Abstract
In Escherichia coli, both secretory and inner membrane proteins initially are targeted to the core SecYEG inner membrane translocase. Previous work has also identified the peripherally associated SecA protein as well as the SecD, SecF and YajC inner membrane proteins as components of the translocase. Here, we use a cross-linking approach to show that hydrophilic portions of a co-translationally targeted inner membrane protein (FtsQ) are close to SecA and SecY, suggesting that insertion takes place at the SecA/Y interface. The hydrophobic FtsQ signal anchor sequence contacts both lipids and a novel 60 kDa translocase-associated component that we identify as YidC. YidC is homologous to Saccharomyces cerevisiae Oxa1p, which has been shown to function in a novel export pathway at the mitochondrial inner membrane. We propose that YidC is involved in the insertion of hydrophobic sequences into the lipid bilayer after initial recognition by the SecAYEG translocase.
Keywords: membrane protein/Oxa1p/SRP/targeting/translocase
Introduction
Two main targeting pathways with different substrate specificity direct proteins to the translocase (the SecAYEG complex) in the Escherichia coli inner membrane (Valent et al., 1998). The SecB pathway utilizes the cytosolic chaperone SecB to transfer periplasmic and outer membrane protein precursors to the peripheral translocase component SecA at the inner membrane (reviewed in Driessen et al., 1998). The signal recognition particle (SRP) pathway is specialized mainly in the co-translational targeting of inner membrane proteins (MacFarlane and Muller, 1995; de Gier et al., 1996; Ulbrandt et al., 1997; Koch et al., 1999), although the exact subset of inner membrane proteins that depends on the SRP has yet to be defined.
In eukaryotic cells, a related but more complex SRP mediates co-translational targeting of both secreted and membrane proteins to the membrane of the endoplasmic reticulum (ER; reviewed in Rapoport et al., 1996). Mammalian SRP is a ribonucleoprotein complex that binds via its 54 kDa subunit (SRP54) to hydrophobic targeting signals in short nascent polypeptides. Interaction of the SRP with the α-subunit of the SRP receptor (SR) allows insertion of the nascent protein into the translocation channel of the ER membrane. The translocation channel is an oligomeric structure that is formed by subunits of the Sec61 complex, of which the Sec61α and Sec61γ subunits are related in sequence to E.coli SecY and SecE, respectively. Nascent membrane proteins are distinguished from secreted proteins at the translocase, which opens laterally towards the lipid bilayer in response to the insertion of a transmembrane domain (Martoglio et al., 1995). During co-translational insertion of a membrane protein, different proteinaceous components are contacted prior to its eventual release into the lipid bilayer (Do et al., 1996; Laird and High, 1997).
The E.coli SRP consists of 4.5S RNA and a 48 kDa GTPase designated P48 (or Ffh for fifty-four homologue), which show homology to the eukaryotic SRP 7S RNA and SRP54, respectively (reviewed in Luirink and Dobberstein, 1994; Wolin, 1994). In addition, an E.coli homologue of SRα has been identified (the GTPase FtsY), based on sequence similarity, affinity for SRP in vitro and defective targeting upon depletion in a conditional mutant strain. The E.coli SRP interacts in vitro with nascent proteins that expose a particularly hydrophobic targeting sequence, which in part explains its preference for inner membrane proteins (Valent et al., 1995, 1997). Upon addition of FtsY, GTP and inner membranes, the nascent chain is released from the SRP and inserts into the membrane in the vicinity of the translocase components SecA and SecY (Valent et al., 1998).
In the present study, we have used bifunctional and photoreactive cross-linking reagents to analyse in detail the molecular environment of a nascent inner membrane protein (FtsQ) in the E.coli inner membrane after co-translational targeting by the SRP. Our data indicate that the signal anchor sequence of a 108mer insertion intermediate is close to both lipids and YidC, whereas its hydrophilic regions are still adjacent to both SecA and SecY. YidC is a newly characterized polytopic inner membrane protein that is homologous to Oxa1p, a component shown to function in inner membrane protein assembly in mitochondria (He and Fox, 1997; Hell et al., 1997, 1998). YidC was co-purified with the core SecYEG translocase and its expression was up-regulated upon overexpression of translocase components. Based on these observations, we suggest that YidC is a novel translocase component that functions in the transfer of hydrophobic polypeptide segments from the SecAYEG complex into the lipid bilayer.
Results
Model protein and general experimental strategy
The E.coli protein FtsQ was used as a model substrate to investigate the early stages of inner membrane protein insertion in vitro. FtsQ is a monotopic type II membrane protein that interacts with the SRP in vitro (Valent et al., 1997) and depends on SRP for efficient targeting in vivo (J.W.de Gier, unpublished observation). It is involved in cell division and contains a signal anchor (SA) sequence between residues 24 and 49 (Carson et al., 1991).
Site-specific cross-linking approaches were employed to examine in detail the molecular environment of the SA sequence and flanking hydrophilic regions of membrane-inserted nascent FtsQ 108mer (108FtsQ). The insertion intermediates were generated by in vitro translation of truncated mRNA in a homologous cell-free translation system in the presence of inverted inner membrane vesicles (IMVs) to allow targeting, and in the presence of [35S]methionine to label the nascent chains (Valent et al., 1998). Since the truncated mRNAs lack a termination codon, the targeted nascent chains remain attached to the ribosome as peptidyl-tRNA.
Nascent FtsQ inserts into the membrane at the SecA/Y interface
Using lysine-specific bifunctional cross-linkers, we have shown previously that FtsQ nascent chains contact E.coli SRP (Valent et al., 1997). Addition of IMVs during translation induces the release of the SRP in a process that depends upon the SRP receptor FtsY and requires GTP (Valent et al., 1998). The released nascent chains then associate with the membrane in close proximity to the translocase components SecA and SecY and acquire a sodium carbonate-resistant conformation in the membrane (Valent et al., 1998; Scotti et al., 1999).
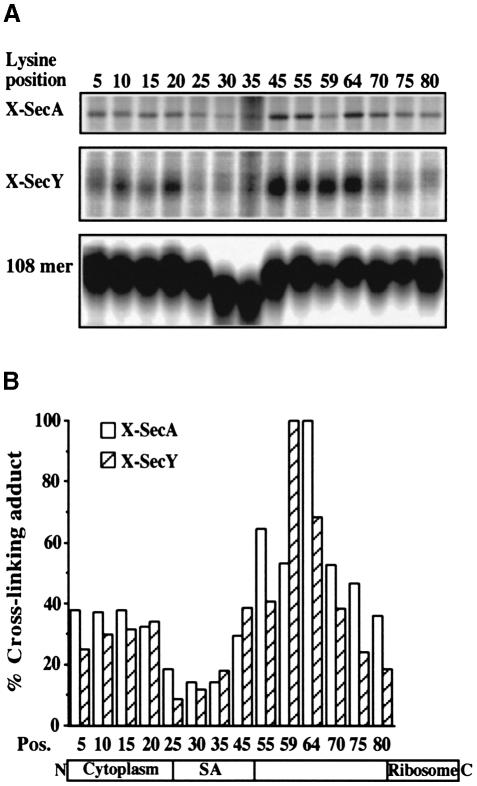
To examine in detail how the targeted 108FtsQ interacts with the inner membrane Sec complex, a series of single lysine 108FtsQ mutants was generated with the lysines introduced ∼5 residues apart, excluding the ribosome-embedded region of the nascent chain. This allowed us to scan the molecular environment throughout the exposed region of the membrane-targeted nascent chain using the membrane-permeable lysine-specific reagent disuccinimidyl suberate (DSS).
Each mutant protein was synthesized in vitro in the presence of IMVs and then subjected to extraction with sodium carbonate and cross-linking. Two mutants (lysines at position 40 and 49) were hardly expressed for reasons that are unclear and were not analysed further. All other mutants were expressed and acquired carbonate resistance to ∼40% (data not shown), very similar to the wild-type protein, which contains a single lysine at position 59 (Valent et al., 1998). Given the relatively low intrinsic efficiency of the E.coli in vitro translocation system, this result indicates that the mutant proteins analysed were targeted and inserted into the membrane properly. Apparently, a single lysine in the strongly hydrophobic SA sequence is tolerated at certain positions. It should be noted that a comparable mutant membrane protein harbouring two lysines in the SA sequence is still capable of SRP-mediated targeting and subsequent insertion into the ER membrane (High et al., 1991).
Analysis of the resulting cross-links showed that all positions gave adducts at ∼120 and ∼42 kDa, but with different efficiencies (Figure 1A). The adducts were identified by immunoprecipitation using anti-SecA and anti-SecY sera, respectively, as previously observed with wild-type 108FtsQ (not shown; Valent et al., 1998). In Figure 1B, a quantitation of the cross-linked products is shown, normalized for differences in translation efficiencies. The efficiency of cross-linking to SecA and SecY is largely similar, indicating that membrane insertion takes place at an interface between SecA and SecY. Cross-linking was strongest in the region around position 60, which is exposed just outside the ribosome. The region between positions 25 and 45 that corresponds to the SA showed only weak cross-linking to SecA/Y, suggesting that it is no longer juxtaposed to lysine residues in SecA/Y and may have left the translocase at this stage of insertion. The residual cross-linking to SecA/Y detected with these mutants may be attributed to the N–terminal amino group of 108FtsQ since similar results were obtained using a mutant 108FtsQ that lacks any lysine residues (data not shown). Weak but significant cross-linking to SecA/Y was detected in the N–terminal region (positions 5–20), which is exposed to the cytoplasm in the native protein (Carson et al., 1991). Apparently, this region is still close to the translocation site.
Fig. 1. Scanning cross-linking of single lysine 108FtsQ mutants to SecA and SecY. (A) Mutant 108FtsQ was synthesized in the presence of IMVs. The samples were treated with the bifunctional cross-linker DSS and extracted with sodium carbonate. Panels corresponding to SecA and SecY cross-linking adducts (X-SecA and X-SecY) and nascent chains (108mer) are shown. The latter panel is derived from a separate 15% gel to allow a better resolution and quantification of small polypeptides (see Materials and methods). (B) SecA and SecY cross-linking adducts from (A) were quantified by phosphoimaging and each signal was corrected for the respective translation efficiency. Highest values for cross-linking efficiency were taken as 100%. Average values of three independent experiments are shown. The positions of the lysine residues in the mutant 108FtsQ as well as their location in the different domains of 108FtsQ are indicated.
The signal anchor sequence of nascent FtsQ interacts with both YidC and lipids
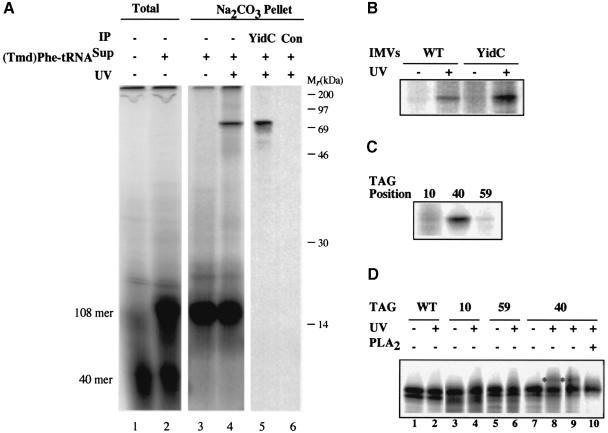
From analogous studies of membrane protein insertion at the ER of eukaryotes (Martoglio et al., 1995; Do et al., 1996; Mothes et al., 1997), we anticipated that the SA sequence of 108FtsQ might have left the protein-conducting channel and be in contact with other membrane proteins or lipids, consistent with the sodium carbonate-resistant nature of the membrane-inserted nascent chains. To explore this possibility, we used an alternative, site-specific, cross-linking approach that allows cross-linking to both proteins and lipids (Brunner, 1996). A stop codon (TAG) was introduced at position 40 in the SA region of 108FtsQ and suppressed during in vitro synthesis by addition of 4-(3-trifluoromethyl-3-diazirinyl)phenylalanyl-tRNASup [(Tmd)Phe-tRNASup], which carries a highly reactive carbene-generating photoreactive probe.
In the absence of (Tmd)Phe-tRNASup, 40mer nascent chains were produced (Figure 2A, lane 1), which did not achieve carbonate resistance (Figure 2A, lane 3). Addition of (Tmd)Phe-tRNASup suppressed the TAG40 mutation, resulting in significant amounts of nascent 108FtsQ (Figure 2A, lane 2). This species was targeted normally and became membrane inserted, as judged by its resistance to sodium carbonate extraction (Figure 2A, lane 3) and the DSS-dependent cross-linking of its endogenous lysine to SecA/Y (data not shown). UV irradiation resulted in the appearance of a major photocross-linking product of ∼69 kDa (Figure 2A, lane 4), which was not seen in a non-irradiated control sample (Figure 2A, lane 3). Since each 108mer nascent chain contains the (Tmd)Phe, the cross-linking efficiency could be quantified and amounted to 13%. Considering this relatively high efficiency and the short spacer arm (7 Å) of the photo probe, such cross-linking can be expected to reflect an extensive and close contact. Subtraction of the 12 kDa contribution of the nascent FtsQ chain leaves a cross-linking partner of ∼57 kDa. After screening a number of antisera to candidate E.coli inner membrane proteins, we could positively identify YidC, a polytopic inner membrane protein with a molecular mass of 60 kDa (Sääf et al., 1998), as this major cross-linking partner (Figure 2A, lanes 5 and 6). The amount of the ∼69 kDa cross-linking product increased when E.coli IMVs were prepared from a strain that overproduces YidC, confirming the identity of the cross-linked partner (Figure 2B).
Fig. 2. The SA sequence of targeted 108FtsQ interacts with both YidC and lipids. (A) In vitro translation of 108FtsQTAG40 was carried out in the presence of IMVs and in the absence or presence of (Tmd)Phe-tRNASup as indicated. Aliquots were TCA precipitated (Total). Upon translation in the presence of (Tmd)Phe-tRNASup, samples were UV irradiated or kept in the dark for 10 min at 4°C and extracted with sodium carbonate as indicated. UV-irradiated samples were immunoprecipitated using antiserum raised against YidC or control pre-immune serum as indicated. (B) IMVs from a strain that overproduces YidC were compared with wild-type IMVs in the photocross-linking reaction described in (A). YidC adducts in the sodium carbonate pellet are shown. (C) 108FtsQTAG10 and 59 were compared with 108FtsQTAG40 in the photocross-linking reaction described in (A). The amounts of suppressed 108FtsQ produced during translation were equalized to enable direct visual comparison of cross-linking efficiencies. YidC cross-linking adducts in the sodium carbonate pellet are shown. (D) 108FtsQ nascent chains were produced and cross-linked as described in (A) with (Tmd)Phe incorporated at positions 10, 40 and 59 as indicated. As a control, wild-type 108FtsQ (WT) was produced under the same translation conditions. Prior to sodium carbonate extraction, samples were UV irradiated or kept in the dark as indicated. As a control, 108FtsQTAG40 nascent chains were treated with bee venom phospholipase (PLA2; lane 10) or mock treated in incubation buffer (lane 9). Lipid cross-linking adducts are indicated by an asterisk.
In addition to YidC, we found that residue 40 of 108FtsQ was photocross-linked to a low molecular weight component (∼0.5–1 kDa) (Figure 2D, cf. lanes 7 and 8). Based on its size, we presumed this to be phospholipid and confirmed this by cleavage with phospholipase A2 (PLA2) (Figure 2D, lanes 9 and 10).
Specific and periodic contact between the signal anchor sequence of nascent FtsQ and YidC
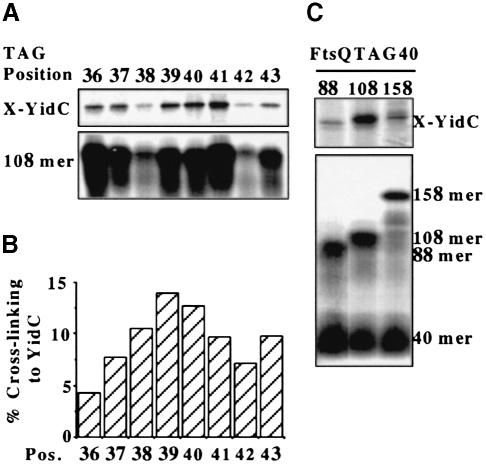
To investigate the interaction of the SA sequence of 108FtsQ with YidC in more detail, TAG mutations were introduced at positions 36–43 and the constructs were analysed for photocross-linking to YidC (Figure 3A). Quantitation of the results of this scanning photocross-linking approach revealed a helical periodicity (Figure 3B), suggesting that the SA has a helical structure with one side interacting with YidC, possibly via helix–helix contacts. The analysed SA region may also be in contact with lipids since small mobility shifts of 108FtsQ were observed upon irradiation at each position but without an obvious periodic pattern (data not shown). YidC was most likely not cross-linked with DSS via the lysines engineered into the SA of 108FtsQ because the transmembrane domains of YidC contain very few lysine residues (Sääf et al., 1998). TAG mutations at positions 10 and 59 in the hydrophilic regions that flank the FtsQ SA did not show significant cross-linking to YidC (Figure 2C) nor to lipids (Figure 2D, lanes 3–6), suggesting that the contact with YidC and lipids is restricted to the SA sequence. The proximity of these positions to SecA and SecY as shown by bifunctional cross-linking (Figure 1) was not detected by the Tmd probe, probably due to quenching of the photo probe by the aqueous environment.
Fig. 3. Scanning photo cross-linking of targeted FtsQ mutants to YidC. (A) TAG codons at positions 36–43 in the SA sequence of 108FtsQ were suppressed as described in the legend to Figure 2. Half of each sample was kept in the dark and extracted with sodium carbonate to determine the amount of suppressed 108FtsQ present in each membrane fraction (108mer panel). The other half was UV irradiated for 10 min prior to extraction. YidC cross-linking adducts in sodium carbonate-extracted membranes are shown (X-YidC panel). (B) YidC cross-linking adducts from (A) were quantified and expressed relative to the amount of suppressed 108FtsQ present prior to cross-linking in the sodium carbonate-extracted membranes. The average values of three independent experiments are shown. (C) FtsQ constructs of different length with a TAG codon at position 40 were suppressed as described in the legend to Figure 2. After translation, aliquots were taken and either directly TCA precipitated to determine suppression and translation efficiencies (lower panel) or extracted with sodium carbonate to determine the integration efficiency of each construct into the membrane (not shown). The rest of the translation reactions were UV irradiated for 10 min (the same amount of nascent chains was present in each sample prior to cross-linking). YidC cross-linking adducts in sodium carbonate-extracted membrane pellets are shown (X-YidC panel).
To analyse the interaction of the SA sequence with YidC during membrane insertion, two additional FtsQ insertion intermediates of 88 and 158 amino acids with a TAG40 mutation were investigated. The 88, 108 and 158mers achieved similar resistance to sodium carbonate extraction (not shown), but cross-linking to YidC was significantly reduced for the 88 and 158mers (Figure 3C), indicating that the SA–YidC contact occurs transiently at a very specific stage of FtsQ membrane insertion.
YidC is associated with the SecYEG translocase
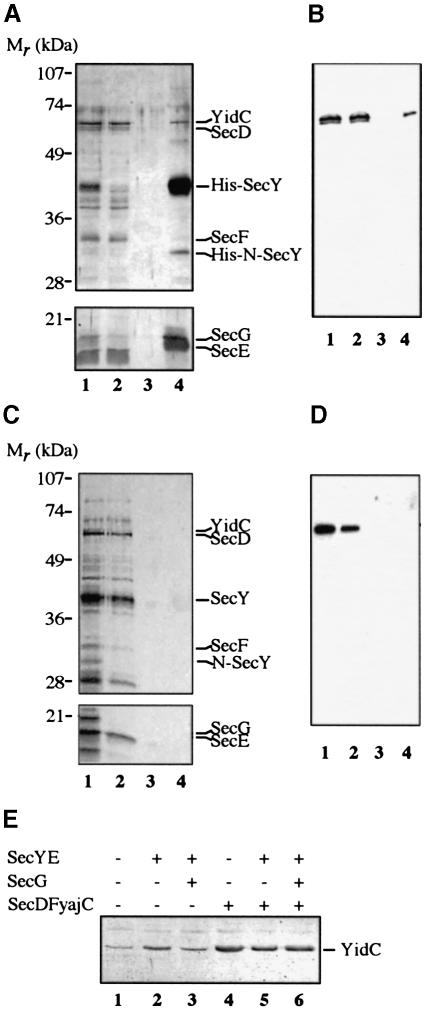
The simultaneous cross-linking of SecA/Y and YidC to different regions of the same 108FtsQ insertion intermediate suggests that YidC is, at least transiently, positioned in the vicinity of the translocase. To examine a possible direct association of YidC with translocase components, we purified the translocase from a strain that overproduces all known components of the translocase, SecY, E, G, D and F and YajC, one of which (SecY) carries a histidine tag to facilitate purification under native conditions (van der Does et al., 1998). IMVs from this strain were purified and subjected to solubilization using the relatively mild detergent dodecylmaltoside (DDM). The translocase was purified from the detergent extract using ion exchange chromatography and subsequently Ni+–NTA chromatography. Purified fractions were analysed initially by SDS–PAGE and silver staining (Figure 4A). The identity of the protein bands in the purified fractions was verified using polyclonal antibodies specific for the known translocase components (data not shown).
Fig. 4. YidC is associated with the SecYEG complex. YajCSechisYEGDF (A and B) and YajCSecYEGDF (C and D) IMVs solubilized with DDM and partially purified by DEAE anion exchange chromatography (lane 1) loaded on an Ni+–NTA column (lane 2, flow-through), washed (lane 3, wash) and eluted with imidazole (lane 4, elution). Samples were analysed by SDS–PAGE and silver staining (A and C). YidC was detected using Western blotting with the YidC polyclonal antiserum (B and D). The positions of the various proteins are indicated. His-N-SecY is a proteolytic N–terminal fragment of SecY. (E) IMVs derived from the wild-type cells (lane 1) and cells overproducing SecYE (lane 2), SecYEG (lane 3), YajCSecDF (lane 4), YajCSecYEDF (lane 5) or YajCSecYEGDF (lane 6) were isolated. Samples were loaded on SDS–PAGE gels and the levels of YidC were detected with the YidC polyclonal antiserum after Western blotting.
Upon ion exchange chromatography, all overproduced translocase components co-eluted in a single fraction, as shown previously (van der Does et al., 1998) (Figure 4A, lane 1). A 60 kDa protein, identified as YidC by immunoblotting, was co-purified significantly with the translocase components (Figure 4A and B, lane 1). In contrast to SecD and SecF, part of YidC was co-purified further with SechisYEG by Ni+–NTA chromatography (Figure 4A and B, lanes 2–4). As a negative control, the procedure was repeated with YajCSecYEGDF complex without a histidine tag on SecY. During anion exchange chromatography, this YajCSecYEGDF complex eluted in fractions different from the His tag-containing YajCSecYEGDF complex, resulting in several additional contaminating bands (compare Figure 4A, lane 1 and C, lane 1). In the absence of the histidine tag on SecY, neither the SecYEG complex nor the YidC protein was found to bind to the Ni+–NTA column (Figure 4C and D).
Plasmid-directed overproduction of Sec protein has been reported to result in the simultaneous overproduction of chromosomally encoded Sec proteins due to a stabilizing effect of the specific interactions among the translocase components (Matsuyama et al., 1990; Sagara et al., 1994). The levels of chromosomally encoded YidC were determined by immunoblotting in IMVs derived from cells that overproduce various combinations of Sec proteins (van der Does et al., 1998) and compared with the wild type (Figure 4E). Overproduction of SecYE, or YajCSecDF with or without the SecYEG complex caused a dramatic increase in the level of YidC. The overexpression of the various Sec proteins was evident from gels stained with Coomassie Brilliant Blue (data not shown). Taken together these data tentatively identify YidC as a component that is associated with the translocase.
Discussion
In this work, four novel and important observations are reported. First, we show that a polytopic E.coli inner membrane protein of unknown function, YidC, interacts with the SA sequence of a co-translationally targeted inner membrane protein (FtsQ) at an early and very specific stage of FtsQ membrane insertion. Secondly, YidC can be partly co-purified with the SecYEG translocase and its expression is up-regulated in response to overexpression of translocase components. Thirdly, in addition to YidC, lipids associate with the SA sequence while FtsQ is still attached to the ribosome. Fourthly, the hydrophilic regions of targeted FtsQ that flank the SA sequence show extensive contact to both SecA and SecY. Based on these data, we propose a model for the molecular environment of membrane-inserted nascent 108FtsQ (Figure 5).
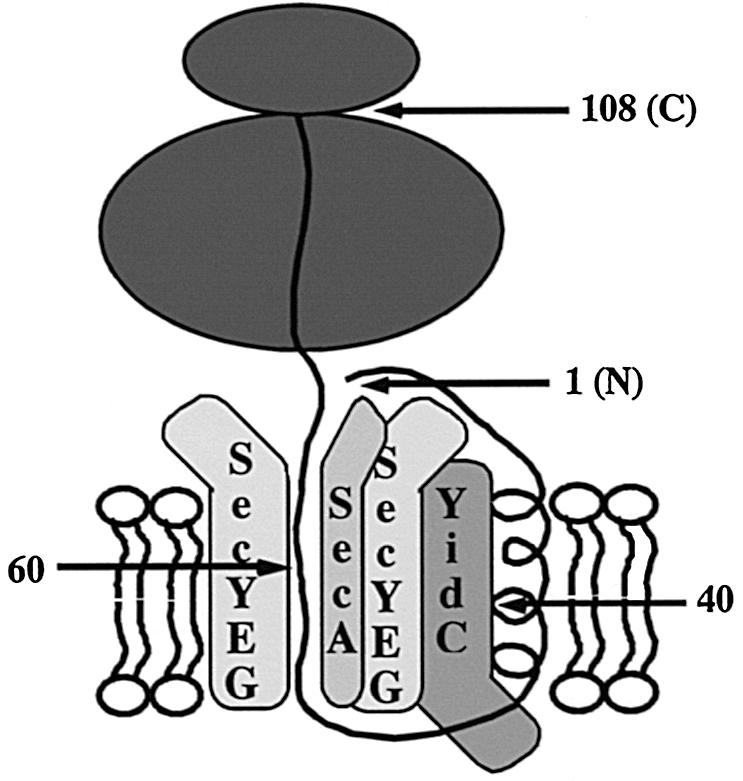
Fig. 5. Model for the molecular environment of membrane-inserted nascent 108FtsQ.
In a previous study, we have described a cross-linking approach to demonstrate that the SRP pathway delivers nascent FtsQ at the membrane (Valent et al., 1998). The SRP receptor FtsY, GTP and inner membranes were shown to be required for release of nascent FtsQ from the SRP. Upon release of the SRP at the membrane, targeted nascent FtsQ was shown to insert into the membrane in a carbonate-resistant conformation in the vicinity of SecA and SecY. These data were interpreted to suggest that the SecB and SRP targeting mechanisms deliver a variety of precursor proteins to a related or identical translocase complex of the E.coli inner membrane. Here, we have used a scanning bifunctional and photocross-linking approach to analyse how nascent FtsQ inserts into the membrane as a first step towards its functional assembly. The combination of these techniques has proven invaluable for the analysis of interactions in aqueous and hydrophobic compartments of the membrane (Mothes et al., 1997; Plath et al., 1998). It should be noted that the approach is unbiased in the sense that a completely homologous in vitro assay was used in which none of the components was overproduced.
The SA sequence of 108FtsQ was found to interact with both lipids and YidC by photocross-linking (Figure 2). Apparently, the SA sequence exits the translocase laterally and enters the lipid environment before translation of the protein is terminated, similar to membrane protein integration into the ER in eukaryotes (Martoglio et al., 1995; Mothes et al., 1997). Assuming that the ribosome covers the C–terminal 35 amino acids of nascent FtsQ, it follows that lipids and YidC are encountered when the SA is exposed only 25–50 amino acids outside the ribosome and that YidC must be close to the translocase, which is cross-linked to the hydrophilic regions of the same, which insertion intermediate.
Both SecA and SecY were found in proximity to the complete exposed hydrophilic region of 108FtsQ, which flanks the SA sequence using the bifunctional cross-linker DSS (Figure 1). SecA is a hydrophilic protein that is able to penetrate deeply into the membrane during its catalysis of pre-protein translocation (reviewed in Driessen et al., 1998). Our data suggest that SecA inserts likewise during membrane protein insertion close to SecY (which is bound by SecA with high affinity), thus lining the channel that initially receives the nascent chain (Figure 5). Interestingly, recent targeting studies that involved proteoliposomes instead of crude IMVs have shown that SecA is not required for the initial insertion of 108FtsQ in the vicinity of SecY (Scotti et al., 1999). SecA may have been recruited at the 108FtsQ insertion site to play a role later in the assembly process when the large periplasmic C–terminal tail of the monotopic type II protein FtsQ is translocated into the periplasm.
Optimal cross-linking to SecA/Y took place when the cross-linking residue was located around position 60 (∼15 residues from the ribosomal nascent chain exit site). Apparently, the nascent chain exit site of the ribosome is very close to the cytosolic side of the translocase. A possible direct contact between the translocase and co–translationally targeted ribosomes, as observed in eukaryotes (Kalies et al., 1994), awaits further analysis.
No function for YidC has been reported to date, and it has not been identified previously as a component of the membrane insertion machinery of E.coli. Our results suggest that YidC is a translocase component that is associated with the SecYEG heterotrimer (Figure 4). The nature and extent of the connection between YidC and the translocase remain to be established. Possibly, YidC participates in the formation of a translocase that is specific for proteins targeted co-translationally via the SRP. This translocase would differ from the translocase approached post-translationally via SecB in its exact composition but would share common core elements (SecYEG). A similar specialization of translocase complexes has been observed in Saccharomyces cerevisiae (reviewed in Stirling, 1999). Alternatively, YidC may be recruited specifically at the translocase upon the co-translational insertion of inner membrane proteins.
It should be noted that upon Ni+–NTA chromatography, only part of YidC was co-purified with SechisYEG, whereas part remained in the unbound fraction with SecD, SecF and YajC (Figure 4A and B). This may reflect a relatively weak association of YidC with SecYEG or the existence of different subcomplexes that contain YidC. In this respect, it is noteworthy that overexpression of SecYE but also of YajCSecDF results in up-regulation of YidC expression (Figure 4E). Duong and Wickner (1997) have identified two heterotrimeric translocase subcomplexes, SecYEG and YajCSecDF, based on co-immunoprecipitation from detergent-solubilized membranes. Interestingly, an unidentified 60 kDa protein was co-immunoprecipitated with both subcomplexes and we surmise that this is YidC.
Significantly, YidC is homologous to the S.cerevisiae mitochondrial inner membrane protein Oxa1p, which has been reported to play a crucial role in the biogenesis of N-tail proteins, a subset of inner membrane proteins with a long exported N–terminal region (He and Fox, 1997; Hell et al., 1997, 1998). Cross-linking studies suggested that Oxa1p interacts transiently with nascent species of N-tail proteins such as CoxII (Hell et al., 1998). By analogy, it has been suggested that Oxa1p homologues function in a novel export mechanism for the N–terminal tails of inner membrane proteins in bacteria and chloroplasts that both contain an Oxa1p homologue (Sundberg et al., 1997; Hell et al., 1998). Our data clearly indicate that the role of YidC is not restricted to N-tail proteins since it is found in contact with both nascent FtsQ and nascent leader peptidase (data not shown), inner membrane proteins that differ in topology and carry no large translocated N-tails.
The precise function of YidC remains to be determined. However, we speculate that after the initial integration of SA sequences within the E.coli translocase, YidC acts as a receptor for SA sequences that have exited the membrane insertion site laterally. Strikingly, a similar function has been proposed for the polytopic ER membrane protein TRAM, which has no known homologue in bacteria, and also associates with SA sequences and cleavable signal sequences during their co-translational membrane insertion at the translocon (Görlich et al., 1992; Do et al., 1996). We cannot entirely exclude the possibility that YidC acts in parallel rather than sequentially to SecA/Y. However, the simultaneous cross-linking of 108FtsQ to SecA/Y (from position 59) and YidC (from position 40) and preliminary cross-linking data using even shorter nascent FtsQ chains make such a scenario less likely. The data presented also do not rule out the possibility that YidC plays some role during the membrane insertion of cleavable signal sequences present on secretory proteins. Finally, our identification of YidC as a component of the E.coli SecYEG translocase, which is absent from S.cerevisiae mitochondria (Glick and von Heijne, 1996), raises intriguing questions concerning the evolution of the mitochondrial inner membrane translocation machinery.
Materials and methods
Enzymes and materials
Restriction enzymes, Taq polymerase and bee venom PLA2 were from Roche Molecular Biochemicals GmbH (Mannheim, Germany). Megashortscript T7 transcription kit was from Ambion Inc. (Austin, TX). [35S]Methionine and protein A–Sepharose were from Amersham International (Buckinghamshire, UK). DSS was from Pierce (Rockford, IL). T4 RNA ligase was from Epicentre Technologies (Madison, WI). All other chemicals were supplied by Sigma Chemical Co. (St Louis, MO).
Strains and growth conditions
Strain MC4100 was used to obtain translation lysates and IMVs [both prepared as described in De Vrije et al. (1987)]. Strain MRE600 was used to prepare translation lysate for suppression of TAG stop codons in the presence of (Tmd)Phe-tRNASup (Ellman et al., 1991). Strain Top10F′ was used as expression host for plasmid pAra14-YidC and for routine maintenance of plasmid constructs (Valent et al., 1997). For overexpression of YidC, cells were grown in LB medium containing 0.2% fructose to mid-log phase. Expression of YidC was induced by adding l-arabinose to 0.2%. Growth was continued for 2 h and IMVs were prepared as described (De Vrije et al., 1987). Strain SF100 was used as expression host for plasmids pET320 (van der Does et al., 2000), pET605 (van der Does et al., 1998), pET610 (van der Does et al., 1998) and pET606 (A.Kaufmann, in preparation) to allow the overexpression of SecYE, SecYEG, SechisYEG and YajCSecDF, respectively. For overproduction, cells were induced at an OD660 of 0.8 by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside and growth was continued for 2 h. IMVs were isolated as described (Van der Does et al., 1998).
Plasmid constructs
To introduce single lysines in nascent 108FtsQ, pC4Meth108FtsQ was mutated in a two-step PCR procedure to replace the endogenous lysine codon at position 59 by an arginine codon (Valent et al., 1997). The resulting pC4Meth108FtsQK– plasmid was used as a template to introduce single lysine codons at positions 5, 10, 15, 20, 25, 30, 35, 40, 45, 49, 55, 64, 70, 75 and 80 by PCR. pC4Meth108FtsQ was used as a template to introduce TAG stop codons at positions 10, 36, 37, 38, 39, 40, 41, 42, 43 and 59 by the same procedure. Plasmids pC4Meth88FtsQ and pC4Meth158FtsQ were obtained by PCR using pUC18-FtsQ as a template (Valent et al., 1995). A TAG mutation at position 40 was introduced into both pC4Meth88FtsQ and pC4Meth158FtsQ.
For overexpression of YidC, yidC was cloned into the expression vector pAra14 (Cagnon et al., 1991) by PCR on MC4100 cells introducing NcoI and HindIII sites at the 5′ and 3′ ends, respectively.
The nucleotide sequences of all cloned (mutant) genes were confirmed by DNA sequencing.
In vitro transcription, translation, targeting and cross-linking
To prepare truncated mRNA, pC4MethFtsQ derivative plasmids were linearized with HindIII and transcribed using T7 polymerase as described by the manufacturer (Ambion Inc., Austin, TX). For bifunctional cross-linking, in vitro translation and targeting of FtsQ constructs were carried out as described (Valent et al., 1997). Prior to bifunctional cross-linking, 3-(cyclohexylamino)propanesulfonic acid (CAPS) was added (50 mM final concentration; pH 9.0) to minimize cross-linking from the N–terminal amino group. Cross-linking was induced with 0.2 mM DSS for 10 min at 25°C and quenched at 4°C by adding 1/10 volume of quench buffer (1 M glycine, 100 mM NaHCO3 pH 8.5). Soluble and peripheral membrane proteins were extracted with 0.18 M Na2CO3 pH 11.3 for 15 min on ice. Membrane fractions containing integral membrane proteins were pelleted by ultracentrifugation (10 min at 110 000 g).
For photocross-linking, (Tmd)Phe was site-specifically incorporated into FtsQ nascent chains by suppression of TAG stop codons using (Tmd)Phe-tRNASup in an in vitro translation system essentially as described (Ellman et al., 1991). Translation was carried out for 20 min at 37°C at 6 mM Mg(OAc)2. (Tmd)Phe-tRNASup was prepared as described (High et al., 1993). Suppression strongly depended on the position of the TAG codon. Samples were irradiated for 10 min on ice at 15 cm from a Spectroline model B100/F black light lamp equipped with a 100 W mercury bulb and a 365 nm filter.
Antisera
The YidC polyclonal antiserum was raised in rabbit against a peptide that consisted of the 17 C–terminal amino acids of YidC by Agrisera (Umeå, Sweden). The antisera directed against SecA and SecY were a kind gift of W.Wickner.
Phospholipid analysis
PLA2 treatment was carried out essentially as described (Martoglio et al., 1995). Membrane fractions of targeted FtsQ nascent chains were prepared as described above and resuspended in 100 μl of 100 mM Tris pH 8, 10 mM CaCl2 and 1% Triton X-100 per 22 μl targeting reaction. Ten units of bee venom PLA2 were added and samples were incubated for 10 min at 41°C. Digestion was stopped by adding 10% trichloroacetic acid (TCA), 25% acetone.
Purification of the translocase complex
YajCSecYEGDF IMVs with or without a His6 tag at the N–terminus of SecY were solubilized in 2% DDM and partially purified by DEAE anion exchange chromatography in 10 mM Tris pH 8.0, 20% glycerol, 0.03% DDM using a linear gradient of 0–300 mM KCl. This sample was loaded onto a HiTrap chelating column (Pharmacia Biotech, Uppsala, Sweden) equilibrated with 10 mM imidazole pH 8.0, 20% glycerol and 0.03% DDM, washed in the same buffer and eluted with 100 mM imidazole pH 7.0, 20% glycerol, 0.03% DDM (van der Does et al., 1998, 2000).
Sample analysis and quantification
Samples were analysed on 10, 12 or 15% SDS–polyacrylamide gels. Lipid cross-linking adducts were characterized on 16.5% tricine gels. Radiolabelled proteins were visualized by phosphoimaging using a Molecular Dynamics PhosphorImager 473 and quantified using the Imagequant software from Molecular Dynamics.
Acknowledgments
Acknowledgements
We thank Corinne ten Hagen-Jongman, Quido Valent and Jelto Swaving for technical support. Steve High and Nellie Harms are thanked for continuous discussion and critical reading of the manuscript. Irmi Sinning, Niek Dekker and William Wickner are acknowledged for generous supply of materials. This work was supported by a TMR project grant from the European Commission (to P.S. and J.L.), a TMR fellowship from the E.C. (to J.W.de G.), grants from the Swedish Natural and Technical Sciences Research Councils and the Swedish Cancer Foundation (to G.v.H.), the Swiss National Science Foundation (to J.B.) and a PIONIER grant of The Netherlands Organization for Scientific Research (NWO) (to A.D.)
References
- Brunner J. (1996) Use of photocrosslinkers in cell biology. Trends Cell Biol., 6, 154–157. [DOI] [PubMed] [Google Scholar]
- Cagnon C., Valverde, V. and Masson, J.-M. (1991) A new family of sugar-inducible expression vectors for Escherichia coli.Protein Eng., 4, 843–847. [DOI] [PubMed] [Google Scholar]
- Carson M.J., Barondess, J. and Beckwith, J. (1991) The FtsQ protein of Escherichia coli: membrane topology, abundance and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol., 173, 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gier J.W.L., Mansournia, P., Valent, Q.A., Phillips, G.J., Luirink, J. and von Heijne, G. (1996) Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett., 399, 307–309. [DOI] [PubMed] [Google Scholar]
- De Vrije T., Tommassen, J. and de Kruijff, B. (1987) Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vesicles requires both ATP and the proton motive force. Biochim. Biophys. Acta, 900, 63–72. [DOI] [PubMed] [Google Scholar]
- Do H., Falcone, D., Lin, J.L., Andrews, D.W. and Johnson, A.E. (1996) The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell, 85, 369–378. [DOI] [PubMed] [Google Scholar]
- Driessen A.J.M., Fekkes, P. and van der Wolk, J.P.W. (1998) The Sec system. Curr. Opin. Microbiol., 2, 216–222. [DOI] [PubMed] [Google Scholar]
- Duong F. and Wickner, W. (1997) Distinct catalytic roles of the SecYE, SecG and SecDFyaiC subunits of preprotein translocase holoenzyme. EMBO J., 16, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman J., Mendel, D., Anthony-Cahill, S., Noren, C.J. and Schultz, P.G. (1991) Biosynthetic method for introducing unnatural amino acids site-specifically into proteins. Methods Enzymol., 202, 301–337. [DOI] [PubMed] [Google Scholar]
- Glick B.S. and von Heijne, G. (1996) Saccharomyces cerevisiae mitochondria lack a bacterial-type Sec machinery. Protein Sci., 5, 2651–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Hartman, E., Prehn, S. and Rapoport, T. (1992) A protein of the endoplasmic reticulum involved in polypeptide translocation. Nature, 357, 47–52. [DOI] [PubMed] [Google Scholar]
- He S. and Fox, T.D. (1997) Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell, 8, 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Herrmann, J., Pratje, E., Neupert, W. and Stuart, R.A. (1997) Oxa1p mediates the export of the N- and C–termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett., 418, 367–370. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann, J.M., Pratje, E., Neupert, W. and Stuart, R.A. (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA, 95, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S., Görlich, D., Wiedmann, M., Rapoport, T.A. and Dobberstein, B. (1991) The identification of proteins in the proximity of signal-anchor sequences during their targeting to and insertion into the membrane of the ER. J. Cell Biol., 113, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S., et al. (1993)Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem., 268, 26745–26751. [PubMed] [Google Scholar]
- Kalies K.U., Görlich, D. and Rapoport, T.A. (1994) Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol., 126, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H.-G., Hengelage, T., Neumann-Haefelin, C., MacFarlane, J., Hoffschulte, H.K., Schimz, K.-L., Mechler, B. and Müller, M. (1999) In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/B as constituents of two independent protein-targeting pathways of Escherichia coli.Mol. Biol. Cell, 10, 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird V. and High, S. (1997) Discrete cross-linking products identified during membrane protein biosynthesis. J. Biol. Chem., 272, 1983–1989. [DOI] [PubMed] [Google Scholar]
- Luirink J. and Dobberstein, B. (1994) Mammalian and Escherichia coli signal recognition particles. Mol. Micobiol., 11, 9–13. [DOI] [PubMed] [Google Scholar]
- MacFarlane J. and Müller, M. (1995) Functional integration of a polytopic membrane protein of E.coli requires the bacterial signal recognition particle. Eur. J. Biochem., 223, 766–771. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann, M.W., Brunner, J. and Dobberstein, B. (1995) The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell, 81, 207–214. [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Akimaru, J. and Mizushima, S. (1990) SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett., 269, 96–100. [DOI] [PubMed] [Google Scholar]
- Mothes M., Heinrich, S.U., Graf, R., Nilsson, I.M., von Heijne, G., Brunner, J. and Rapoport, T.A. (1997) Molecular mechanism of membrane protein integration in the endoplasmic reticulum. Cell, 89, 523–533. [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes, W., Wilkinson, B.M., Stirling, C. and Rapoport, T.A. (1998) Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell, 94, 795–807. [DOI] [PubMed] [Google Scholar]
- Rapoport T.A., Jungnickel, B. and Kutay, U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Sääf A., Monne, M., de Gier, J.W. and von Heijne, G. (1998) Membrane topology of the 60-kDa Oxalp homologue from Escherichia coli.J. Biol. Chem., 273, 30415–30418. [DOI] [PubMed] [Google Scholar]
- Sagara K., Matsuyama, S. and Mizushima, S. (1994) SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J. Bacteriol., 176, 4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti P.A., Valent, Q.A., Manting, E.H., Urbanus, M.L., Driessen, A.J.M., Oudega, B. and Luirink, J. (1999) SecA is not required for SRP-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem., 274, 29883–29888. [DOI] [PubMed] [Google Scholar]
- Stirling C.J. (1999) Protein targeting to the endoplasmic reticulum in yeast. Microbiology, 145, 991–998. [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter, J.G., Fridborg, I., Cleary, S.P., Robinson, C. and Coupland, G. (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell, 9, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt N.D., Newitt, J.A. and Bernstein, H.D. (1997) The E.coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell, 88, 187–196. [DOI] [PubMed] [Google Scholar]
- Valent Q.A., Kendall, D.A., High, S., Kusters, R., Oudega, B. and Luirink, J. (1995) Early events in preprotein recognition in E.coli: interactions of SRP and trigger factor with nascent polypeptides. EMBO J., 14, 5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent Q.A., de Gier, J.-W.L., von Heijne, G., Kendall, D.A., ten Hagen-Jongman, C.M., Oudega, B. and Luirink, J. (1997) Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol. Microbiol., 25, 53–64. [DOI] [PubMed] [Google Scholar]
- Valent Q.A., Scotti, P.A., High, S., de Gier, J.W.L., von Heijne, G., Lentzen, G., Wintermeyer, W., Oudega, B. and Luirink, J. (1998) The E.coli SRP and SecB targeting pathways converge at the translocon. EMBO J., 17, 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does C., Manting, E.H., Kaufmann, A., Lutz, M. and Driessen, A.J.M. (1998) Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry, 37, 201–210. [DOI] [PubMed] [Google Scholar]
- Van der Does C., Swaving,J., van Klompenburg,W. and Driessen,A.J.M. (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Wolin S.L. (1994) From the elephant to E.coli: SRP-dependent protein targeting. Cell, 77, 787–790. [DOI] [PubMed] [Google Scholar]