Abstract
Some proteins have been shown to mimic the overall shape and structure of nucleic acids. For some of the proteins involved in translating the genetic information into proteins on the ribosome particle, there are indications that such observations of macromolecular mimicry even extend to similarity in interaction with and function on the ribosome. A small number of structural results obtained outside the protein biosynthesis machinery could indicate that the concept of macromolecular mimicry between proteins and nucleic acids is more general. The implications for the function and evolution of protein biosynthesis are discussed.
Keywords: elongation factor G/elongation factor Tu/protein–nucleic acid interactions/ribosomal function/RNA world
Introduction
For the purpose of this mini-review, we will define the concept ‘macromolecular mimicry’ as the phenomenon that large parts of proteins (domains or subdomains) resemble in shape (and possibly also in function) a large part of a nucleic acid. This definition stems from the observation by us (Nissen et al., 1995) that the structure of elongation factor G (EF–G) in complex with GDP (Czworkowski et al., 1994; Al-Karadaghi et al., 1996) has the same overall shape as the structure of the ternary complex of elongation factor Tu (EF–Tu). At the same time, a functional macromolecular mimicry was observed by Cech et al. (Doudna et al., 1995), who showed that monoclonal antibodies raised against an autoantigenic epitope of human insulin receptor also reacted with a selected RNA sequence. It has since been proposed that protein–RNA mimicry may be the background for several auto-immunological disorders, where autoantibodies bind to self-RNA (Keene, 1996a,b).
The concept of ‘molecular mimicry’ is not new, and in the literature covers diverse fields such as the recognition of cell surfaces by viruses and other parasites, the binding of agonists and antagonists to receptors, and not least the cross-reactions in autoimmune responses (for reviews, see Hall, 1994; Davies, 1997). However, the definition of ‘macromolecular mimicry’ used here is much narrower than that of ‘molecular mimicry’ and even narrower than that used in a recent review (Pedersen et al., 1999). It only includes examples of mimicry for which there is either solid structural or reasonably solid biochemical evidence for a similarity in shape between proteins and nucleic acids. Within the definition is the assumption that the protein (or protein domain) that is mimicking the nucleic acid will, during its biological function, bind to a binding site that is constructed such that it will also bind the nucleic acid. Excluded from our definition are the many examples of RNA molecules that mimic tRNA. These have been described in a recent review (Giege et al., 1998) and in a recent book chapter (Springer et al., 1998). Implicit in the present use of the concept is the assumption of a pre-biotic RNA world, which has evolved into the biological complexity of present times by initially letting the much more versatile proteins mimic the restricted functions of replicative and enzymatic RNA. However, the concept as presented here arose solely on the basis of a few examples of structural similarity in the biological processes of genetic control and translation of genetic information.
Here we describe some details of the few known examples of ‘macromolecular mimicry’, with the main emphasis on our own work in protein biosynthesis. We will expand the concept slightly in describing how parts of some protein factors involved in protein biosynthesis are most likely to mimic RNA, although this is not at the moment supported by structural evidence.
Mimicry of single nucleotides
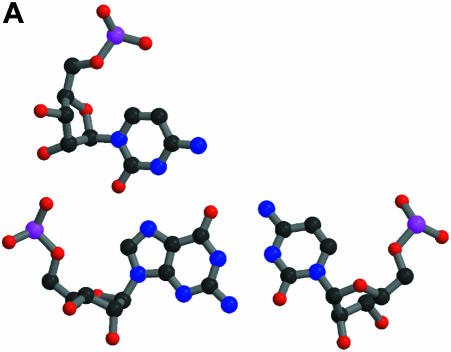
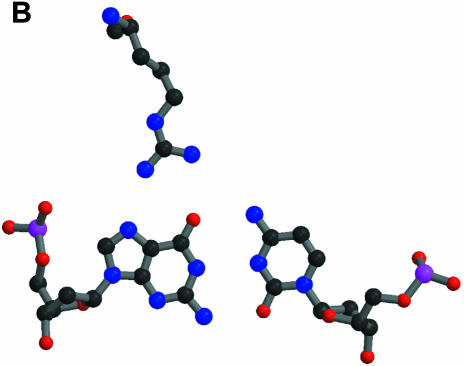
The definition of ‘macromolecular mimicry’ does not necessarily imply that the mimicry exists at an atomic or residue level. However, it is a well established fact that proteins do mimic nucleic acids down to the level of single residues. A possible scheme for the sequence-specific recognition of double-helical nucleic acids by proteins was postulated as early as 1976 (Seeman et al., 1976) on the basis of the observation of triple base interactions in tRNA. Several examples of specific interactions between transcription factors and DNA have been found (some initial examples are found in Pavletich and Pabo, 1991; Fairall et al., 1993). Figure 1 shows some more recent structures where a triple base interaction found in the P4–P6 domain of the ribozyme of the Tetrahymena thermophila intron (Cate et al., 1996) is compared with the specific binding of transcription factor TFIIIA to DNA (Nolte et al., 1998). It is obvious from this figure that the protein residue of TFIIIA in its recognition of a standard Watson–Crick DNA base pair utilizes the same hydrogen bond pattern as the extra base of the triple base pair of the P4–P6 domain in its interaction with a standard RNA base pair.
Fig. 1. Specific interactions with single-standard base pairs. The atomic colouring is: C, black; N, blue; O, red; P, magenta. (A) A triple base pair from the P4–P6 domain of the Tetrahymena ribozyme (Cate et al., 1996). At the bottom is shown the standard base pair of G108 and C213, with C260 at the top making hydrogen bonds to G108. The PDB code is 1GID. (B) At the top is shown Arg154 of transciption factor TFIIIA interacting with the standard base pair G9 and C56 of its cognate DNA (Nolte et al., 1998). The PDB code is 1TF6. This and the following figures were made with MOLSCRIPT (Kraulis, 1991) and Raster3D (Merrit and Murphy, 1994).
In the specific recognition of some tRNAs by their cognate tRNA synthetases, the anticodon interacts directly with the protein. One example of this has been shown (Cusack et al., 1998) in the structure of the complex of tRNAPro and its tRNA synthetase, where protein side chains use a hydrogen bonding scheme similar to that in the standard DNA base pair. Often acidic residues and arginines are involved in this type of direct specific interaction with a nucleotide base. The protein side chains can thus be seen as mimicking the base-specific hydrogen bonds in double-stranded DNA. Furthermore, in some of the examples described below, acidic residues of the mimicking proteins simulate the electrostatic environment of the phosphate backbone of DNA. It is thus likely that detailed atomic simulations of macromolecular interactions will be found in proteins that need to mimic specific recognitions of nucleic acids.
Macromolecular mimicry of nucleic acids
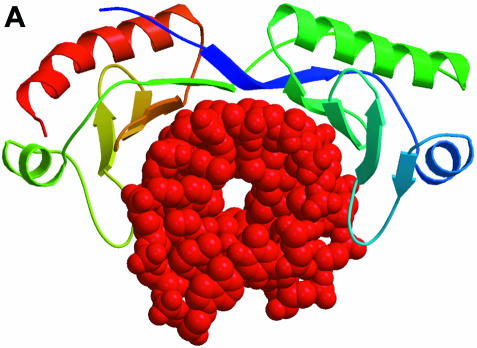
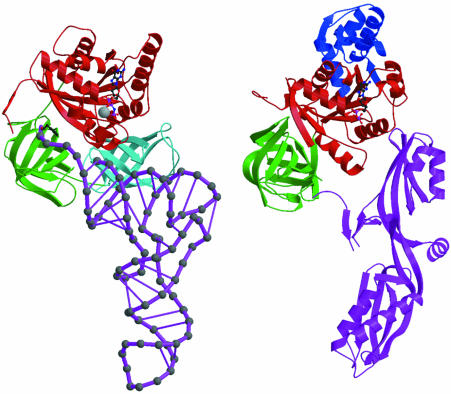
The TATA box-binding protein (TBP) is a transcription factor that binds specifically to the TATA box element upstream of the transcription initiation site of eukaryotic genes, and thus plays a central role in the positive or negative regulation of transcription. The structure of the TBP–TATA box complex (Kim et al., 1993; Kim and Burley, 1994) shows a large concave surface on TBP formed by a curved β–sheet that makes minor groove and backbone contacts with a partially unwound and bent form of the TATA box element. Recently, the solution structure of an N-terminal fragment of the TBP-associated factor TAFII230 in complex with TBP was determined (Liu et al., 1998). The structure reveals that TAFII230 binds to the concave DNA-binding surface of TBP. The TBP-binding surface of TAFII230 mimics the minor groove of the partially unwound and bent TATA box element, thus providing a negative control of the TATA box-binding activity of TBP (Figure 2). The binding surface of TAFII230 contains a large number of hydrophobic methyl-containing and phenylalanine residues mimicking the bases of the DNA, while a number of acidic residues are found on the rim of the hydrophobic patch in a position to interact with the basic residues of TBP, which in the TBP–TATA box complex are found to interact with the DNA backbone. Thus, the TAFII230 protein is a nearly perfect macromolecular structural mimic of the DNA surface that is formed by TBP after specific recognition of the TATA box element.
Fig. 2. Interactions of the TATA box-binding protein (TBP). The TBP at the top is shown with a colour ramp starting with blue at the N–terminus and ending with red at the C-terminus. The interacting partners are shown in red. (A) TBP is shown in its interaction with the DNA of the TATA box element (Kim and Burley, 1994). The PDB code is 1CDW. (B) A domain of TAFII230 occupies the TBP-binding site for DNA in the TBP–TAFII230 complex (Liu et al., 1998). The PDB code is 1TBA.
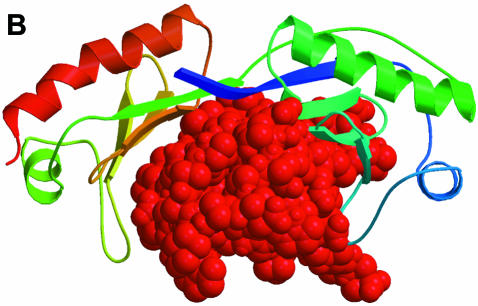
The uracil-DNA glycosylase inhibitor (Ugi) found in a Bacillus subtilis bacteriophage protects the uracilcontaining phage DNA from the host DNA repair enzyme uracil-DNA glycosylase (UDG). The structure of Ugi complexed with human UDG has been determined (Mol et al., 1995). It shows the compact barrel-type structure of Ugi of dimensions similar to a DNA helix bound to the conserved DNA-binding groove of UDG (Figure 3). Although the structure of a complex between UDG and a DNA is not known, from biochemical evidence it is very likely that a well conserved leucine residue of UDG intercalates the DNA bases, and that the repair enzyme forces an expulsion of the uracil from the DNA helix for specific interaction and for excision and repair. The interacting surfaces of Ugi and UDG display electrostatic and shape complementarity, such that Ugi mimics the surface of a DNA molecule. A negatively charged ridge on the edge of the Ugi β–sheet fits nicely into the positively charged active site groove of UDG. A hydrophobic pocket of Ugi surrounds the conserved leucine residue. The structure has served as a model for the binding of UDG to uracil-containing DNA (Mol et al., 1995).
Fig. 3. The structure of uracil-DNA glycosylase inhibitor (Ugi) in complex with the DNA repair enzyme uracil-DNA glycosylase (UDG). UDG is shown in a colour ramp as in Figure 2. Ugi is shown in red at the top occupying the DNA-binding site of UDG (Mol et al., 1995). The PDB code is 1UGH.
In the solution structure of the U1A protein of the U1 splicesome in complex with RNA (Avis et al., 1996), a C–terminal helix is found, which in the crystal structure of the free protein interacts very intimately with the RNA-binding site (Jovine et al., 1996). Thus, in the complex between U1A and its specific RNA, this helix is replaced by the RNA. Therefore, this C–terminal helix of U1A can be regarded as a cis–acting RNA mimic that possibly prevents unspecific binding of RNA to U1A. This could well be a general mechanism in spliceosomal proteins.
A recent structural study of karyopherin α (a nuclear import factor) has revealed a structure of 10 tandem repeats, which form a twisted, helical shape with a large groove (Conti et al., 1998). The structure does have the appearance of a DNA double helix, but with a much larger rise per helical turn. Furthermore, the structure of karyopherin α in complex with two peptides with a nuclear localization signal (NLS) was presented in the same publication. Karyopherin α binds to ‘classical’ NLSs, which are characterized by several lysine and arginine residues in an as yet unknown context with other amino acids, and that direct the tagged proteins to nuclear import. The NLS can occupy one or two binding sites in karyopherin α. Interestingly, NLS peptides are not excised after proper nuclear import, in contrast to many other localization signal sequences, and the NLSs of several nuclear proteins are known to be involved in DNA binding in the nucleus. The binding of the NLS peptides to many acidic residues at the rim of the large groove of karyopherin α is strikingly similar to the specific binding of the NLS of transcription factor LEF–1 to the major groove of its cognate DNA (Love et al., 1995). Therefore, it can be proposed that the nuclear import factors present NLS-binding sites that mimic the DNA targets in the nucleus. It is entirely possible that a specificity of binding between import factors and NLSs select functional transcription factors for import into the nucleus.
Macromolecular mimicry in translation
The translation of genetic information into fully functional proteins happens on the ribosomal particle. The process of translation is divided into three phases: initiation, elongation and termination. During the initiation phase, an initiation factor IF–2 in complex with initiator tRNA and GTP recognizes the start codon of the mRNA and delivers the initiator tRNA at the ribosomal P–site. IF–2:GDP is released from the ribosome after formation of this initiation complex. During the elongation phase, three elongation factors, EF–Tu, EF–Ts and EF–G, are active. EF–Tu forms a complex with aminoacylated tRNA (aa–tRNA) and GTP, protects the amino acid ester from hydrolysis and assists in the correct interaction between the anticodon on the tRNA and the codon on mRNA presented by the ribosome particle at its A–site. EF–Tu:GDP is released after such a cognate interaction, and aa–tRNA is brought to the peptidyl transferase centre of the ribosome where a new peptide bond is formed between the incoming amino acid and the peptide linked to the tRNA in the P–site. EF–Ts catalyses a nucleotide exchange on EF–Tu in order to reactivate this factor. EF–G bound to GTP performs translocation on the ribosome, such that the tRNA in the A–site is brought into the P–site, and such that the next codon on mRNA is exposed in the A–site. The termination phase involves at least three proteins (in bacteria). Two of those, RF1 and RF2, recognize a stop codon in the ribosomal A–site and hydrolyse the ester bond between the peptide and the tRNA in the P–site. This reaction is stimulated by RF3 in complex with GTP.
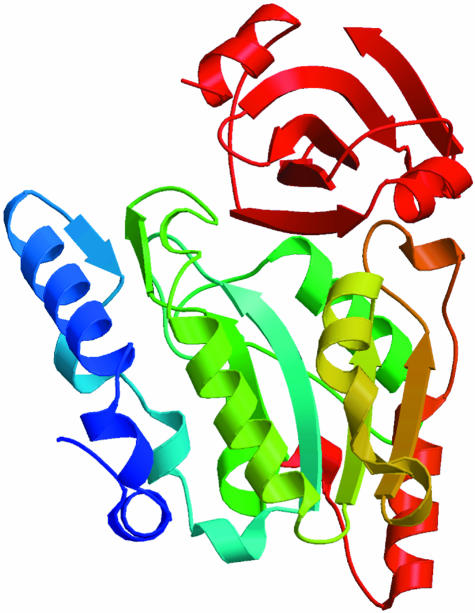
Many structural details of the translation factors and their interaction with the ribosome have been obtained during recent years. Most studies have been concentrated on the elongation phase. Thus, almost all functional states of the elongation factors have been structurally characterized. The structure of the inactive EF–Tu:GDP from several prokaryotes (Abel et al., 1996; Polekhina et al., 1996; Song et al., 1999) and from bovine mitochondria (Andersen et al., 1999) has been determined. Crystal structures and one model of the active EF-Tu:GDPNP, where GDPNP is a non-hydrolysable analogue of GTP, have also been published (Berchtold et al., 1993; Kjeldgaard et al., 1993; Krásny et al., 1998). All this information provides a solid basis for the remarkable conformational change of EF–Tu during its activation. EF–Tu is apparently composed of two functional parts, one of which is the nucleotide-binding domain (domain 1) and the other is domains 2 and 3 together. The two parts of the molecule rotate by ∼90° when GDP in its nucleotide-binding pocket is replaced by GTP. At the same time, two so-called switch regions, found in many other G–proteins on the surface of their nucleotide-binding domains, change their local secondary structures. The large structural change and the nucleotide exchange are catalysed by EF–Ts. Structures of the EF–Tu:EF–Ts complex are also known (Kawashima et al., 1996; Wang et al., 1997).
Two structures of the ternary complex of EF–Tu have now been determined. The first is of yeast Phe–tRNAPhe in complex with Thermus aquaticus EF–Tu:GDPNP (Nissen et al., 1995), and the second is of Escherichia coli Cys–tRNACys in complex with T.aquaticus EF-Tu:GDPNP (Nissen et al., 1999). Both structures show that the complex is very elongated, with the anticodon of the tRNA pointing away from EF–Tu. Only the T–stem helix, the free 5′-phosphate and the CCA-amino acid part of the tRNAs are in intimate contact with the protein. The structures of EF–Tu:GDPNP and free tRNA are not changed to any large degree when forming the complex. The structure of the ternary complex of EF–Tu has been compared (Nissen et al., 1995) with that of EF-G:GDP (Czworkowski et al., 1994; Al-Karadaghi et al., 1996). The observation is that the overall shape of the ternary complex of EF-Tu is surprisingly similar to the shape of EF–G:GDP (Figure 4). Domains 1 and 2 of EF–Tu and EF–G have almost identical folds, apart from an insertion of a helical domain in domain 1 of EF–G. Domains 3, 4 and 5 of EF–G have folds reminiscent of some of the ribosomal proteins (Ævarsson et al., 1994). Moreover, the spacial organization of these domains is such that they mimic the structure of a tRNA molecule almost perfectly.
Fig. 4. Structural comparison of elongation factors. On the left is the ternary complex of Phe-tRNA:EF-Tu:GDPNP (Nissen et al., 1995) and on the right is EF-G:GDP (Al-Karadaghi et al., 1996). The PDB codes are 1TTT and 1DAR, respectively. In both factors, domain 1 is red and domain 2 is green. EF–G has a blue helical insertion in domain 1. Nucleotides bound to domain 1 are shown in ball-and-stick models, and an Mg2+ ion in EF–Tu is shown as a grey ball. Domain 3 of EF–Tu is cyan. The tRNA and domains 3, 4 and 5 of EF–G are magenta. The anticodon of tRNA is at the bottom of the ternary complex, and Phe attached to the terminal ribose is seen in black between domains 1 and 2 of EF–Tu.
This ‘macromolecular mimicry’ of a tRNA by the three domains of the protein EF–G must have a functional importance for their interaction with the ribosome. The functional cycle of the elongation phase is such that EF–G, after it has catalysed the translocation of tRNAs and mRNA on the ribosome, is released from the ribosome as EF–G:GDP. The next functional event during the cycle is the binding of a new aa–tRNA:EF–Tu:GTP complex. Thus, the minimum prediction one can make from the mimicry is that EF–G:GDP is forming a binding site for the ternary complex on the ribosome (Liljas, 1996; Nyborg et al., 1996). The release of EF–G:GDP and the binding of the ternary complex are thus closely linked in the elongation cycle, while the binding of EF–G:GTP, the structure of which is not yet known, is preceded and followed by several functional and structural states of the ribosome compared with these events. However, the cryo-electron microscopic reconstruction of the ribosome bound to EF–G (Agrawal et al., 1998) most probably shows a GTP-like form of EF–G. Interestingly, the anticodon stem mimicking part of EF–G in this reconstruction is moved by ∼10 Å compared with the GDP form. The length of a three base codon is about the same. Finally, from sequence comparisons of EF–Tu, EF–G, IF–2 and RF3, based on the known structures of EF–Tu and EF–G, it is likely that they all contain domains 1 and 2 with very similar folds (Ævarsson, 1995). This implies that they all bind to the ribosome in similar modes, and that the GTPase-activating centre on the ribosome is the same for all of them (Nissen et al., 1995). Whether they also evolved from an ancestral G–protein, and whether that ancestral G–protein evolved by mimicking an RNA, is much more speculative (Nissen et al., 1995).
The function of the release factors that specifically recognize stop codons is such that a ‘macromolecular mimicry’ is almost obvious. Furthermore, it has been know for many years that special tRNAs with mutations in their anticodons work as suppressors of stop codons. Release factors have been shown to compete with suppressor tRNAs for the stop codon (Drugeon et al., 1997). Some publications have indicated that such a mimicry could be found by sequence comparisons between RFs and EF–G (Ito et al., 1996; Buckingham et al., 1997) supported by further biochemical experiments (Buckingham et al., 1997; Pavlov et al., 1998). Structural information on a release factor is not yet available to support this view. However, it is very plausible that the specific recognition of the stop codon by release factors involves side chains mimicking nucleic acid base–base interaction, as has been seen in the tRNA synthetases that recognize anticodons. The re-initiation of protein biosynthesis after termination has been studied extensively in recent years. Experiments indicate that the ribosome recycling factor (RRF or RF4) is involved in the dissociation of mRNA and tRNA from the ribosome (Janosi et al., 1996, 1998; Kaji et al., 1998). Dissociation of the 50S subunit from the 70S post-termination complex is thus catalysed by RRF in a process that involves RRF and that requires GTP hydrolysis (Karimi et al., 1999). The crystal structure of RRF from Thermogata maritima was determined recently. It contains two domains, of which one is globular and the other has an extended three-helix bundle. The shape of the RRF is very similar to that of a tRNA. The implication is that RRF binds to the ribosomal A–site and thus induces disassambly of the post-termination complex (Selmer et al., 1999). This is the newest example of macromolecular mimicry between a protein and a tRNA. Whether ‘macro– molecular mimicry’ is also found in part of IF–2 is more uncertain. The ternary complex of initiator tRNA and IF–2:GTP must interact with the P–site of the ribosome, and this interaction can be very different from the interaction of the ternary complex of EF–Tu with the A–site. However, one possibility exists that a part of IF–2, which is a larger molecule than EF–Tu or EF–G, is mimicking a tRNA bound to the A–site in order to ensure the exclusive binding of initiator tRNA to the P–site.
Astonishing progress has been made regarding structural information about the ribosome itself. Not only have we seen ever increasing resolutions of molecular reconstructions of the ribosome based on cryo-electron microscopy in recent years (Frank et al., 1995; Stark et al., 1995, 1997a,b; Agrawal et al., 1996, 1998), but also very recently the crystal structures of the 30S and 50S ribosomal subunits were published in the same issue of Nature (Ban et al., 1999; Clemons et al., 1999), followed by the crystal structure of the whole ribosomal particle in Science (Cate et al., 1999). For the subject of this mini-review, the most interesting information comes from cryo-electron microscopic reconstructions of ribosomes bound to the ternary complex of EF–Tu and to EF–G (Stark et al., 1997b; Agrawal et al., 1998). These complexes are both formed with the help of antibiotics, kirromycin and fusidic acid, respectively, which both prevent the release of the GDP form of the factors. Although the binding of the antibiotics could conceivably induce small alterations in the specific interaction of the factors with the ribosome, a possibility indicated in the complex with EF–Tu (Stark et al., 1997b), the two structures nevertheless support the notion of one common binding mode. The structures also indicate that domains 1 and 2, which are common to EF–Tu and EF–G, have extensive interactions with the 50S and 30S subunits, respectively. The publication of the crystal structure of the 50S subunit of the ribosome (Ban et al., 1999) in addition provides models for the binding site of EF–G and the ternary complex of EF–Tu. This model provides an overview of ribosomal proteins, which could be in contact with the elongation factors, and postulates the close interaction of the ricin/sarcin loop with the switch region 1 of these factors. Such an interaction could possibly lead to the induced GTPase activity of EF–Tu and EF–G after productive binding to the ribosome.
Conclusion
The few examples mentioned above represent very different levels of complexity of ‘macromolecular mimicry’, from very localized mimicry of a few residues (uracil-DNA glycosylase inhibitor, karyopherin α), to vast mimicry of entire domains or macromolecular shapes and surfaces (TAFII230 and translation factors). The general picture emerges that mimicry has been employed as a way to impose stringent control of the interactions of nucleic acids with a given binding site.
As an illustration of this view, a possible functional description of EF–G on the ribosome could be the following: EF–G:GTP enters the ribosome after peptide formation, where the tRNA is left in the A/P state, with the codon–anticodon interaction at the A–site of the 30S subunit, and the newly formed peptidyl-tRNA with its CCA end at the P–site of the 50S subunit (Moazed and Noller, 1989). The cryo-electron microscopic reconstruction of EF–G on the ribosome (Agrawal et al., 1998) shows that the tRNA-mimicking part occupies the A–site on the 30S subunit, as judged from a comparison with a reconstruction of ribosome and tRNA. A possible function of EF–G is that it will mechanically force the tRNA out of the A–site (Abel and Jurnak, 1996), and thus provide translocation. However, EF–G could, by binding to the A–site of the 30S subunit via its ‘macromolecular mimicry’, prevent the peptidyl-tRNA from re-binding to the A–site, and thereby control the correct progress of the elongation cycle.
A very puzzling observation is the very large conformational change of EF–Tu after GTP hydrolysis. At present this is unique among the G–proteins (Kjeldgaard et al., 1996). It is very unlikely that such a large change is needed in order to release the tRNA from the ternary complex into the A–site, or to release EF–Tu itself from its initial binding site at the ribosome. If one assumes that the conformational change of EF–Tu happens while it is still in contact with the ribosome, and not merely in the cytoplasm after release, then this could perhaps be a signal for the large displacement of the CCA end of tRNA from the initial codon–anticodon testing site to the peptidyl transferase site. The conformational change could, furthermore, induce the ribosome to progress into a new functional and structural state.
As seen from this overview, ‘macromolecular mimicry’ seems to be widely adopted as a theme in DNA repair, nuclear localization, transcription, mRNA splicing and in translation, which suggests that this is a very general concept.
Acknowledgments
Acknowledgements
We would like to express our gratitude to Professor Brian F.C.Clark for his continued support for many years and for his enduring willingness to share with us his knowledge of protein biosynthesis and his large scientific network. Our work has been financially supported by the Programme for Biotechnology Research of the Danish Natural Science Research Council and by the EU Research Project ‘Inhibition of transitions in protein synthesis’ (Contract No. BIO4CT972188). The Danish Natural Science Research Council has provided a Post-Doctoral fellowship to P.N., while M.K. has received a Hallas-Møller senior research fellowship from the Novo-Nordisk Foundation.
References
- Abel K. and Jurnak, F. (1996) A complex profile of protein elongation: translating chemical energy into molecular movement. Structure, 4, 229–238. [DOI] [PubMed] [Google Scholar]
- Abel K., Yoder, M.D., Hilgenfeld, R. and Jurnak, F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. [DOI] [PubMed] [Google Scholar]
- Ævarsson A. (1995) Structure-based sequence alignment of elongation factors Tu and G with related GTPases involved in translation. J. Mol. Evol., 41, 1096–1104. [PubMed] [Google Scholar]
- Ævarsson A., Brazhnikov, E., Garber, M., Zheltonosova, J., Chirgadze, Y., Al-Karadaghi, S., Svensson, L.A. and Liljas, A. (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus.EMBO J., 13, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek, P., Grassucci, R.A., Li, Y., Leith, A., Nierhaus, K.H. and Frank, J. (1996) Direct visualization of A-, P- and E-site transfer RNAs in the Escherichia coli ribosome. Science, 271, 1000–1002. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek, P., Grassucci, R.A. and Frank, J. (1998) Visualization of the elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karadaghi S., Ævarsson, A., Garber, M., Zheltonosova, J. and Liljas, A. (1996) The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange. Structure, 4, 555–565. [DOI] [PubMed] [Google Scholar]
- Andersen G.R., Thirup,S., Spremulli,L.L. and Nyborg,J. (1999) Crystal structure of bovine mitochondrial elongation factor Tu. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Avis J.M., Allain, F.H., Howe, P.W., Varani, G., Nagai, K. and Neuhaus, D. (1996) Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol., 257, 398–411. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen, P., Hansen, J., Capel, M., Moore, P.B. and Steitz, T.A. (1999) Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature, 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova, L., Reiser, C.O.A., Schirmer, N.K., Sprinzl, M. and Hilgenfeld, R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Buckingham R.H., Grentzmann, G. and Kisselev, L. (1997) Polypeptide chain release factors. Mol. Microbiol., 24, 449–456. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R. and Doudna, J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N. and Noller, H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Clemons W.M., May, J.L.C., Wimberley, B.T., McCutcheon, J.P., Capel, M.S. and Ramakrishnan, V. (1999) Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature, 400, 833–840. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy, M., Leighton, L., Blobel, G. and Kuriyan, J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk, A., Krikliviy, I. and Tukalo, M. (1998) tRNAPro anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure, 6, 101–108. [DOI] [PubMed] [Google Scholar]
- Czworkowski J., Wang, J., Steitz, T.A. and Moore, P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.M. (1997) Molecular mimicry: can epitope mimicry induce autoimmune disease?Immunol. Cell Biol., 75, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Cech, T.R. and Sullenger, B.A. (1995) Selection of an RNA molecule that mimics a major autogenic epitope of human insulin receptor. Proc. Natl Acad. Sci. USA, 92, 2355–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugeon G., Jean-Jean, O., Frolova, L., Le Goff, X., Philippe, M., Kisselev, L. and Haenni, A.L. (1997) Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res., 25, 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Schwabe, J.W., Chapman, L., Finch, J.T. and Rhodes, D. (1993) The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature, 366, 483–487. [DOI] [PubMed] [Google Scholar]
- Frank J., et al. (1995)A model of protein synthesis based on a new cryo-electron microscopy reconstruction of the E.coli ribosome. Nature, 376, 441–444. [DOI] [PubMed] [Google Scholar]
- Giege R., Frugier, M. and Rudinger, J. (1998) tRNA mimics. Curr. Opin. Struct. Biol., 8, 286–293. [DOI] [PubMed] [Google Scholar]
- Hall R. (1994) Molecular mimicry. Adv. Parasitol., 34, 81–132. [DOI] [PubMed] [Google Scholar]
- Ito K., Ebihara, K., Uno, M. and Nakamura, Y. (1996) Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA–protein mimicry hypothesis. Proc. Natl Acad. Sci. USA, 93, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L., Ricker, R. and Kaji, A. (1996) Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie, 78, 959–969. [DOI] [PubMed] [Google Scholar]
- Janosi L., et al. (1998)Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J., 17, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L., Oubridge, C., Avis, J.M. and Nagai, K. (1996) Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy. Structure, 4, 621–631. [DOI] [PubMed] [Google Scholar]
- Kaji A., Teyssier, E. and Hirokawa, G. (1998) Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—a possible new target for antibacterial agents. Biochem. Biophys. Res. Commun., 250, 1–4. [DOI] [PubMed] [Google Scholar]
- Karimi R., Pavlov, M.Y., Buckingham, R.H. and Ehrenberg, M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kawashima T., Berthet-Colominas, C., Wulff, M., Cusack, S. and Leberman, R. (1996) The structure of the Escherichia coli EF-Tu:EF-Ts complex at 2.5 Å resolution. Nature, 379, 511–518. [DOI] [PubMed] [Google Scholar]
- Keene J.D. (1996a) RNA recognition by autoantigens and autoantibodies. Mol. Biol. Rep., 23, 173–181. [DOI] [PubMed] [Google Scholar]
- Keene J.D. (1996b) RNA surfaces as functional mimetics of proteins. Chem. Biol., 3, 505–513. [DOI] [PubMed] [Google Scholar]
- Kim J.L. and Burley, S.K. (1994) 1.9 Å resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nature Struct. Biol., 1, 638–653. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger, J.H., Hahn, S. and Sigler, P.B. (1993) Crystal structure of yeast TBP/TATA-box complex. Nature, 365, 512–520. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen, P., Thirup, S. and Nyborg, J. (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure, 1, 35–50. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nyborg, J. and Clark, B.F.C. (1996) The GTP-binding motif—variations on a theme. FASEB J., 10, 1347–1368. [PubMed] [Google Scholar]
- Krásny L., Mesters, J.R., Tieleman, L.N., Kraal, B., Fucík, V., Hilgenfeld, R. and Jonák, J. (1998) Structure and expression of elongation factor Tu from Bacillus stearothermophilus.J. Mol. Biol., 283, 371–381. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Liljas A. (1996) Protein synthesis: imprinting through molecular mimicry. Curr. Biol., 6, 247–249. [DOI] [PubMed] [Google Scholar]
- Liu D., Ishima, R., Tong, K.I., Bagby, S., Kokubo, T., Muhandiram, D.R., Kay, L.E., Nakatani, Y. and Ikura, M. (1998) Solution structure of a TBP–TAFII230 complex: protein mimickry of the minor groove surface of the TATA box unwound by TBP. Cell, 94, 573–583. [DOI] [PubMed] [Google Scholar]
- Love J.J., Li, X., Case, D.A., Giese, K., Grosschedl, R. and Wright, P.E. (1995) Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Murphy, M.E.P. (1994) Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Moazed D. and Noller, H.F. (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature, 342, 142–148. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Arvai, A.S., Sanderson, R.J., Slupphaug, G., Kavli, B., Krokan, H.E., Mosbaugh, D.W. and Tainer, J.A. (1995) Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell, 82, 701–708. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F.C. and Nyborg, J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Thirup, S., Kjeldgaard, M. and Nyborg, J. (1999) The crystal structure of Cys-tRNACys:EF-Tu:GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure, 7, 143–156. [DOI] [PubMed] [Google Scholar]
- Nolte R.T., Conlin, R.M., Harrison, S.C. and Brown, R.S. (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyborg J., Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Clark, B.F.C. and Reshetnikova, L. (1996) Structure of the ternary complex of EF-Tu: macromolecular mimicry in translation. Trends Biochem. Sci., 21, 81–82. [PubMed] [Google Scholar]
- Pavletich N.P. and Pabo, C.O. (1991) Zinc finger–DNA recognition: crystal structure of a Zif268–DNA complex at 2.1 Å. Science, 252, 809–817. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer, D.V., Dincbas, V., MacDougall, J., Buckingham, R.H. and Ehrenberg, M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Pedersen G.N., Nyborg, J. and Clark, B.F.C. (1999) Macromolecular mimicry of nucleic acid and protein. IUBMB Life, 48, 1–6. [DOI] [PubMed] [Google Scholar]
- Polekhina G., Thirup, S., Kjeldgaard, M., Nissen, P., Lippmann, C. and Nyborg, J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu:GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Seeman N.C., Rosenberg, J.M. and Rich, A. (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA, 73, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M., Al-Karadaghi, S., Hirokawa, G., Kaji, A. and Liljas, A. (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science, 286, 2349–2352. [DOI] [PubMed] [Google Scholar]
- Song H., Parsons, M.R., Rowsell, S., Leonard, G. and Philips, S.E.V. (1999) Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol., 285, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Springer M., Portier,C. and Grunberg-Manago,M. (1998) RNA mimicry in the translational apparatus. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 377–413. [Google Scholar]
- Stark H., Müller, F., Orlova, E.V., Schatz, M., Dube, P., Erdemir, T., Zemlin, F., Brimacombe, R. and Van Heel, M. (1995) The 70S Escherichia coli ribosome at 23 Å resolution: fitting the ribosomal RNA. Structure, 3, 815–821. [DOI] [PubMed] [Google Scholar]
- Stark H., Orlova, E.V., Rinke-Appel, J., Jünke, N., Mueller, F., Rodnina, M., Wintermeyer, W., Brimacombe, R. and van Heel, M. (1997a) Arrangement of tRNAs in pre- and posttranslational ribosomes revealed by electron cryomicroscopy. Cell, 88, 19–28. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina, M.V., Rinke-Appel, J., Brimacombe, R., Wintermeyer, W. and van Heel, M. (1997b) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature, 389, 403–406. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang, Y., Meyering-Voss, M., Sprinzl, M. and Sigler, P.B. (1997) Crystal structure of the EF-Tu:EF-Ts complex from Thermus thermophilus.Nature Struct. Biol., 4, 650–656. [DOI] [PubMed] [Google Scholar]