Abstract
TLS (also known as FUS) is an RNA-binding protein that contributes the N-terminal half of fusion oncoproteins implicated in the development of human liposarcomas and leukemias. Here we report that male mice homozygous for an induced mutation in TLS are sterile with a marked increase in the number of unpaired and mispaired chromosomal axes in pre-meiotic spermatocytes. Nuclear extracts from TLS–/– testes lack an activity capable of promoting pairing between homologous DNA sequences in vitro, and TLS–/– mice and embryonic fibroblasts exhibit increased sensitivity to ionizing irradiation. These results are consistent with a role for TLS in homologous DNA pairing and recombination.
Keywords: fusion oncoproteins/homologous recombination/spermatogenesis/TLS deficiency
Introduction
The TLS gene (translocated in liposarcoma) was first identified as encoding the N-terminus of TLS–CHOP, a fusion oncoprotein that is expressed as a consequence of the t(12;16) translocation, which is invariably associated with human myxoid and round cell liposarcomas (Crozat et al., 1993; Rabbitts et al., 1993). In other human sarcomas and leukemias, chromosomal translocations fuse either TLS or the related EWS gene to a set of unrelated transcription factors. The common feature of these diverse fusion oncoproteins is the presence of a TLS/EWS-type N-terminal domain (reviewed in Rabbitts, 1994; Kuroda et al., 1998; Ron, 1998). This domain plays an essential role in transformation by the fusion oncoproteins (May et al., 1993; Zinszner et al., 1994; Kuroda et al., 1997), but the nature of the biochemical processes involved remains a mystery. Our understanding of the relationship between the transforming properties of TLS/EWS fusion oncoproteins and the functions of the corresponding normal TLS and EWS gene products has been significantly hampered by lack of a read-out for the biological activities of TLS or EWS.
The TLS gene product and the related EWS have features typical of RNA-binding proteins: their C-termini, deleted from the aforementioned fusion oncoproteins, contain R-G-G repeats flanking a highly conserved RNA binding domain of the RRM type (Delattre et al., 1992; Crozat et al., 1993; Rabbitts et al., 1993); TLS binds RNA in vitro and in vivo (Crozat et al., 1993; Prasad et al., 1994; Zinszner et al., 1997); and the protein engages in nucleo-cytoplasmic shuttling (Zinszner et al., 1994, 1997). These features, together with the relative abundance of TLS and the fact that the protein can be purified in a complex with known hnRNPs, suggest that TLS may be implicated in chaperoning mRNA or pre-mRNA (Zinszner et al., 1994, 1997; Calvio and Lamond, 1995). The Drosophila homolog of TLS/EWS, a protein known as SARFH (or CABEZA), is rapidly recruited to actively transcribed regions of chromatin, and this association does not have a discernible degree of target gene specificity (Immanuel et al., 1995). TLS and the related EWS and TAFII68 proteins have recently been found to interact with a variety of cellular targets. Notable among these are transcription factors, such as nuclear receptors of thyroid and steroid hormones (Powers et al., 1998), and components of the basal transcriptional machinery (Bertolotti et al., 1996, 1998; Petermann et al., 1998). The physiological significance of these associations or their potential role in transformation by the derivative oncogenic fusion proteins are not known. In order to study the cellular functions of TLS we disrupted the gene in the mouse germline. Surprisingly, analysis of TLS-deficient animals and cells points to a possible role for the protein in homologous DNA pairing and recombination.
Results
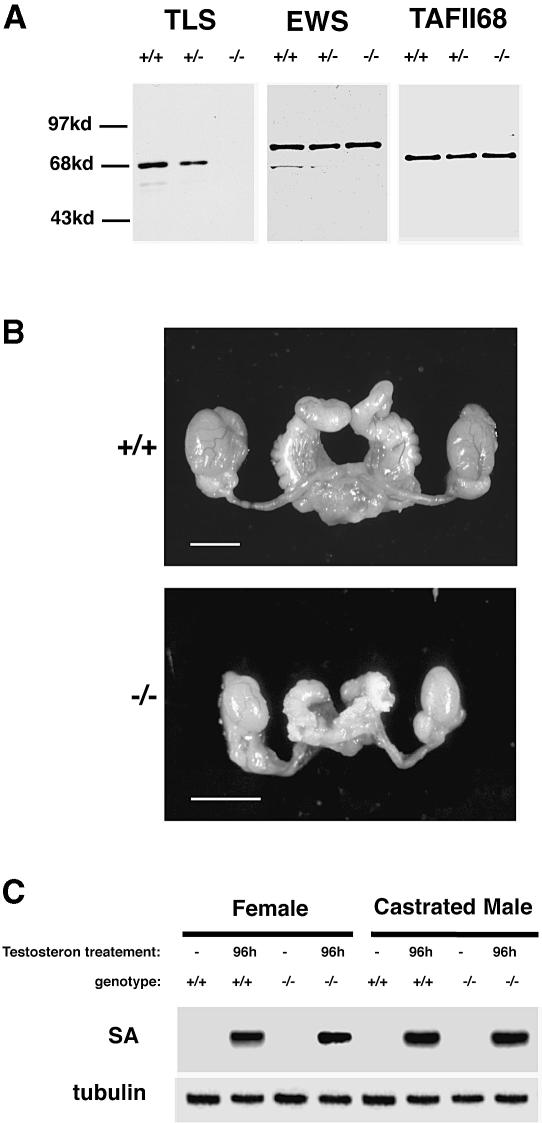
The mouse TLS gene was disrupted in embryonic stem (ES) cells. The TLS coding region was interrupted inside exon 8 (immediately upstream of the RNA recognition motif) by a promoterless insertion cassette that creates an allele encoding a truncated TLS–NEO fusion protein that is expressed at low levels (data not shown). This approach was instrumental in ‘trapping’ the locus at a very high frequency. Cells derived from the TLS–/– animals express no intact TLS protein (Figure 1A) and the TLS–NEO fusion protein encoded by the targeted allele is expressed at very low levels (supplementary Figure 1; the supplementary data are available in The EMBO Journal Online). Mice heterozygous for the targeted allele are indistinguishable from wild-type litter-mates. This finding, together with the very low level of expression of the TLS–NEO fusion protein, implies that the latter does not have discernible neomorphic features. TLS–/– offspring of heterozygote matings are represented at the expected ratio at weaning (96+/+, 200+/–, 85–/–). They are slightly smaller at birth and by the time of weaning, 3 weeks later, mutant animals of both sexes are ∼70% in size and readily distinguishable from wild-type or TLS+/– litter-mates [weight at weaning of TLS–/– males 9 ± 2 g versus wild type 15 ± 2 g (n = 25); TLS–/– females 8 ± 2 g versus wild type 14 ± 2 g (n = 25)]. Other than their reduced size, the mutant animals appear developmentally normal. In a specific-pathogen-free animal facility, the survival of TLS–/– animals of partially outbred background (with equal contribution of genes from the 129svev and CD1 strains) is virtually unimpaired. In the inbred, 129svev background, rare mutant animals are alive at weaning, but none reach adulthood. The cause of this perinatal attrition of TLS–/– animals in the inbred background is not known.
Fig. 1. Normal androgen action and internal genitalia of TLS–/– mice. (A) TLS, EWS and TAFII68 Western blots of whole-cell extracts from mouse embryonic fibroblasts with the TLS genotypes indicated. (B) Photomicrograph of the internal genitalia of adult (9–week-old) wild-type and TLS–/– male sibling mice. The bar corresponds to 5 mm. (C) Northern blot of kidney mRNA from female and castrated male mice injected with testosterone (4 μg/g/day) and hybridized with the androgen-responsive SA gene (upper panel) and β–tubulin (lower panel). Note the normal androgen responses of the mutant mice.
Mating of TLS–/– animals to wild-type counterparts reveals complete male sterility and reduced fertility of females. The latter is reflected in a litter size that is approximately half of that normally observed in this mixed 129svev;CD1 background. The phenotype in the females was not pursued further. The sterile males exhibited normal mounting behavior and produced coital plugs. The size of the accessory sexual organs was reduced in proportion to the total body size of the mutant mice. The anatomy of the internal genitalia was normal and the seminal vesicle contained a normal amount of seminal fluid (Figure 1B). These anatomical features and the normal mounting behavior of the TLS–/– mice suggest that androgen function was not compromised by the mutation. However, because of the reported association of TLS protein with nuclear hormone receptors (Powers et al., 1998), a direct assessment of androgen action was carried out. Females and castrated males were injected with testosterone and the induction of SA, an androgen-responsive marker gene, was measured in the kidney (Melia et al., 1998). Wild-type and TLS–/– mice had indistinguishable responses (Figure 1C). Furthermore, testosterone levels in wildtype and mutant mice were indistinguishable [TLS–/–, mean = 6 ng/ml, SD = 7.5 ng/ml (n = 11); wild type, mean = 4.5 ng/ml, SD = 5 ng/ml (n = 9)]. These results provide evidence against a general deficit in androgen-dependent gene expression; they do not, however, exclude subtler or cell-type-specific defects in androgen function.
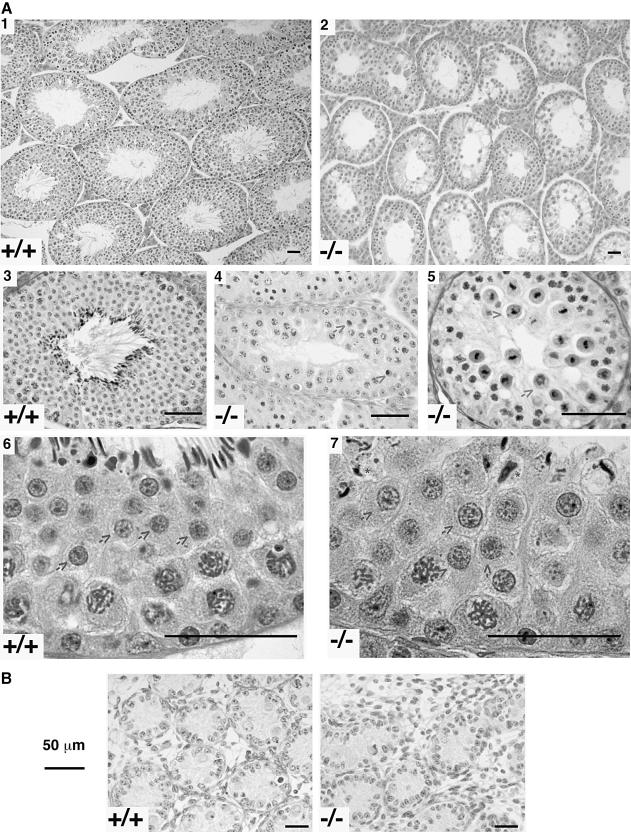
Mutant mice had testes that were two-thirds to half of the normal size. Histological examination of 13 adult mutant and 13 wild-type siblings revealed this to be caused by a selective reduction in the volume of the semini– ferous epithelium. Spermatogonia were well preserved (Figure 2A, panels 4, 5 and 7) and were the predominant cells in 3/13 mutant mice examined (see supplementary Figure 2). In the 10 others, spermatocytes were present in most tubules but in epithelial stages III–VIII apoptosis of pachytene cells was observed (Figure 2A, panel 4). Because of this, there were many more leptotene/zygotene spermatocytes than pachytene/diplotene spermatocytes in epithelial stage IX–XI tubules (see supplementary Figure 2). Furthermore, numerous examples of death during meiotic division were observed in stage XII tubules (Figure 2A, panel 5). These early defects were partial in that some round spermatids and even some abnormally shaped condensing forms were occasionally observed (Figure 2A, panel 7). Interestingly, many, and in some mutant mice all, of these round spermatids were much larger than those of wild-type siblings (Figure 2A, compare images from panels 6 and 7 that were obtained at the same magnification). This result suggests that the mutant round spermatids may be diploid (de Boer et al., 1986). In newborn animals, in which spermatogenesis had not yet progressed beyond the earliest stages, the wild-type and mutant testes were similar in appearance (Figure 2B), indicating the initial testicular development to be normal. These results indicate that absence of TLS leads to a significant defect in the meiotic process. The apparent increase in proportion of Leydig cells (Figure 2A, panel 2) is likely to be due to loss of tubular volume rather than a true increase in the number or size of these cells. Comparable levels of testosterone in wild-type and mutant mice attest to the normal function of these Leydig cells.
Fig. 2. TLS deficiency is associated with defective spermatogenesis. (A) Photomicrographs of representative sections of mouse testes with the TLS genotype indicated. The arrows in panels 4 and 5 point to degenerating pre-meiotic spermatocytes, and in panels 6 and 7 arrows point to round spermatids that are conspicuously larger in the mutant testes. The asterisks in panel 7 point to deformed elongated spermatids. (B) Photomicrographs of representative sections of newborn mouse testes with the TLS genotype indicated.
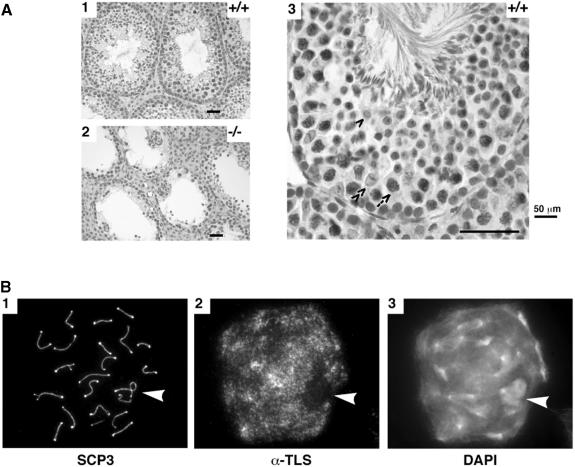
The expression pattern of TLS in the testes of wild-type mice was examined by in situ immunohistochemistry (Figure 3A). TLS was abundantly expressed in early stages of spermatogenesis, particularly in the large pachytene spermatocytes, which are conspicuous by their size and loose organization of their chromatin (Figure 3A, panel 3, arrow). TLS immunoreactivity was present but somewhat weaker in the round spermatids (Figure 3A, panel 3, single arrowhead) and was undetectable in condensing forms or sperm. Spermatogonia had low but detectable levels of TLS and near background levels of staining were noted in Sertoli cells (Figure 3A, panel 3, double arrowhead). As expected, staining of the TLS–/– testes was negative (Figure 3A, panel 2). The immunohistochemical analysis shown here was obtained on formalin-fixed samples, but similar results were observed in frozen sections (see supplementary Figure 3). EWS and TAFII68 signals were present at indistinguishable levels in wild-type and mutant mice (data not shown).
Fig. 3. TLS immunostaining and meiotic progression in wild-type and mutant testes. (A) Formalin-fixed and paraffin-embedded 5 μm sections of wild-type (panels 1 and 3) and mutant testes (panel 2) were reacted with the 4H11 anti-TLS monoclonal antibody, a horseradish peroxidase-conjugated secondary antibody, and the stain developed with diaminobenzidine and counterstained with hematoxylin. Specific cell types in the seminiferous epithelium are identified in panel 3: arrow, pachytene spermatocyte; single arrowhead, round spermatid; double arrowhead, Sertoli cell. (B) Chromosomal spreads of a pachytene/early diplotene spermatocyte fixed and stained for a marker of the axial elements (SCP3), TLS or a chromatin-binding dye (DAPI). The arrowhead points to the sex body that contains the partially synapsed X–Y chromosome pair and is a region of the nucleus with very little TLS staining.
High-level expression of TLS in pachytene spermatocytes, and histological evidence suggesting a meiotic defect, led us to characterize further the normal subcellular localization of TLS. Spermatocytes were swollen in hypotonic buffer, fixed and stained with 4′,6–diamidine-2–phenylindole (DAPI) and antibodies to TLS and SCP3 (the latter is a component of the axial elements). TLS immunoreactivity was highest in pachynema/early diplonema (Figure 3B), but could be detected as early as zygonema (not shown). TLS immunostaining was not specifically associated with the axial elements, but was found throughout the autosomal chromatin. Staining was conspicuously absent over the sex body, the chromosomal domain of the X–Y pair (arrowhead in Figure 3B, panels 2 and 3). The absence of a TLS signal over the sex body is particularly noteworthy since this region of the nucleus remains transcriptionally inactive during meiotic prophase (Monesi, 1965). Similar spreads from TLS–/– testes had no TLS signal, confirming the specificity of the antibody (data not shown).
Progression through meiotic prophase can be followed by SCP3 staining that is present both on unsynapsed axes and at synaptonemal complexes (SCs) that form between homologous chromosomes. In TLS–/– spermatocytes, synapsis was often disrupted, resulting in more than the normal 20 axes or in clusters of mispaired axes. Pachytene nuclei (148 of 839) from five TLS–/– mice had at least one pair of homologous autosomal axes that had failed to synapse (mean 17.6%, range 11.3–25.4%). In comparison, six of 593 pachytene nuclei from the testes of four sibling wild-type mice had autosomal asynapsis (mean 1.0%, range 0–1.5%). Non-homologous synapsis was also more frequent in the mutant mice, occurring in 33 of 839 nuclei (3.9%) compared with three of 593 nuclei (0.5%) in the control sibling population.
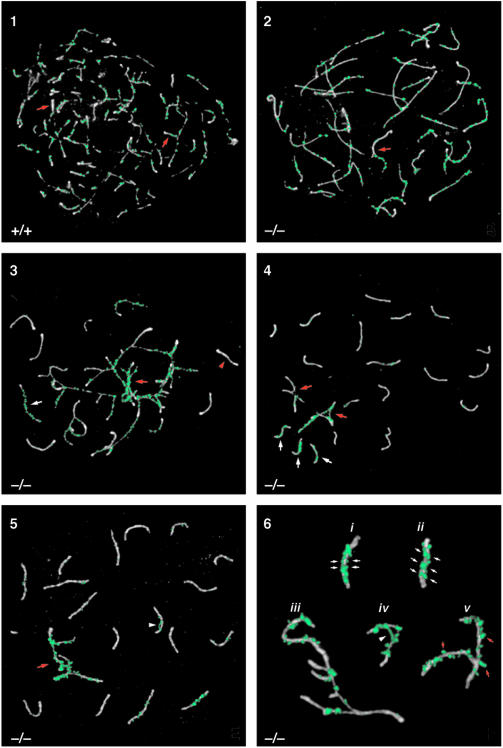
The defect in TLS–/– mice was further characterized using an antibody against RAD51, a RecA homolog that is involved in synapsis (Plug et al., 1996, 1998). RAD51 localizes to unsynapsed axes during zygonema, begins to disappear once synapsis is completed, and is no longer evident by mid-pachynema (Plug et al., 1998). Zygonema in normal mice is characterized by the formation of axial elements and subsequent synapsis of homologous axes. In wild-type spermatocytes, synapsis begins while axial element formation is still occurring, resulting in synapsis of some axes long before axial element formation is complete (Figure 4, panel 1). In the TLS–/– mice, however, almost full axial element formation occurred prior to synapsis (Figure 4, panel 2). RAD51 localized to sites along these long unsynapsed axes (Figure 4, panel 2, red arrow). Zygotene spermatocytes in mutant mice often had clusters of chromosomes with both asynapsed and non-homologously synapsed axes (Figure 4, panel 3). The fully synapsed axes and the synapsed portions of the non–homologously synapsed axes lost most of their RAD51 foci, while the unsynapsed portions of the non–homologously synapsed axes retained numerous abnormal RAD51 foci.
Fig. 4. Analysis of synaptonemal complex formation in TLS–/– spermatocytes by immunolocalization of SCP3 (white) and RAD51 (green) in chromosomal spreads. Panel 1: normal zygotene nucleus from wild-type mouse. Note that synapsis is occurring (red arrows) even as axial elements are still forming. RAD51 localizes to unsynapsed axes and begins to disappear once synapsis has occurred. Panel 2: zygotene nucleus exhibiting synaptic defects. Note the long axial elements that have almost completely formed with little synaptic activity. Some synapsis appears to be non-homologous, based on the differences in length of axial elements (red arrow). Panel 3: zygotene nucleus with major synaptic problems. Synapsed axes show a loss of RAD51 (red arrowhead), while the non-homologously synapsed axes (red arrow) and unsynapsed axes (white arrow) are coated with RAD51. Panel 4: pachytene nucleus with unsynapsed axes and non-homologous synapsis. Considering that the non-homologously synapsed axes involve more than one SC (red arrows), there are >20 axes present in this nucleus. The single axes with a RAD51 coating (white arrows) did not synapse with their homologs. Panel 5: pachytene nucleus containing non-homologously synapsed axes (red arrow). The non-homologously synapsed axes are emphasized by the large amount of RAD51. Also note the abnormal RAD51 bridge connecting two parts of one SC (white arrowhead). Panel 6: high magnification views of abnormal SCs from mutant pachytene nuclei. Incomplete synapsis of two SCs is evident from the double rows of RAD51 along the SCs' lengths (small white arrows) (i and ii). SCP3 staining of these SCs shows that they are thicker than normal, a characteristic of axes that are not fully synapsed. Several axes with partial non-homologous synapses are shown (iii). An abnormal RAD51 bridge has formed between non-homologous regions of the same bivalent (white arrowhead) (iv). SCs containing irregularly shaped RAD51 foci that ‘hang off’ the edges of the SCs, a configuration not observed in normal spermatocytes, are shown (small red arrows) (v).
The normal mice pachynema is characterized by the formation of 19 fully synapsed autosomal bivalents and the partially synapsed X and Y chromosomes. In pachynema of the TLS–/– mice, residual abnormalities resulting from the synaptic defects were evident. Homologous axes that failed to synapse in zygonema remained as univalents in pachynema, and the univalents had abnormal RAD51 staining (Figure 4, panel 4, white arrows). In addition to asynapsed axes, non-homologously synapsed axes were also found in pachynema in mutant mice (Figure 4, panels 4 and 5, red arrows). These axes also exhibited heavy staining with RAD51. In wild-type mice, a single row of round RAD51 foci was evident on recently synapsed SCs (Figure 4, panel 1). In TLS–/– mice, several aspects of RAD51 localization were abnormal. RAD51 bridges were found connecting non-homologous portions of the same SC (Figure 4, panels 5 and 6, white arrowheads). RAD51 staining also identified SCs that, although synapsed, were apparently not paired properly. These SCs had a double row of RAD51 foci along the length of each homologous axial element, suggesting that synapsis was incomplete (Figure 4, panel 6, i and ii). A closer examination of the SCP3 staining of these SCs shows that they are thicker than normal, a characteristic of asynapsed axes (de Boer and de Jong, 1989; Speed, 1989). Furthermore, RAD51 foci on axes with non-homologous synapsis were often irregularly shaped, abnormally large and seemed to hang off the edges of the axes (Figure 4, panel 6, red arrows). Such RAD51 foci are not observed in normal spermatocytes (Plug et al., 1996, 1998). Collectively, these observations point to a role for TLS in the early phases of meiotic prophase. In TLS–/– testes, most homologous chromosomes do synapse; however, the few that fail to execute this essential step properly are sufficient to disrupt spermatogenesis and cause sterility.
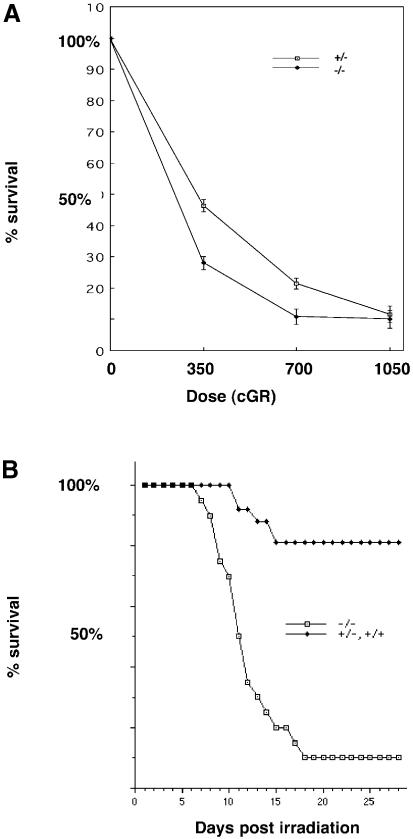
Homologous recombination is a programmed event in meiotic prophase, but also plays a role in repair of certain forms of DNA damage, most notably double-strand breaks induced by ionizing irradiation (reviewed in Stahl, 1994; Thompson, 1996). To explore further a possible link between TLS and homologous recombination we compared the sensitivity of TLS+/– and TLS–/– primary mouse embryonic fibroblasts (MEFs) to ionizing irradiation. The survival of the mutant cells was reproducibly lower than that of wild-type cells across a range of radiation doses (Figure 5A). Even more striking differences in the sensitivity to the effects of irradiation were also observed at the level of the whole animal: 18/20 TLS–/– mice irradiated with 700 cGR were dead by day 18, whereas 21/26 TLS+ mice irradiated with the same dose survived past 28 days (Figure 5B). No differences in radiation sensitivity were observed between TLS+/– and TLS+/+ animals, suggesting that the TLS–NEO allele has no important neomorphic features revealed by this assay. The responses of both TLS+/– and TLS+/+ animals were therefore grouped together in this analysis.
Fig. 5. Increased sensitivity of TLS–/– cells and animals to ionizing irradiation. (A) Day 7 counts of MEFs with the TLS genotypes indicated. The cells were irradiated at day zero with the indicated dose of γ–rays. The number of cells on the plate at day 7 is expressed as a fraction of the cell count on an identical plate at 24 h after plating. Shown are means and SEM of a representative experiment, performed in triplicate and reproduced three times using different pools of MEFs. (B) Survival curve of a cohort of mice discordant for TLS genotype that had received 700 cGR of γ–rays on day 1. Each of the 20 TLS–/– mice was matched with one or more siblings of TLS+/+ or TLS+/– genotype.
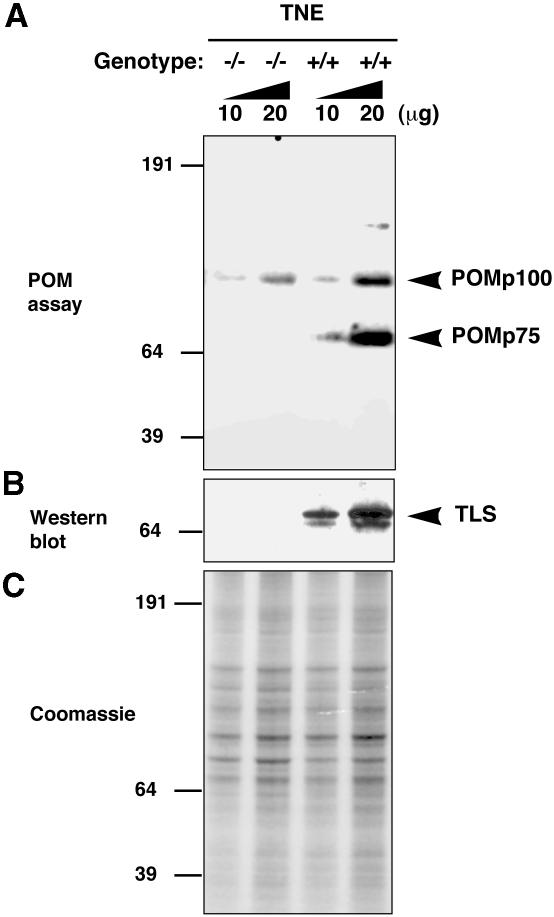
Homologous recombination requires pairing of hom– ologous DNA sequences. An in vitro assay, known as the ‘pairing on membrane assay’ (POM; Bertrand et al., 1993; Akhmedov et al., 1995; Thyagarajan and Campbell, 1997), mimics certain aspects of this essential process. Cellular proteins, separated by size on SDS–PAGE and blotted onto a nitrocellulose membrane, are tested for their ability to promote pairing between a soluble double-stranded DNA sequence labeled on one strand and its single-stranded homolog that has been immobilized to the membrane. Pairing activity occurs in situ and is detected by an autoradiographic signal on the membrane that localizes to the active protein. The assay identifies two major proteins in mammalian nuclei that promote this activity, one migrating at ∼100 kDa and the other at ∼75 kDa (Akhmedov et al., 1995). We sought to determine whether the profile of POM positivity was different in wild-type and TLS–/– testes. Nuclear extracts prepared from TLS–/– adult male mice lack the ∼75 kDa POM protein that is very conspicuous in the wild-type testes (Figure 6A). This deficiency in POMp75 correlates with the absence of TLS protein that normally migrates at an identical position on SDS–PAGE (Figure 6B). The POMp100 band was present in extracts of both wild-type and mutant testes. Its lower levels in the latter may reflect the differences in distribution of cell types in the wild-type and mutant testes. These findings are consistent with TLS participating directly in POMp75 activity and further support a role for TLS in homologous DNA pairing.
Fig. 6. TLS contributes to the in vitro pairing–promoting activity present in nuclei of testicular cells. (A) Proteins extracted from nuclei of testicular cells with the TLS genotypes indicated were resolved by SDS–PAGE, blotted onto nitrocellulose and the blot was subjected to a pairing on membrane assay (POM assay). (B) TLS Western blot performed on a parallel sample. (C) Coomassie staining of a parallel-run gel to control for equal loading of nuclear proteins. Note the absence of the major POMp75 species in the extracts from TLS–/– testes.
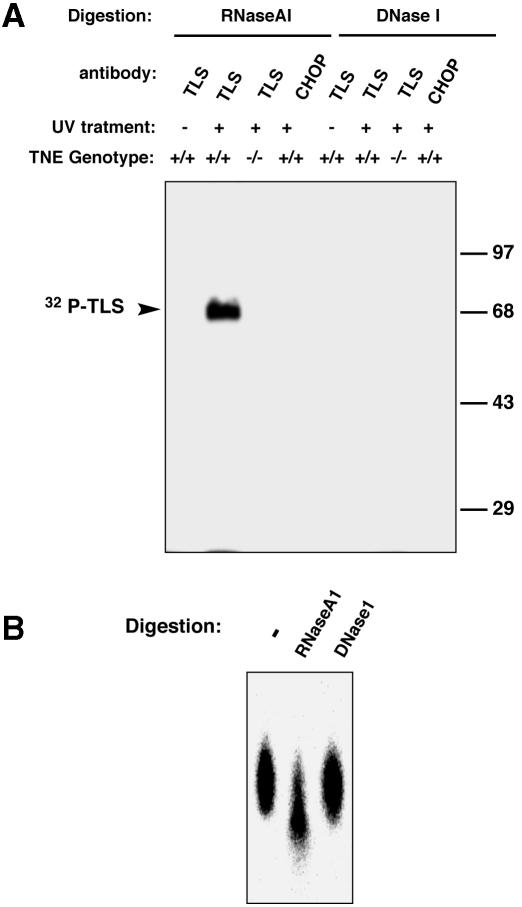
Absence of TLS immunostaining over the sex body (Figure 3B), a chromosomal region that is not transcribed in meiotic prophase, is consistent with TLS protein binding to RNA in spermatocytes. Cells from collagenase-treated testes of wild-type and TLS–/– mice were isolated and irradiated with UV light ex vivo in an effort to cross-link TLS protein to associated polynucleotides; we have previously determined that under these experimental conditions the cross-linked TLS is insoluble but can be released into the soluble phase by treatment with nuclease. Such treatment liberates TLS cross-linked to a protected remnant of the polynucleotide that can then be labeled in vitro (Zinszner et al., 1997). The solubilized TLS was immunoprecipitated and the protein–polynucleotide adduct was end-labeled by T4 kinase and resolved by SDS–PAGE. A labeled species migrating at the size of TLS was observed only in the wild-type testes and only in the lysates from which soluble TLS was recovered by treatment with RNase A1 but not DNase I (Figure 7A). Recovery of the labeled species from the gel by excision of the labeled band and proteinase K digestion revealed it to be sensitive to subsequent digestion with RNase A (Figure 7B). While this assay does not exclude the possibility that a fraction of TLS associates with DNA, it suggests that in the testes the major polynucleotide bound by TLS is RNA.
Fig. 7. Testicular TLS associates with RNA. (A) In vivo UV cross-linking of RNA species to TLS in testicular cells. Suspensions of freshly isolated testicular cells with the indicated TLS genotypes were irradiated with UV and the cellular TLS was solubilized by RNase A or DNase I treatment as indicated and immunoprecipitated with anti-TLS or control (anti-CHOP) antibodies. Polynucleotides in the immunoprecipitates were end-labeled with 32P and the TLS-labeled polynucleotide complex was resolved by 10% SDS–PAGE and subjected to autoradiography. (B) The labeled material in (A) was excised from the gel, digested with proteinase K, recovered by ethanol precipitation, divided into three aliquots that were subjected to repeat digestion with RNase A or DNase I and resolved on a 20% acrylamide–8 M urea gel. Note that DNase I digestion fails to degrade the labeled species whereas RNase A degrades it.
Discussion
TLS deficiency causes a profound defect in spermatogenesis and a mild defect in somatic growth, and enhances sensitivity to ionizing irradiation. TLS is normally expressed at high levels during meiotic prophase I and TLS deficiency is associated with the absence of an activity in testicular extracts that promotes pairing between homologous DNA molecules in vitro. This activity resides in the TLS protein itself, since purified POMp75 contains TLS peptide sequences (Bertrand et al., 1999) and purified recombinant TLS promotes homologous DNA pairing in vitro (Baechtold et al., 1999). Although phenotypic analysis does not permit us to pinpoint the primary defect(s) in TLS–/– mice, the observed perturbations in meiotic synapses are consistent with interruption of a preparatory or early step in homologous recombination. In male mammals, asynapsis of even part of one pair of homologs generally leads to apoptosis of the affected spermatocyte by around mid-pachynema, and results in reduced fertility, or even sterility, depending on the number of spermatocytes affected (de Boer and de Jong, 1989; Speed, 1989). Yet despite the fact that many TLS–/– spermatocytes are partially asynaptic, most proceed through meiosis without an arrest in pachynema. Nonetheless, TLS–/– males are sterile. Since many spermatocytes survive until late pachynema, and some even divide and form spermatids, asynapsis cannot be the sole cause of sterility. TLS may therefore also have additional roles in spermatogenesis.
TLS's role in homologous pairing and recombination may depend on its single-stranded polynucleotide-binding activity and interactions with DNA rather than RNA—presumably this is the basis of its ability to promote pairing on membranes in the in vitro POM assay. Although TLS is strongly associated with RNA in vivo (Zinszner et al., 1997; Figure 4B), in vitro it can also bind single-stranded DNA (Prasad et al., 1994; Perrotti et al., 1998). It is, however, intriguing to consider an alternative whereby TLS plays out its essential role in meiosis as an RNA-binding protein. This might come about if an RNA transcript, acting in cis, plays a role in homolog recognition or pairing. From studies of Drosophila larvae we know that TLS's homolog is rapidly associated with nascent RNA, binding to it while it is still in close proximity to the transcribed gene (Immanuel et al., 1995). Perhaps TLS's RNA chaperone activity is required for a step in pairing that involves the nascent transcript, not as a precursor of mRNA but rather as a polynucleotide that hybridizes with sequences on the homologous chromosome. In vitro studies have demonstrated a possible role for an RNA transcript in promoting homologous pairing catalyzed by the Escherichia coli RecA protein (Kotani and Kmiec, 1994).
Although male sterility has been attributed to defective function of several genes presumed to encode RNA-binding proteins (reviewed in Cooke and Elliott, 1997), the best characterized defects lead to widespread depletion of pre-meiotic germ cells (Ruggiu et al., 1997; Beck et al., 1998) and thus have little in common with the phenotype uncovered here. Hence, we favor a role for TLS in repair of DNA damage or in meiosis that is a consequence of its widespread and ‘non-gene-specific’ association with RNA. A recent study suggests that a signaling pathway downstream of the c-ABL kinase positively regulates the nucleic acid binding properties of TLS (Perrotti et al., 1998). Given the evidence that c-ABL is itself activated by DNA damage (Kharbanda et al., 1995), it is intriguing to speculate on the possibility that TLS is a regulated effector of c–ABL that participates in the response to genotoxic stress. c–ABL is expressed at high levels in spermatocytes, and c–ABL–/– mice exhibit defects in spermatogenesis (Kharbanda et al., 1998). These findings are consistent with both genes functioning in the same pathway during meiosis and repair of DNA damage. By this highly speculative model, TLS–CHOP (and the related EWS–CHOP) may perturb some aspect of TLS function related to repair of DNA damage. TLS–CHOP may thus represent not only a neomorphic mutation in CHOP (which deregulates downstream signaling pathways; Kuroda et al., 1999), but also one that leads to the acquisition of a subtle mutator phenotype that may promote secondary genetic changes relevant to progression of liposarcoma.
Materials and methods
Gene knock-out, histological and histochemical evaluation of testes
Production of the mutant TLS allele has been described in detail elsewhere (Kuroda et al., 1999). Briefly, a knock-in of a cassette containing the Neo coding region (with its stop codon intact) surrounded by loxP sites and followed by a CHOP coding sequence and terminator sequences from the TK gene was used to replace exons 8 and 9 of TLS in W4 ES cells. The targeted allele was studied before Cre-mediated Neo excision. In this state it encodes a fusion protein in which Neo replaces the RNA-binding C–terminal portion of TLS at amino acid 274 (the CHOP coding sequence, which was introduced into the replacement cassette for other reasons, is not expressed before Neo excision; Kuroda et al., 1999). Three identically targeted clones were transmitted through the mouse germline. Founder chimeras were mated to outbred CD1 mice (Charles River Laboratories) or to 129svev mice (Taconic) and the heterozygotes were intercrossed to produce TLS–/– offspring. MEFs were produced from day 13.5 mouse embryos, and whole-cell extracts of early passage MEFs were analyzed by Western blotting for TLS, EWS and TAFII68 protein as previously described (Zinszner et al., 1994, 1997; Bertolotti et al., 1996). Testes were fixed in Bouin's solution, embedded in paraffin and sectioned at 5 μm for standard histological evaluation. TLS immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections using the 4H11 anti-TLS C–terminus monoclonal antibody (Zinszner et al., 1997). Evaluation of androgen responses in kidneys of wild-type and TLS–/– mice was carried out using the SA gene as a marker (Melia et al., 1998). Preparation and staining of spermatocyte nuclei from wild-type and TLS–/– mouse testes followed established protocols (Peters et al., 1997; Plug et al., 1998).
Radiation sensitivity of mice and cells
Irradiation of mice was carried out using a cobalt source at a dose rate of 340 cGR/min, following a published protocol (Nussenzweig et al., 1997). First passage MEFs with TLS+/– and TLS–/– genotypes were trypsinized and irradiated in suspension on ice using the same source. Cells were subsequently plated at a density of 2–5 × 104 cells per well in six-well plates, and cell counts were performed at 1 and 7 days after plating (Patel et al., 1998). The count at day 1 was used to normalize for plating efficiency of the irradiated cells of both genotypes.
Pairing on membrane and in vivo RNA cross-linking assays
Nuclei were isolated from adult testes (Lan et al., 1997) and nuclear proteins were extracted in 0.5 M NaCl, resolved by SDS–PAGE, transferred to a nitrocellulose membrane, and a POM assay was performed as previously described (Bertrand et al., 1993).
In vivo cross-linking of TLS to polynucleotides was performed by a modification of a previously described procedure (Zinszner et al., 1997). Briefly, cells were isolated by treating testicular fragments with collagenase type III (1 testis in 5 ml of 0.25 mg/ml collagenase at 37°C for 30 min). The recovered cells were resuspended in phosphate-buffered saline and placed in a 60 mm tissue-culture dish and exposed on ice to UV light (900 mJ/cm2) using a Stratalinker™ cross-linking oven. The cells were lysed, soluble nuclear proteins extracted under high-salt conditions, and the lysate was diluted to physiological salt concentrations. It was then treated with RNase or DNase and clarified of insoluble complexes by ultracentrifugation. TLS was immunoprecipitated from the supernatant using the 4H11 monoclonal antibody and the covalently attached polynucleotide was end-labeled by T4 kinase using [γ–32P]ATP as a phosphate donor. The labeled species were resolved by 8% SDS–PAGE and revealed by autoradiography.
Supplementary data
Supplementary data to this paper (supplementary Figures 1, 2 and 3) are available in The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank C.Campbell, H.Cooke, D.Elliot, M.Hardy, M.Jasin, P.Morris, A.Pierce, R.Reijo, M.Ruggeo, P.Walden and A.Zinn for useful discussions; A.Meseguer, A.Bertolotti and L.Tora for cDNA probes and antibodies; M.Hardy for testosterone measurements; and N.Tanese for careful review of the manuscript. Supported by NIH grant CA60945. Basel Institute for Immunology was founded and is supported by F.Hoffmann-La Roche Ltd. D.R. is a Stephen Birnbaum Scholar of the Leukemia Society of America.
References
- Akhmedov A.T., Bertrand, P., Corteggiani, E. and Lopez, B.S. (1995) Characterization of two nuclear mammalian homologous DNA-pairing activities that do not require associated exonuclease activity. Proc. Natl Acad. Sci. USA, 92, 1729–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechtold H., Kuroda, M., Sok, J., Ron, D., Lopez, B.S. and Akhmedov, A.T. (1999) Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem., 274, 34337–34342. [DOI] [PubMed] [Google Scholar]
- Beck A.R.P., Miller, I.J., Anderson, P. and Streuli, M. (1998) RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl Acad. Sci. USA, 95, 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Lutz, Y., Heard, D.J., Chambon, P. and Tora, L. (1996) hTAFII68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J., 15, 5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Melot, T., Acker, J., Vigneron, M., Delattre, O. and Tora, L. (1998) EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68 and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol., 18, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P., Corteggiani, E., Dutreix, M., Coppey, J. and Lopez, B.S. (1993) Homologous pairing between single-stranded DNA immobilized on a nitrocellulose membrane and duplex DNA is specific for RecA activity in bacterial crude extract. Nucleic Acids Res., 21, 3653–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P., Akhmedov, A.T., Delacote, F., Durrbach, A. and Lopez, B.S. (1999) Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene, 18, 4515–4521. [DOI] [PubMed] [Google Scholar]
- Calvio C. and Lamond, A. (1995) Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA, 1, 724–733. [PMC free article] [PubMed] [Google Scholar]
- Cooke H.J. and Elliott, D.J. (1997) RNA-binding proteins and human male infertility. Trends Genet., 13, 87–89. [DOI] [PubMed] [Google Scholar]
- Crozat A.Y., Åman, P., Mandahl, N. and Ron, D. (1993) Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma with t(12;16)(q13;p11). Nature, 363, 640–644. [DOI] [PubMed] [Google Scholar]
- de Boer P. and de Jong,J. (1989) Chromosome pairing and fertility in mice. In Gillies,C. (ed.), Fertility and Chromosome Pairing: Recent Studies in Plants and Animals. CRC Press, Boca Raton, FL, pp. 37–76. [Google Scholar]
- de Boer P., Searle, A.G., van der Hoeven, F.A., de Rooij, D.G. and Beechey, C.V. (1986) Male pachytene pairing in single and double translocation heterozygotes and spermatogenic impairment in the mouse. Chromosoma, 93, 326–336. [DOI] [PubMed] [Google Scholar]
- Delattre O. et al. (1992) Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumors. Nature, 359, 162–165. [DOI] [PubMed] [Google Scholar]
- Immanuel D., Zinszner, H. and Ron, D. (1995) Association of SARFH (sarcoma-associated RNA-binding fly homolog) with regions of chromatin transcribed by RNA polymerase II. Mol. Cell. Biol., 15, 4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S., Ren, R., Pandey, P., Shafman, T.D., Feller, S.M., Weichselbaum, R.R. and Kufe, D.W. (1995) Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature, 376, 785–788. [DOI] [PubMed] [Google Scholar]
- Kharbanda S. et al. (1998) Functional role for the c-Abl tyrosine kinase in meiosis I. Oncogene, 16, 1773–1777. [DOI] [PubMed] [Google Scholar]
- Kotani H. and Kmiec, E.B. (1994) A role for RNA synthesis in homologous pairing events. Mol. Cell. Biol., 14, 6097–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Ishida, T., Takanashi, M., Satoh, M., Machinami, R. and Watanabe, T. (1997) Oncogenic transformation and inhibition of adipocytic conversion of preadipocytes by TLS/FUS–CHOP type II chimeric protein. Am. J. Pathol., 151, 735–744. [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Sok,J. and Ron,D. (1998) The TLS–CHOP oncogene and human liposarcoma. In Cooper,C. (ed.), Translocations in Solid Tumors. Landes Bioscience, Georgetown, TX. [Google Scholar]
- Kuroda M. et al. (1999) Induction of a novel secreted protein by the myxoid liposarcoma oncogene. Proc. Natl Acad. Sci. USA, 96, 5025–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Z.J., Palladino, M.A., Rudolph, D.B., Labus, J.C. and Hinton, B.T. (1997) Identification, expression and regulation of the transcriptional factor polyomavirus enhancer activator 3, and its putative role in regulating the expression of γ-glutamyl transpeptidase mRNA-IV in the rat epididymis. Biol. Reprod., 57, 186–193. [DOI] [PubMed] [Google Scholar]
- May W.A., Lessnick, S.L., Braun, B.S., Klemsz, M., Lewis, B.C., Lunsford, L.B., Hromas, R. and Denny, C.T. (1993) The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol., 13, 7393–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia M.J., Bofill, N., Hubank, M. and Meseguer, A. (1998) Identification of androgen-regulated genes in mouse kidney by representational difference analysis and random arbitrarily primed polymerase chain reaction. Endocrinology, 139, 688–695. [DOI] [PubMed] [Google Scholar]
- Monesi V. (1965) Differential rate of ribonucleic acid synthesis in the autosomes and sex chromosomes during male meiosis in the mouse. Chromosoma, 17, 11–21. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., Sokol, K., Burgman, P., Li, L. and Li, G.C. (1997) Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival and development. Proc. Natl Acad. Sci. USA, 94, 13588–13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.J. et al. (1998) Involvement of Brca2 in DNA repair. Mol. Cell, 1, 347–357. [DOI] [PubMed] [Google Scholar]
- Perrotti D. et al. (1998) TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J., 17, 4442–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann R., Mossier, B.M., Aryee, D.N., Khazak, V., Golemis, E.A. and Kovar, H. (1998) Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene, 17, 603–610. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Plug, A.W., van Vugt, M.J. and de Boer, P. (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res., 5, 66–68. [DOI] [PubMed] [Google Scholar]
- Plug A.W., Xu, J., Reddy, G., Golub, E.I. and Ashley, T. (1996) Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc. Natl Acad. Sci. USA, 93, 5920–5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plug A.W., Peters, A.H., Keegan, K.S., Hoekstra, M.F., de Boer, P. and Ashley, T. (1998) Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci., 111, 413–423. [DOI] [PubMed] [Google Scholar]
- Powers C.A., Mathur, M., Raaka, B.M., Ron, D. and Samuels, H.H. (1998) TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone and retinoid receptors. Mol. Endocrinol., 12, 4–18. [DOI] [PubMed] [Google Scholar]
- Prasad D., Ouchida, M., Lee, L., Rao, V. and Reddy, E. (1994) TLS/FUS fusion domain of TLS/FUS–erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene, 9, 3717–3729. [PubMed] [Google Scholar]
- Rabbitts T.H. (1994) Chromosomal translocations in human cancer. Nature, 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H., Forster, A., Larson, R. and Nathan, P. (1993) Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nature Genet., 4, 175–180. [DOI] [PubMed] [Google Scholar]
- Ron D. (1998) TLS–CHOP and the role of RNA-binding proteins in oncogenic transformation. In Vogt,P. and Rauscher,F. (eds), Chromosomal Translocations and Oncogenic Transcription Factors. Springer-Verlag, Berlin, Germany, Vol. 220, pp. 131–142. [DOI] [PubMed] [Google Scholar]
- Ruggiu M., Speed, R., Taggart, M., McKay, S.J., Kilanowski, F., Saunders, P., Dorin, J. and Cooke, H.J. (1997) The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature, 389, 73–77. [DOI] [PubMed] [Google Scholar]
- Speed R. (1989) Heterologous pairing and fertility in humans. In Gillies,C. (ed.), Fertility and Chromosome Pairing: Recent Studies in Plants and Animals. CRC Press, Boca Raton, FL. [Google Scholar]
- Stahl F.W. (1994) The Holliday junction on its thirtieth anniversary. Genetics, 138, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H. (1996) Evidence that mammalian cells possess homologous recombinational repair pathways. Mutat. Res., 363, 77–88. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B. and Campbell, C. (1997) Elevated homologous recombination activity in Fanconi anemia fibroblasts. J. Biol. Chem., 272, 23328–23333. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Albalat, R. and Ron, D. (1994) A novel effector domain from the RNA-binding proteins TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev., 8, 2513–2526. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Sok, J., Immanuel, D., Yin, Y. and Ron, D. (1997) TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci., 110, 1741–1750. [DOI] [PubMed] [Google Scholar]