA-binding properties. Moreover, one of these repeats, referred to as the R1 domain, also displayed an RNA-binding ability. To get insights into the mode of RNA–protein recognition, we elucidated the three-dimensional structure of this novel RNA-binding domain. We present here the solution structure of R1b, the second unit of the repeats of hamster EPRS. This represents the first available structure of such a repeated unit and allows the function of this recurrent motif to be addressed.
Results
RNA binding to single or plural elements of the repeats
The polypeptide domain (designated R3) that covalently links the glutamyl- and prolyl-tRNA synthetase domains of hamster EPRS is made of three imperfectly repeated units designated R1a, R1b and R1c (Figure 1). The repeated units of hamster EPRS were cloned into the pET28b expression vector that provided a C–terminal His6 tag. The 210 amino acid R3 protein, made of the three repeats, was purified to homogeneity on a Ni–NTA affinity column followed by Source–15S chromatography. This last purification step was required to remove nucleic acids that were co-eluted with the protein. The contaminating nucleic acid fraction, which could be digested with RNase A but not with DNase I, was mainly composed of RNA. The purified R3 fraction displayed an A280/A260 ratio of 1.8, typical of a protein fraction free of nucleic acids.
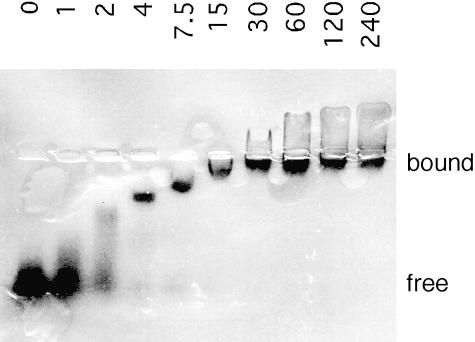
The purified recombinant R3 protein was used in a gel retardation assay to test its ability to bind various RNA substrates. The four homopolymers of 5′–32P–labeled ribonucleotides poly(A), poly(C), poly(U) and poly(G) were tested for their capacity to form RNA–protein complexes with R3. As shown in Figure 2 with poly(G), R3 binds single-stranded RNA ligands with an apparent dissociation constant of ∼2.5 μM. All the four homopolymers contributed essentially similar patterns, with the exception of poly(U), which displayed an ∼10–fold reduced affinity for R3. In addition, we observed binding of R3 to in vitro transcribed tRNAs, an RNA ligand rich in secondary structure, with apparent KDs in the 1–10 μM range (results not shown). Hence, the ability of R3 to bind RNA is neither sequence nor structure specific.
Fig. 2. Gel retardation experiment exemplifying the general RNA-binding capacity of the repeats. RNA homopolymers of guanylate nucleotide were end-labeled with 32P using polynucleotide kinase. R3 (0–240 μM) was incubated with 10 nM [32P]poly(G) in the incubation buffer at 25°C for 15 min. After electrophoresis at 4°C on a native agarose gel, the mobility shift of poly(G) was visualized by autoradiography.
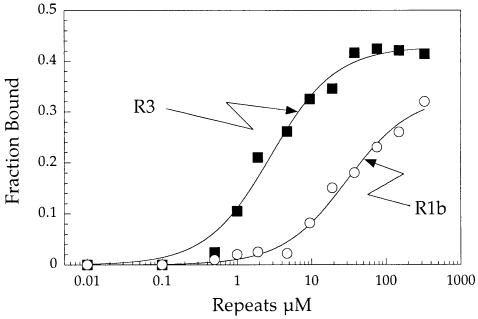
We also produced in Escherichia coli a recombinant protein, designated R1b (with a His6 tag), made of the second repeated unit of R3. We observed that R1b can also bind to single- and double-stranded RNA ligands. The RNA-binding properties of R3 and R1b for poly(G) were compared in a filter binding assay (Figure 3). The isolated single-domain R1b showed a reduced affinity for RNA (apparent KD of 30 ± 4 μM) compared with the tri-repeated domain R3 (apparent KD of 2.9 ± 0.4 μM). The stronger RNA-binding capacity of R3 can be explained by the synergistic effect contributed by the presence of three independent RNA-binding domains of the R1b type. The RNA-binding affinities of R3 and R1b are comparable to those determined for other well documented RNA-binding domains associated to eukaryotic aminoacyl-tRNA synthetases. The p43 component of the mammalian multisynthetase complex and the N–terminal appended domain of yeast glutaminyl-tRNA synthetase bind tRNA with apparent KDs of ∼40 and 1 μM, respectively (Quevillon et al., 1997; Wang and Schimmel, 1999). In contrast, the yeast and bacterial counterparts of mammalian p43, Arc1p (Simos et al., 1998) and Trbp111 (Morales et al., 1999), bind much more strongly to RNA (KD in the 10–30 nM range).
Fig. 3. Analysis of the association of poly(G) to the repeats by a filter binding assay. R3 or R1b was incubated at increasing concentrations (0.01–330 μM) with 30 nM [32P]poly(G) as described in the legend to Figure 2. The incubate was applied to nitrocellulose filters (Millipore) to recover radioactivity associated to the proteins. The apparent dissociation constants for poly(G) were determined by non-linear regression of a binding equation to the experimental data. Values of 2.9 and 30 μM were obtained for R3 and R1b, respectively.
Determination of the three-dimensional structure of R1b
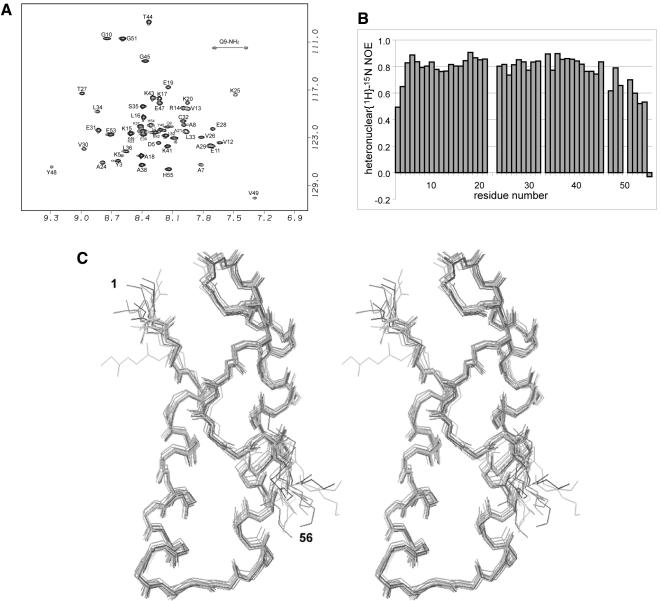
To investigate the mode of RNA–protein interaction that contributes an essentially non-specific association of the repeats with RNA ligands, the solution structure of Cricetulus griseus R1b was determined. Two-dimensional and three-dimensional heteronuclear NMR spectra were obtained using unlabelled and uniformly 15N–labelled protein samples. The spectra were well resolved (Figure 4A) and allowed for almost complete assignments of the 1H and 15N resonances. We have also undertaken a brief study of the dynamics of the protein by measuring {1H}–15N steady state heteronuclear NOEs. The N- and C–terminal (1–2 and 56–59) regions of R1b give poor NMR signals. This lack of signal, and hence of structural constraints, is associated with a high level of the dynamics of the neighbouring NH vectors (Figure 4B) and results from disordered regions. The rest of the molecule is more rigid and its structure could be determined to high resolution (Figure 4B, C and Table I).
Fig. 4. (A) 1H–15N HSQC spectrum of R1b. The assignment is labelled with individual amino acid and residue number. (B) Heteronuclear 1H–15N NOE measurement on R1b, showing the relatively high mobility of the Ω–loop and C–terminus. High values of the heteronuclear NOE reflect decreased mobility, typical values for rigid proteins are above 0.7. (C) Stereo view of the 15 calculated structures of R1B (1–56). Backbone atoms are superimposed to the mean structure from residues 3 to 53.
Table I. Structural statistics of the R1B structure calculation.
| R.m.s. deviations from experimental restraints (Å) | |
| NOE (730)a | 0.029 ± 0.001 |
| H-bonds (65) | 0.030 ± 0.006 |
| R.m.s. deviations from experimental dihedral | |
| restraints (°) (40) | 0.875 ± 0.089 |
| Deviation from idealized covalent geometry | |
| bonds (pm) | 0.310 ± 0.006 |
| angles (°) | 0.570 ± 0.018 |
| impropers (°) | 0.764 ± 0.028 |
| van der Waals energy (kJ/mol)b | –281 ± 5.3 |
| Coordinate precision (Å) | |
| backbone atoms (residues 3–53) | 0.316 ± 0.093 |
| all heavy atoms (residues 9–56) | 0.858 ± 0.131 |
aIncludes 307 intraresidual, 181 sequential, 135 short-range (1 < |i–j| < 5) and 107 long-range NOEs.
bThis term is the van der Waals energy computed with Lennard–Jones parameters in X-PLOR (Brünger, 1993). It was not included in the simulated annealing or restrained minimization protocols.
R1b exhibits a well defined structure from residue 3 to 52. The main structural element is an antiparallel coiled-coil constituting two α–helices located from residue Asp4 to Glu19 and from Lys23 to Tyr40. These helices are amphipathic, with one side being polar and the other side highly hydrophobic. They present an angle of 14° between them. The first helix ends with Lys20 and Pro22 being in αl and trans conformations respectively, as shown by specific NOE cross-peaks. A seven residue Ω–loop connects Tyr40 to Tyr48, allowing the three tyrosine side-chains (Tyr3, Tyr40 and Tyr48) to be in close proximity. This structure is made possible by the presence of a small very conserved glycine residue at position 45. The R1b domain is thus organized around a long hydrophobic core composed of the hydrophobic side of each of the helices and the three aromatic Tyr residues that lock the coiled-coil conformation. Finally, the peptidic chain folds back onto helix1 in an extended conformation.
The three-dimensional structure suggests the possible formation of some intrahelical salt bridges or hydrogen bonds involving side-chains that stabilize the helical conformations (i–i + 3 or i–i + 4). Among these, we can identify the Glu11–Arg14, Lys15–Glu19, Thr27–Glu31 and Lys25–Glu28 bonds. An interhelical salt bridge between Lys5 and Glu39 is also highly probable. This is further supported by the good conservation of the first three pairs of amino acids (Figure 1). Lys20 and Lys41 are also candidates for C–capping helix1 and helix2, respectively.
Residues Leu52 and Glu53 as well as the six C–terminal histidine residues have been added to the R1b domain for its expression in E.coli and purification (see Materials and methods). The side-chain of Leu52 is connected to the hydrophobic core of the protein as shown by NOE contacts between Leu52 and Val13 side-chains. As this residue is non-native but seems to dictate the back-folding of the chain by hydrophobic interactions, the structure of the C–terminus of the protein (after Val49) may not represent the native structure. We presume instead that the well conserved Pro50–Gly51 motif, very often associated with β turns II (Yang et al., 1996), would drive the peptidic chain away from the coiled-coil domain in the native protein. Furthermore, an increased local mobility of the protein backbone after Tyr48, and especially after Gly51, is shown by the {1H}–15N steady state heteronuclear NOEs (Figure 4B).
Conserved residues and comparison with other repeated motifs of synthetases
In eukaryotes, several members of the aaRS family contain polypeptide domains homologous to R1b (Figure 1). Sequence homology between them is high and concerns two categories of residues. The hydrophobic residues that stabilize the coiled-coil structure constitute the first one. The second one gathers the highly conserved amino acids that do not seem to play a significant structural role in the overall fold and are thought to be functionally important: they are either basic (Arg14, Lys17, Lys20, Lys23, Lys37) or aliphatic (Val30, Leu34). Figure 5A displays these residues on the R1b structure and it is interesting to note that they are spatially close. They lie on one face of the helical domain. This structural motif, comprising residues Asp4 to Tyr40, is well conserved among all the members of this family.
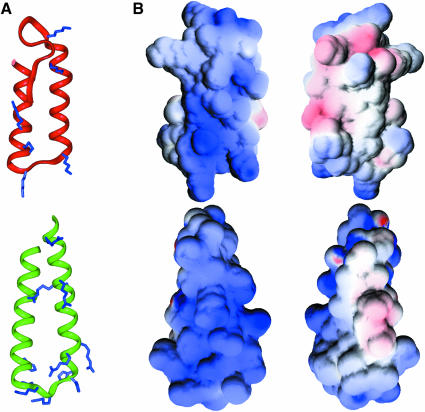
Fig. 5. (A) Ribbon diagram of the lowest energy structure of R1B (3–51) (top), compared with another RNA-binding protein, S15 (24–71) (bottom). The conserved basic residues in both families are indicated. (B) Electrostatic surface potential of R1B [in front view, same orientation as in (A)] and S15 [slightly rotated compared with (A)], and 180° rotated view. Positive and negative charges are shown in blue and red, respectively. The figures were generated using GRASP (Nicholls et al., 1991).
According to the sequence alignment of Figure 1, we can also distinguish two sub-classes of motifs: the first one, which contains R1b, is characterized by the presence of aromatic residues at positions 40 and 48 and a glycine at position 45; while the second sub-class, corresponding to the repeats of MetRS, GlyRS and HisRS, lacks these residues. The structure of the 40–48 region seems to be dictated by the close packing of Tyr40 and Tyr48 with Tyr3, so we can presume the Ω–loop to be only present in the first sub-class of repeated elements. The sequences that build these loops are well conserved (six residues out of nine). The structural organization in the second family found in MetRS, GlyRS and HisRS remains to be determined.
Comparison with other RNA-binding motifs
Unlike DNA binding proteins, a very small number of RNA–protein complex structures (apart from those involving tRNA) have been solved so far. For most of the RNA-binding proteins, little is known on the molecular recognition of RNA, and the wide variety of structural patterns adopted by RNA molecules does not ease the construction of models. The solution structure of R1b shares a helix–turn–helix architecture with a variety of protein sub-domains that can be superimposed to R1b with a root mean square deviation (r.m.s.d.) under 2 Å: ribosomal protein S15 (Berglund et al., 1997; Clemons et al., 1998) (residues 14–48 compared with residues 4–20 and 22–39 of R1b); influenza A virus NS1 protein (Chien et al., 1997; Liu et al., 1997) (residues 11–46 compared with residues 4–39 of R1b); tRNA-binding helical arm of seryl-tRNA synthetase (Biou et al., 1994) (residues 45–61 and 66–83 compared with residues 4–20 and 22–39 of R1b). Despite this structural homology, no sequence homology can be recovered from these proteins. In the examples mentioned above, the RNA-binding areas have been putatively assigned (S15 or NS1) or identified in RNA–protein complexes (SerRS) as regions of high positive potential exposed at the surface of the proteins.
The S15 protein is an 89–residue protein constitutive of the small, 30S subunit of the ribosome. Its central domain exhibits the same helix–turn–helix fold as R1b (Figure 5). The connection between the two helices contains, like R1b, a high number of basic residues that are conserved among bacterial species, and a more hydrophobic domain at the other end. This protein interacts with the 16S ribosomal RNA at a three-way helical junction, but the basis of the RNA recognition is not yet known (Clemons et al., 1998). The basic patch is supposed to contribute an RNA-binding site (Figure 5). Similarly, the structure of seryl-tRNA synthetase from Thermus thermophilus bound to tRNASer (Biou et al., 1994) shows that the N–terminus of the protein is a coiled-coil domain that interacts with the variable arm of tRNASer. This association is essentially non-specific and is mediated mostly by interactions between lysine and arginine side-chains and the phosphate–sugar backbone of tRNA. The contact area produces a patch of positive potential.
The global helix–turn–helix fold of R1b allows the clustering of the basic residues that are strictly conserved in all the members of that family. As shown in Figure 5B, they form a continuous area of positive potential exposed on the surface of R1b that builds a putative RNA-binding site. R1b thus displays an electrostatic plot similar to that found at the RNA-binding sites of the prokaryotic S15 ribosomal protein and the helical arm of seryl-tRNA synthetase.
Discussion
Our results show that the linker polypeptide from the bifunctional EPRS is composed of several tandemly repeated RNA-binding domains built around an antiparallel coiled-coil architecture. The R1b topology resembles that of bona fide RNA-binding domains, such as the helical arm of seryl-tRNA synthetase (Biou et al., 1994), the helical folds of ribosomal protein S15 (Berglund et al., 1997; Clemons et al., 1998) or viral NS1 protein (Chien et al., 1997; Liu et al., 1997). In accordance with the finding that R3 or R1b binds non-selectively to RNA targets, the general RNA-binding capacity seems to be controlled by the arrangement of the conserved basic residues at the surface of the domain. The putative RNA-binding site identified by the analysis of surface electrostatic properties of the R1b structure is mainly composed of conserved lysine and arginine residues. Therefore, the binding activity of R1b almost certainly involves non-specific interactions between side-chains of the residues from the conserved basic patches and the phosphate–sugar backbone of RNA. Our RNA-binding experiments show that the affinity of an isolated domain for RNA is low (30 μM) compared with the complete domain made of three repeated units (2.9 μM). Thus, our results suggest that the occurrence of repeated units has been selected in evolution to enhance the RNA-binding ability of this recurrent domain.
Several monofunctional eukaryotic aaRSs also share a similar structural unit (Figure 1). ProRS, TrpRS, GlyRS and HisRS are dimers, and therefore possess two identical repeats; GluRS is a monomer, but the enzyme from Caenorhabditis elegans has a C–terminal extension made of six repeats; human MetRS possesses a single repeat and is a monomer, but is associated within the multisynthetase complex that also comprises the repeated units of EPRS. Thus, none of the registered motifs can be ascribed to an isolated domain. This observation is also consistent with the interpretation that the function of this RNA-binding domain requires the involvement of several units. It is also noteworthy that these homologous domains are fused to aaRSs of different specificities. Therefore, they are unlikely to be directly involved in the selection of the cognate tRNA by the synthetase. Accordingly, the deletion of the motifs of EPRS had no detectable effect on the activity of the two synthetases (Cerini et al., 1991; Kerjan et al., 1992; Ting et al., 1992; Stehlin et al., 1998), whereas HisRS (Raben et al., 1994) and GlyRS (Wu et al., 1995) partially lost their aminoacylation activity after deletion of their extensions. Although the mode of RNA–protein interaction is not known, the structural study of R1b provides a rational basis for further mutational analysis. The binding activity of the motifs from this family (Figure 1) should then rest on the electrostatic potential of a particular motif, on the number of motifs (1, 3 or 6) associated together, and on the synthetase to which they are associated.
Other putative RNA-binding domains appended to different eukaryotic aaRSs have been characterized. Yeast GlnRS possesses a large 228-amino-acid polypeptide extension ascribed to a cis-acting cofactor of the enzyme catalytic domain (Whelihan and Schimmel, 1997; Wang and Schimmel, 1999). The yeast protein Arc1p has been described as a trans-acting cofactor of MetRS and GluRS (Simos et al., 1998). Interestingly, the non-specific RNA-binding domain of Arc1p can functionally replace the unrelated RNA-binding domain of yeast GlnRS (Wang and Schimmel, 1999). The p43 auxiliary protein from the multisynthetase complex of higher eukaryotes displays an RNA-binding domain homologous to Arc1p (Quevillon et al., 1997) and modulates the activity of ArgRS (Park et al., 1999). A p43-like domain is also appended to the C–terminus of human TyrRS (Kleeman et al., 1997), but its removal had no effect on the activity of the synthetase (Wakasugi and Schimmel, 1999).
If the function of these appended domains is to enhance the activity of aaRSs, the non-specific RNA-binding domains could increase the rate of association of tRNA to the synthetase. On the other hand, the lack of specificity suggests that the general RNA-binding domains appended to eukaryotic aminoacyl-tRNA synthetases are not directly involved in the selection and delivery of tRNA to the enzyme. Deutscher and coworkers have provided experimental evidence in favour of a tRNA channelling cycle in eukaryotic cells (Negrutskii and Deutscher, 1991; Stapulionis and Deutscher, 1995). According to their model, tRNA would be vectorially transferred from the synthetase to the elongation factor and further on to the ribosomes without dilution into the cellular fluid. We suggest that, in vivo, these repeated motifs facilitate the delivery of tRNA molecules to aminoacyl-tRNA synthetases by creating a gradient of positive potential. This would be a means to increase the local concentration of tRNA to improve the efficacy of the protein translation machinery in eukaryotic cells.
Materials and methods
Protein overexpression and purification
The cDNA encoding the second repeated unit (R1b) of hamster EPRS was inserted into the NdeI–XhoI sites of the bacterial expression vector pET–28b (Novagen). The protein encoded by the ensuing plasmid had the sequence Met-Val-(R1b from Tyr261 to Gly309)-Leu-Glu-(His)6. The BL21(DE3) E.coli host strain containing the recombinant plasmid was grown at 37°C in M9 minimal medium supplemented with kanamycin; 15NH4Cl was used for production of the uniformly 15N–labelled sample. Cultures were grown to A600 = 0.75, and expression was induced by addition of 1 mM isopropyl-β–d-thiogalactopyranoside for 5 h at 37°C. Cells were washed with ice-cold extraction buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 10 mM imidazole) and lysed by sonication. The lysate was loaded on a His⋅Bind column. After a two-step wash with extraction buffer containing 30 and 50 mM imidazole, R1b was eluted by a 100 mM imidazole step and further purified on a Source–15S column. After concentration and dialysis against 20 mM potassium phosphate pH 7.0 and 2 mM dithiothreitol, the protein was stored at –80°C at a concentration of 1–2 mM.
The linker domain of EPRS carrying the three repeated units (R3) was expressed in E.coli and purified as described above for R1b.
RNA-binding assay
Homopolymeric ribonucleotides (Amersham Pharmacia Biotech) were end-labeled with [γ–32P]ATP and T4 polynucleotide kinase. Plasmids carrying tRNA genes were linearized and subjected to in vitro transcription according to standard procedures. Homogeneous R3 or R1b was incubated with 32P–labelled RNA in the binding buffer containing 10 mM Tris–HCl pH 8.0, 50 mM NaCl, 5 mM MgCl2, 10 mM 2–mercaptoethanol and 5% glycerol, in a final volume of 10 μl. After incubation at 25°C for 15 min, the mixture was placed on ice.
For the band shift assay, the incubate was loaded on a 1.5% agarose gel run in 0.5× TBE at 4°C. After electrophoresis, the gel was dried and subjected to autoradiography. Free and bound probes were also quantified by using a PhosphorImager (Molecular Dynamics).
For measurement of dissociation constants of the proteins and various RNA by a filter binding assay, the RNA–protein complexes formed during the incubation were recovered by filtration through nitrocellulose filters (HAWP02500; Millipore). The filters were washed twice with 1 ml of ice-cold binding buffer without glycerol, and radioactivity was counted in a scintillation counter.
NMR spectroscopy
R1b was dialysed against 20 mM potassium phosphate pH 7.0, 5 mM dl–1,4–dithiothreitol–d10. The samples (1.5 mM) were made up in 0.5 mM NaN3, 10% D2O and put in sealed NMR tubes. All NMR spectra were recorded on Bruker AMX–600 and DRX–800 spectrometers at 293 K, on uniformly 15N–labelled samples, using States-TPPI quadrature detection in the indirectly observed dimensions. The water suppression was achieved using the WATERGATE sequence (Piotto et al., 1992).
Mixing times used were up to 120 ms for the NOESY spectra. All NMR data were processed and analyzed using the GIFA (Pons et al., 1996) and FELIX (MSI Corp., San Diego) softwares on Silicon Graphics workstations. The assignment was based mainly on the analysis of 2D NOESY and 3D NOESY-HSQC experiments, since TOCSY transfers were poor.
NMR experimental restraints
A total number of 704 NOEs were identified on NOESY and NOESY-HSQC spectra. The NOE cross-peaks were classified in five groups of distances (1.8–2.5, 1.8–3.5, 1.8–4.0, 1.8–4.5 and 1.8–5.0 Å) according to decreasing intensity. Eight hydrogen groups were stereospecifically assigned. Pseudo-atom corrections were added to the upper distance restraints (1 Å for the methyl and degenerate methylene groups and 2.4 Å for the aromatic rings). The restraints on φ angles were based on measurements of the vicinal coupling constants 3JHN–Hα obtained from an HNHA experiment (Vuister and Bax, 1993). In addition, 33 hydrogen bonds identified in the preliminary structures (among which 31 are α–helices backbone H-bonds) were converted into 66 distance restraints; the distance interval between the hydrogen atom (donor heavy atom) and the acceptor atom was set to 1.7–2.3 Å (2.7–3.3 Å), respectively.
Structure calculations and analysis
Forty structures calculated by DIANA (Güntert et al., 1991) using the REDAC strategy (Güntert and Wüthrich, 1991) were transferred to X–PLOR (Brünger, 1993) for refinement by simulated annealing, and the 15 lowest energy structures were taken for the final set. They show no NOE violation higher than 0.4 Å and no dihedral angle violation higher than 7°. Computing was achieved on O2 Silicon Graphics workstations. All plots were generated using InsightII and GRASP (Nicholls et al., 1991).
Protein Data Bank accession number
The R1b co-ordinates and the NMR restraints file have been deposited in the Brookhaven Data Bank under accession codes 1R1B (minimized average structure) and 1D2D (set of 15 structures).
Acknowledgments
Acknowledgements
This work was supported by grants from the ‘Programme Physique et Chimie du Vivant’ from CNRS (E.G., M.M.), the ‘Association pour la Recherche sur le Cancer’ (E.G., M.M.) and the ‘Agence Nationale de Recherches sur le SIDA’ (M.M.).
References
- Alzhanova A.T., Fedorov, A.N., Ovchinnikov, L.P. and Spirin, A.S. (1980) Eukaryotic aminoacyl-tRNA synthetases are RNA binding proteins whereas prokaryotic ones are not. FEBS Lett., 120, 225–229. [DOI] [PubMed] [Google Scholar]
- Arnez J.G. and Cavarelli, J. (1997) Structures of RNA-binding proteins. Q. Rev. Biophys., 30, 195–240. [DOI] [PubMed] [Google Scholar]
- Berglund H., Rak, A., Serganov, A., Garber, M. and Härd, T. (1997) Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus. Nature Struct. Biol., 4, 20–23. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk, A., Tukalo, M. and Cusack, S. (1994) The 2.9 Å crystal structure of T.thermophilus seryl-tRNA synthetase complexed with tRNASer. Science, 263, 1404–1410. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Cerini C., Kerjan, P., Astier, M., Gratecos, D., Mirande, M. and Semeriva, M. (1991) A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J., 10, 4267–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerini C., Semeriva, M. and Gratecos, D. (1997) Evolution of the aminoacyl-tRNA synthetase family and the organization of the Drosophila glutamyl-prolyl-tRNA synthetase gene. Eur. J. Biochem., 244, 176–185. [DOI] [PubMed] [Google Scholar]
- Chien C.Y., Tejero, R., Huang, Y., Zimmerman, D.E., Rios, C.B., Krug, R.M. and Montelione, G.T. (1997) A novel RNA-binding motif in influenza A virus non-structural protein 1. Nature Struct. Biol., 4, 891–895. [DOI] [PubMed] [Google Scholar]
- Clemons W.M. Jr, Davies, C., White, S.W. and Ramakrishnan, V. (1998) Conformational variability of the N–terminal helix in the structure of ribosomal protein S15. Structure, 6, 429–438. [DOI] [PubMed] [Google Scholar]
- Fett R. and Knippers, R. (1991) The primary structure of human glutaminyl-tRNA synthetase. A highly conserved core, amino acid repeat regions and homologies with translation elongation factors. J. Biol. Chem., 266, 1448–1455. [PubMed] [Google Scholar]
- Güntert P. and Wüthrich, K. (1991) Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J. Biomol. NMR, 1, 447–456. [DOI] [PubMed] [Google Scholar]
- Güntert P., Braun, W. and Wüthrich, K. (1991) Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol., 217, 517–530. [DOI] [PubMed] [Google Scholar]
- Kerjan P., Triconnet, M. and Waller, J.P. (1992) Mammalian prolyl-tRNA synthetase corresponds to the 150 kDa subunit of the high-Mr aminoacyl-tRNA synthetase complex. Biochimie, 74, 195–205. [DOI] [PubMed] [Google Scholar]
- Kleeman T.A., Wei, D.B., Simpson, K.L. and First, E.A. (1997) Human tyrosyl tRNA synthetase shares amino acid sequence homology with a putative cytokine. J. Biol. Chem., 272, 14420–14425. [DOI] [PubMed] [Google Scholar]
- Liu J., Lynch, P.A., Chien, C.Y., Montelione, G.T., Krug, R.M. and Berman, H.M. (1997) Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nature Struct. Biol., 4, 896–899. [DOI] [PubMed] [Google Scholar]
- Mirande M. (1991) Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog. Nucleic Acid Res. Mol. Biol., 40, 95–142. [DOI] [PubMed] [Google Scholar]
- Morales A.J., Swairjo, M.A. and Schimmel, P. (1999) Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J., 18, 3475–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B.S. and Deutscher, M.P. (1991) Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl Acad. Sci. USA, 88, 4991–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Park S.G., Jung, K.H., Lee, J.S., Jo, Y.J., Motegi, H., Kim, S. and Shiba, K. (1999) Precursor of pro-apoptotic cytokine modulates aminoacylation activity of tRNA synthetase. J. Biol. Chem., 274, 16673–16676. [DOI] [PubMed] [Google Scholar]
- Piotto M., Saudek, V. and Sklenar, V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR, 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Pons J.L., Malliavin, T.E. and Delsuc, M.A. (1996) Gifa V.4: a complete package for NMR data set processing. J. Biomol. NMR, 8, 445–452. [DOI] [PubMed] [Google Scholar]
- Quevillon S., Agou, F., Robinson, J.C. and Mirande, M. (1997) The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine. J. Biol. Chem., 272, 32573–32579. [DOI] [PubMed] [Google Scholar]
- Raben N., Nichols, R., Dohlman, J., McPhie, P., Sridhar, V., Hyde, C., Leff, R. and Plotz, P. (1994) A motif in human histidyl-tRNA synthetase which is shared among several aminoacyl-tRNA synthetases is a coiled-coil that is essential for enzymatic activity and contains the major autoantigenic epitope. J. Biol. Chem., 269, 24277–24283. [PubMed] [Google Scholar]
- Rho S.B., Lee, J.S., Jeong, E.J., Kim, K.S., Kim, Y.G. and Kim, S. (1998) A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J. Biol. Chem., 273, 11267–11273. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Söll, D. (1979) Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu. Rev. Biochem., 48, 601–648. [DOI] [PubMed] [Google Scholar]
- Simos G., Sauer, A., Fasiolo, F. and Hurt, E.C. (1998) A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell, 1, 235–242. [DOI] [PubMed] [Google Scholar]
- Stapulionis R. and Deutscher, M.P. (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl Acad. Sci. USA, 92, 7158–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlin C., Burke, B., Yang, F., Liu, H.J., Shiba, K. and Musier–Forsyth, K. (1998) Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry, 37, 8605–8613. [DOI] [PubMed] [Google Scholar]
- Ting S.M., Bogner, P. and Dignam, J.D. (1992) Isolation of prolyl-tRNA synthetase as a free form and as a form associated with glutamyl-tRNA synthetase. J. Biol. Chem., 267, 17701–17709. [PubMed] [Google Scholar]
- Vuister G.W. and Bax, A. (1993) Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc., 115, 7772–7777. [Google Scholar]
- Wakasugi K. and Schimmel, P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science, 284, 147–151. [DOI] [PubMed] [Google Scholar]
- Wang C.C. and Schimmel, P. (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Whelihan E.F. and Schimmel, P. (1997) Rescuing an essential enzyme RNA complex with a non-essential appended domain. EMBO J., 16, 2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Nada, S. and Dignam, J.D. (1995) Analysis of truncated forms of Bombyx mori glycyl-tRNA synthetase: function of an N–terminal structure in RNA binding. Biochemistry, 34, 16327–16336. [DOI] [PubMed] [Google Scholar]
- Yang A.S., Hitz, B. and Honig, B. (1996) Free energy determinants of secondary structure formation: III. β-turns and their role in protein folding. J. Mol. Biol., 259, 873–882. [DOI] [PubMed] [Google Scholar]