Abstract
The endoplasmic reticulum quality control (ERQC) system retains and degrades soluble and membrane proteins that misfold or fail to assemble. Vph1p is the 100 kDa membrane subunit of the yeast Saccharomyces cerevisiae V–ATPase, which together with other subunits, assembles into the V–ATPase in the ER, requiring the ER resident protein Vma22p. In vma22Δ cells, Vph1p remains an integral membrane protein with wild-type topology in the ER membrane before undergoing a rapid and concerted degradation requiring neither vacuolar proteases nor transport to the Golgi. Failure to assemble targets Vph1p for degradation in a process involving ubiquitylation, the proteasome and cytosolic but not ER lumenal chaperones. Vph1p appears to possess the traits of a ‘classical’ ERQC substrate, yet novel characteristics are involved in its degradation: (i) UBC genes other than UBC6 and UBC7 are involved and (ii) components of the ERQC system identified to date (Der1p, Hrd1p/Der3p and Hrd3p) are not required. These data suggest that other ERQC components must exist to effect the degradation of Vph1p, perhaps comprising an alternative pathway.
Keywords: degradation/ER quality control/proteasome/ubiquitylation
Introduction
Proteins destined for the secretory pathway are translocated across the endoplasmic reticulum (ER) membrane where they fold and may be modified by disulfide bond or carbohydrate addition, proteolytic processing, oligomerization and/or assembly into multi-subunit complexes. A quality control system associated with the ER (ERQC) monitors these processes and selectively retains and degrades those proteins that have failed to fold, oligomerize or assemble correctly (Bonifacino and Weismman, 1998). ERQC substrates include: CFTR-ΔF508, α1-antitrypsin inhibitor, unassembled T-cell receptor subunits, apoB under conditions of limited lipid availability and CPY*, a mutant form of the yeast vacuolar carboxypeptidase Y (CPY), which is retained in the ER (Bonifacino and Weissman, 1998). The ERQC degradative machinery also acts in a regulatory role to moderate the half-life of correctly folded ER resident proteins in response to cellular signals. The cholesterol biosynthesis enzyme HMG-CoA reductase (HMG–R) is turned over rapidly in response to elevated levels of mevalonate and its metabolites (Hampton et al., 1996).
ERQC substrate degradation occurs independently of the lysosome (or yeast vacuole) and proteolysis proceeds in the absence of traffic from the ER to the Golgi (Finger et al., 1993). Inhibition of ERQC degradation was accomplished with drugs that inactivate the cytosolic proteasome or with mutations that reduce proteosomal activity (Ward et al., 1995; Hiller et al., 1996). Substrates of the 26S proteosome require ubiquitylation, as do a number of ERQC substrates (Bonifacino and Weissman, 1998). In yeast, two ubiquitin-conjugating enzymes, Ubc6p and Ubc7p, are localized to the cytosolic face of the ER membrane, and mutations in UBC6 or UBC7 have been found to reduce the degradation rate of ERQC substrates (Biederer et al., 1996; Hiller et al., 1996; Hampton and Bhakta, 1997; Galan et al., 1998; Loayza et al., 1998). The ERQC system utilizes cytosolic proteasome–ubiquitylation components for degradation and requires ERQC substrates to be exported from the ER to the cytosol. Results of co-immunoprecipitation and genetic data demonstrated that components of the translocon were also involved in the retro-translocation of ERQC substrates (Wiertz, 1996b; Pilon et al., 1997; Plemper et al., 1997).
Several approaches in yeast have identified components of the ERQC machinery. In the ER lumen, the molecular chaperone hsp70 protein Kar2p (BiP homolog), but not cytosolic molecular chaperones, participates in the degradation of the soluble substrates CPY* (Plemper et al., 1997), pro-α-factor and a mutant form of α1-antitrypsin inhibitor (Brodsky et al., 1999). Additionally, three genes encoding membrane-associated ER proteins have been identified: Der1p, required for the turnover of CPY*; Hrd3p, for the degradation of HMG–R; and Hrd1p (also named Der3p), as necessary for the degradation of both CPY* and HMG–R (Hampton et al., 1996; Knop et al., 1996; Bordallo et al., 1998).
Vph1p is a 100 kDa polytopic membrane subunit of the yeast vacuolar H+–ATPase (V–ATPase), which assembles in the ER with other subunits into the Vo sector of the enzyme prior to delivery to the vacuolar membrane where the final 13 subunit V–ATPase complex functions to acidify the organelle (Graham and Stevens, 1999). The absence of any one of three ER assembly factors (Vma22p, Vma21p or Vma12p) results in the rapid and specific degradation of the now unassembled Vph1p. Degradation of unassembled Vph1p was found to occur independently of vacuolar proteases and did not require transport from the ER to the Golgi (Hill and Stevens, 1994, 1995; Jackson and Stevens, 1997).
Here we demonstrate that Vma22p is required for Vph1p assembly into the V–ATPase and characterize the degradation of unassembled Vph1p. Our findings that unassembled Vph1p is an integral membrane protein in the ER prior to its degradation and that its turnover involves an energy-dependent step(s), ubiquitylation and the 26S proteasome, classify Vph1p as an ERQC substrate. We have assessed the role of chaperones on both sides of the ER membrane in degrading Vph1p and found that cytosolic but not ER lumenal chaperones are required. The novel attributes of Vph1p turnover include not only the employment of a different subfamily of Ubc enzymes but also that degradation occurs independently of the previously identified DER/HRD components of the yeast ERQC system.
Results
Vph1p is localized to the ER membrane in vma22Δ cells
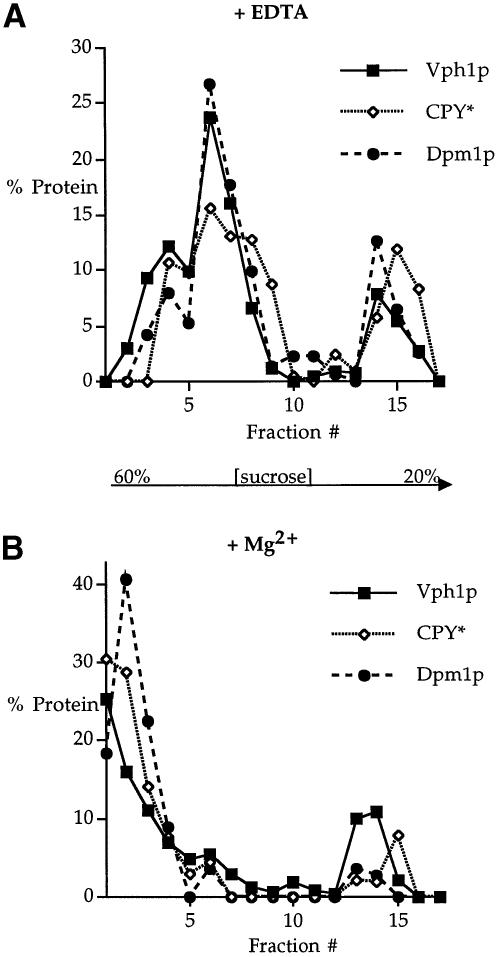
To investigate the subcellular site of Vph1p degradation, we employed sucrose gradient fractionation (Roberg et al., 1997) on whole-cell extracts prepared from vma22Δ cells. Fractions collected from the gradient were resolved by SDS–PAGE and the resulting immunoblots probed for Vph1p, the resident ER membrane protein Dpm1p and CPY*, an ERQC substrate previously localized to the ER (Finger et al., 1993). In EDTA-treated extracts, all three proteins co-fractionated (Figure 1A). The presence of Mg2+ in the fractionation procedure causes ER membranes to shift to fractions of greater buoyant density, probably due to the maintenance of the Mg2+-dependent association of ribosomes with the ER membranes (Roberg et al., 1997), and can be used to demonstrate further a protein's localization to the ER. The parallel fractionation experiment on Mg2+-containing extracts showed that Vph1p, CPY* and Dpm1p again co-fractionated in fractions denser than those obtained in the presence of EDTA (Figure 1B). The co-fractionation of Vph1p with both a well characterized ERQC substrate and a resident ER membrane protein and the collective shift to denser fractions by all three proteins demonstrates that Vph1p is found in the ER of vma22Δ cells.
Fig. 1. Fractionation of Vph1p in vma22Δ cells. Whole-cell lysates from vma22Δ (KHY125/pMM322) cells were fractionated on a density gradient of 20–60% sucrose containing either 10 mM EDTA (A) or 2 mM Mg2+ (B). Relative levels of Vph1p, Dpm1p and CPY* were quantitated by Western blotting and chemifluorescence. The smaller peak of all three proteins higher in the gradient corresponds approximately to the position of the sample loaded on the gradient.
Vph1p is an ERQC substrate in vma22Δ cells
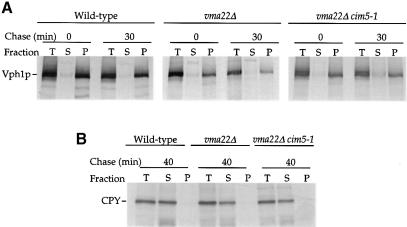
The association of Vph1p with the ER of vma22Δ cells and its rapid degradation suggested that Vph1p is an ERQC substrate. To confirm this, we further examined vma22Δ cells to verify that Vph1p was within the ER membrane, and therefore a bona fide ERQC substrate, instead of peripherally associated with the membrane's cytosolic face, which might result if Vma22p was required for translocation of Vph1p into the ER membrane. Alkaline carbonate extractions (Fujuki et al., 1982) performed on lysates prepared from radiolabeled wild-type and vma22Δ cells indicated that Vph1p fractionated as an integral membrane protein in both (Figure 2A). CPY, a soluble protein localized to the lumen of the secretory pathway, served as a control for the alkaline treatment and was released successfully into the supernatant fraction (Figure 2B).
Fig. 2. Membrane association of unassembled Vph1p. Alkaline carbonate extractions were performed on wild-type (YPH499), vma22Δ (KHY78) and vma22Δ cim5-1 (KHY77) cell extracts either after a 10 min radiolabeling period or following a 30 min chase. Vph1p (A) or CPY (B) was immuno– precipitated from total (T), supernatant (S) and pellet (P) fractions and analyzed by SDS–PAGE and fluorography.
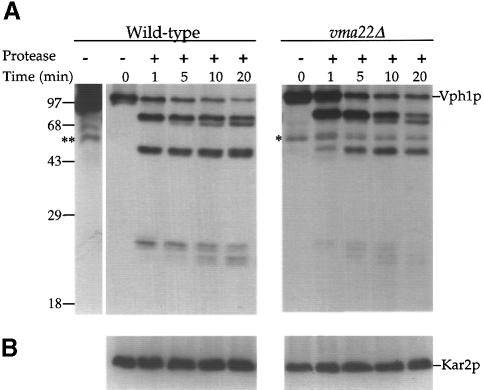
Although Vma22p is an integral membrane protein in vma22Δ cells, it was still possible that it assisted in a membrane insertion of at least one of the six or seven membrane-spanning domains of Vph1p. The absence of Vma22p would result in a form of Vph1p that spans the ER membrane but has an altered topology. To distinguish between a specialized translocation role and the proposed assembly role of Vma22p, a protease shaving assay was performed on Vph1p in microsomes isolated from wild-type or vma22Δ cells. Microsomes were subjected to limiting amounts of protease digestion, the samples prepared for SDS–PAGE and the resulting immunoblots probed with anti-Vph1p antibodies. The protease sensitivity of Vph1p from wild-type and vma22Δ cells was very similar, with both giving a near identical spectrum of cleaved species over the same time period (Figure 3A). The reduced steady-state level of Vph1p in vma22Δ cells required a longer exposure of the immunoblot resulting in the eventual appearance of a non-specific band (*, Figure 3A). This band was also seen in the wild-type extract when the immunoblot was exposed for a similar period of time (**, Figure 3A).Addition of the detergent Triton X-100 resulted in the complete degradation of Vph1p from either strain (data not shown). The microsomes remained intact throughout the course of the protease digestion, as shown by the complete protection of the soluble ER lumenal protein Kar2p (Figure 3B), unless detergent was present in the reaction mix (data not shown).
Fig. 3. Partial proteolysis of Vph1p in wild-type and vma22Δ cells. Microsomes were prepared from wild-type (SNY28) and vma22Δ (KHY38) cells and treated for various times with subtilisin (0.8 μg/ml). Precipitated proteins were resolved by SDS–PAGE and immunoblots probed with either anti-Vph1p (A) or anti-Kar2p (B) antibodies. The single asterisk (*) indicates a non-specific band present in vma22Δ immunoblots and ** indicates the same non-specific band present in wild-type immunoblots when overexposed.
These data indicated that the polytopic Vph1p produced in vma22Δ cells had integrated successfully into the ER membrane and assumed the same membrane topology as that of wild-type Vph1p. However, as our anti-Vph1p antibodies recognize the N–terminal half of Vph1p, we cannot completely eliminate the possibility of differences in the C–terminal insertion pattern. The defect seen in vma22Δ cells is therefore consistent with a failure of Vph1p to assemble with other subunits of the V–ATPase complex. The degradation of a fully translocated but unassembled protein in the ER membrane establishes Vph1p as an ERQC substrate.
Degradation of unassembled Vph1p is an energy-requiring process
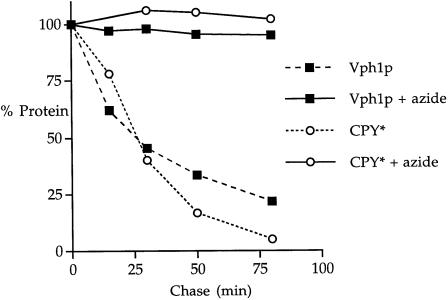
ERQC degradation is an energy-requiring process as ATP is thought to be necessary for retro-translocation, ubiquitylation and proteasome-mediated degradation (McCracken and Brodsky, 1996). To assess whether Vph1p turnover was energy requiring, a radiolabeling kinetic analysis was performed in which energy poison sodium azide was absent or present during the chase period. The half-life of unassembled Vph1p was extended to >800 min in the presence of azide as compared with 25 min in KHY125 cells without azide (Figure 4). The degradation of another ERQC substrate, CPY*, was also greatly stabilized in the presence of azide, extending the half-life of the protein from ∼26 min to >800 min (Figure 4).
Fig. 4. Degradation of unassembled Vph1p is an energy-requiring process. vma22Δ (KHY125/pMM322) cells were radiolabeled, chased in either the absence or presence of 20 mM sodium azide and aliquots harvested at various times. After immunoprecipitation with anti-Vph1p antibodies, the Vph1p-depleted extracts were then used to immunoprecipitate CPY*. After separation by SDS–PAGE, the amount of radioactivity in each sample was quantified using a Storm phosphorimager; the 0 min chase time quantity was designated as 100% and the percentage of protein remaining was plotted against time. To determine half-lives, first order decay curves were generated using average values from at least three independent experiments.
Degradation of unassembled Vph1p involves ubiquitylation and the 26S proteasome
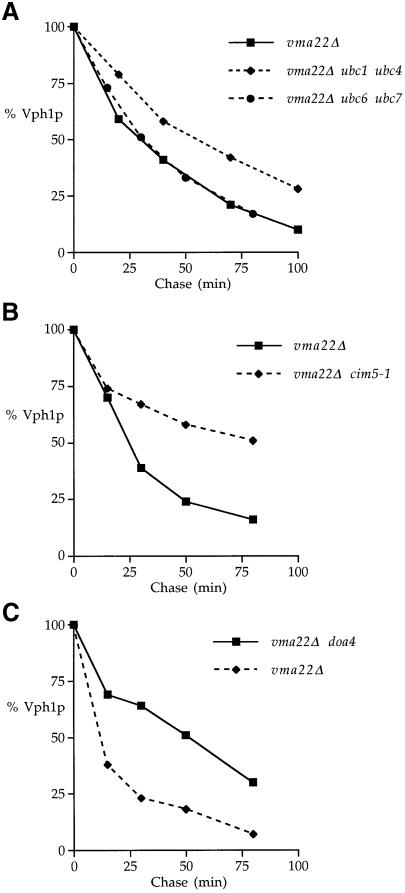
Another hallmark of ERQC–mediated degradation is ubiquitylation involving Ubc enzymes. VMA22 was disrupted in a number of ubcΔ strains to determine whether the absence of a particular Ubc protein retarded the turnover of Vph1p (Table I). Of the ubc mutations tested, the ubc1 ubc4 (Figure 5A) and ubc2 ubc4 double mutantsslowed the degradation of Vph1p ∼2-fold. Surprisingly, Vph1p was not stabilized in a ubc6Δ ubc7Δ double mutant strain (Figure 5A), which lacks the two ER-localized Ubc proteins involved in the ubiquitylation of other yeast ERQC substrates CPY*, Hmg2p, Sec61-2p, Ste6-166p, Pdr5p* and Fur4-430Np (Biederer et al., 1996; Hiller et al., 1996; Hampton and Bhakta, 1997; Galan et al., 1998; Loayza et al., 1998; Plemper et al., 1998).
Table I. Effect of ubc mutations on Vph1p turnover in vma22Δ cells.
| ubc mutation | Stabilization of Vph1p |
|---|---|
| UBC | – |
| ubc1 ubc4 | + |
| ubc2 ubc4 | + |
| ubc6 ubc7 | – |
| ubc1 | – |
| ubc2 | – |
| ubc4 | – |
| ubc5 | – |
| ubc7 | – |
| ubc8 | – |
Fig. 5. The degradation of unassembled Vph1p involves ubiquitylation and the 26S proteasome. (A) KHY172 (vma22Δ), KHY173 (ubc6 ubc7 vma22Δ) and KHY177 (ubc1 ubc4 vma22Δ) cells were radiolabeled, and Vph1p was immunoprecipitated and analyzed as outlined in Figure 4. (B) KHY78 (vma22Δ) and KHY77 (vma22Δ cim5-1) cells. (C) KHY172 (vma22Δ) and KHY51 (vma22Δ doa4Δ) cells.
The involvement of the 26S proteasome in the degradation of unassembled Vph1p was investigated utilizing mutants with defects in proteasome activity. CIM5 encodes a regulatory subunit of the 19S cap of the 26S proteasome, and a cim5-1 mutation previously has been shown to result in the reduced degradation rate of proteasome substrates (Ghislain et al., 1993). VMA22 was deleted in a strain carrying a cim5-1 mutation, and a kinetic analysis was performed to determine the half-life of Vph1p compared with that of a vma22Δ CIM5 parent strain. Figure 5B shows that the cim5-1 mutation significantly reduced the degradation rate of unassembled Vph1p, extending the half-life of the protein from 25 min to 90 min.
DOA4 encodes a proteasome-associated isopeptidase thought to promote proteolysis by cleaving ubiquitin from ubiquitylated substrates during proteasome action (Papa et al., 1999). Introduction of a vma22Δ mutation into a doa4Δ strain was also found to reduce the turnover kinetics of Vph1p (Figure 5C), increasing the half-life of Vph1p ∼3-fold and further implicating ubiquitylation and the 26S proteasome in Vph1p degradation.
Degradation of Vph1p is a concerted process
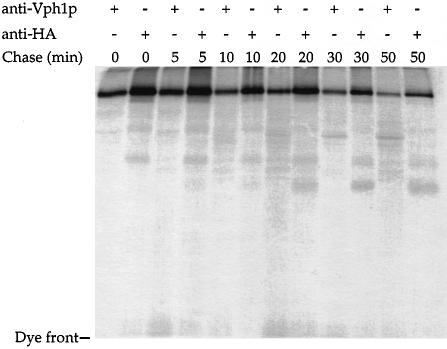
Vph1p consists of an N–terminal half exposed in the cytosol (K.Hill and A.A.Cooper, unpublished data) and a C–terminal half containing six or seven potential membrane-spanning domains. We investigated whether the proteasome degrades only those domains of Vph1p exposed to the cytosol or, alternatively, if Vph1p is extracted from the ER membrane to be fully accessible to the proteasome. To detect any proteolytic fragments that may represent degradative intermediates, we used a functional allele of VPH1 that contained an insertion of three hemagglutinin (HA) epitopes at the C–terminus of Vph1p (Vph1p-HA). This allowed us to monitor the cytosolic N–terminal domain of Vph1p with anti-Vph1p antibodies and the C–terminus with antibodies directed against HA. vma22Δ vph1Δ cells expressing Vph1p-HA were radiolabeled and aliquots harvested at specified time points. The resulting lysates were divided in two and immunoprecipitated with antibodies directed against either Vph1p or HA. Samples were resolved by SDS–PAGE on both 7% (data not shown) and 15% gels to detect both small and large proteolytic fragments. Other than full-length Vph1p, no significant proteolytic species were detected during the course of the degradation using either antibody (Figure 6). Furthermore, the half-life of Vph1p-HA was very similar when calculated from data derived from either antibody. These data suggest that the proteolytic degradation of Vph1p occurs in a concerted process to degrade the entire Vph1p molecule and that Vph1p must be extracted from the ER membrane to be accessible to the proteasome.
Fig. 6. Degradation of unassembled Vph1p is a concerted process. ACY74 (vma22Δ vph1Δ) cells carrying pDJ65C (Vph1p-HA) were radiolabeled and aliquots harvested at various times during the chase period. Lysates were divided in two and Vph1p was immuno- precipitated with either anti-Vph1p or anti-HA antibodies. Samples were resolved by SDS–PAGE on 15% gels.
Unassembled Vph1p is not extracted into the cytosol prior to degradation
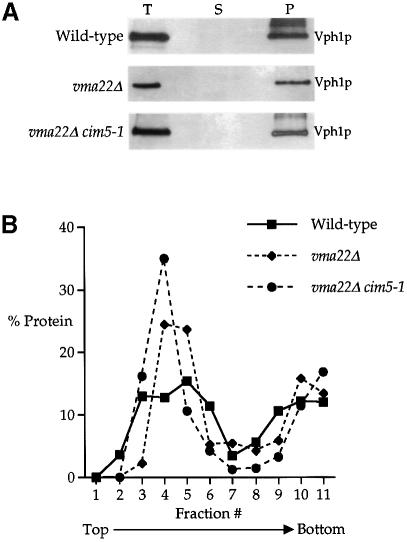
Extraction of Vph1p from the ER membrane for degradation might proceed in one of two ways: complete extraction from the membrane into the cytosol and subsequent degradation by the proteasome or, as each portion of Vph1p is extracted from the membrane, it is degraded immediately by the proteasome. We undertook to determine whether Vph1p was extracted as a full-length molecule from the ER membrane prior to its degradation. Such a species of Vph1p would be expected to accumulate in a supernatant fraction from a strain with reduced proteasome activity. Alkaline carbonate extraction experiments were performed on wild-type, vma22Δ and vma22Δ cim5-1 radiolabeled cell extracts. Vph1p was immunoprecipitated from total, supernatant and pellet fractions after carbonate treatment, the samples resolved by SDS–PAGE and quantified. The fractionation of Vph1p in vma22Δ and vma22Δ cim5-1 cells shown in Figure 2A was very similar to that of wild-type Vph1p, and we could observe no obvious indications of an accumulation of an extracted soluble form of Vph1p. However, this does not completely dispel a two-step membrane extraction/degradation model. The highly hydrophobic character of the C–terminal half of Vph1p might cause a membrane-extracted, pre-degraded form of Vph1p to aggregate in the cytosol with sedimentation properties similar to those of the microsomes used for the carbonate extraction experiments. To distinguish an aggregated extracted form of Vph1p from a membrane-spanning form, we employed the membrane floatation approach of Xiong et al. (1999). Microsomes were first confirmed to contain all of the Vph1p remaining in vma22Δ cells (Figure 7A) and then placed beneath a two-step sucrose gradient prior to centrifugation, during which membranes and associated proteins would be expected to float up through the gradient while aggregates remain pelleted (Xiong et al., 1999). The gradient profiles of wild-type Vph1p and Vph1p from both vma22Δ and vma22Δ cim5-1 cells were very similar and located in the center of the gradient (Figure 7B), indicating that unassembled Vph1p in proteasome-inhibited cells is membrane-associated instead of an aggregate of extracted protein. These data support a model in which Vph1p is extracted and degraded in a concerted process.
Fig. 7. Unassembled Vph1p is not extracted into a soluble fraction prior to degradation. (A) Total cell extracts (T) prepared from wild–type (YPH499), vma22Δ (KHY78) and vma22Δ cim5-1 (KHY77) cells were fractionated into supernatant (S) and microsomal pellet (P) samples, resolved by SDS–PAGE and the resulting immunoblots probed with anti-Vph1p antibodies. (B) Microsomes were layered beneath a discontinuous sucrose gradient and fractionated as described. The relative level of Vph1p in each fraction was determined by Western blotting and chemifluorescence.
Cytosolic but not ER molecular chaperones are required for Vph1p degradation
Molecular chaperones are thought to distinguish native from non-native proteins by binding to hydrophobic stretches of residues that are only exposed in non-native structure. Terminally unassembled or unfolded proteins would continue to display these residues and result in prolonged or repeated interactions with molecular chaperones that may then facilitate their degradation (Hayes and Dice, 1996), potentially by directing a terminally misfolded protein for entry into the ERQC degradative pathway.
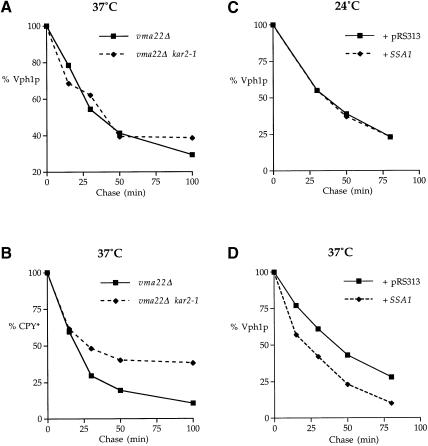
Vph1p has domains exposed on both sides of the ER membrane and so it is possible that both cytosolic and ER lumenal chaperones may contribute to the degradation of unassembled Vph1p. The yeast KAR2 gene encodes the major hsp70 molecular chaperone of the ER lumen, which facilitates both translocation into the ER (Vogel et al., 1990) and the subsequent folding of nascent proteins in the ER lumen (Simons et al., 1995). Kar2p also contributes to the degradation of the soluble ERQC substrates CPY*, pro-α-factor and α1-antitrypsin (Plemper et al., 1997; Brodsky et al., 1999). To determine if Kar2p is involved in Vph1p degradation, we employed the temperature-sensitive kar2-1 allele. At the non-permissive temperature, the mutant Kar2-1p is diminished in its protein-binding capacity but not in its ability to aid in the translocation of proteins into the ER (Brodsky et al., 1999). KHY190 (kar2-1 vma22Δ prc1-1) and KHY181 (KAR2 vma22Δ prc1-1) were radiolabeled at the permissive temperature (24°C) and the chase period was initiated by the addition of heated medium to raise the temperature immediately to 37°C and reduce Kar2-1p's protein-binding capacity. Aliquots were taken at specified time points, the cells were lysed and solubilized Vph1p or CPY* was immunoprecipitated. No difference was observed in the Vph1p turnover rate (Figure 8A), suggesting that Kar2p does not participate in the degradation of Vph1p. To ensure that the rapid temperature shift had inactivated Kar2-1p, we monitored the effect on CPY* turnover using the same cell extracts from which Vph1p had been immunoprecipitated and found that the turnover of CPY* was slowed in kar2-1 cells as compared with that in KAR2 cells (Figure 8B).
Fig. 8. Cytosolic but not lumenal molecular chaperones are involved in the turnover of unassembled Vph1p. KHY190 (vma22Δ kar2-1 prc1-1) and KHY181 (vma22Δ KAR2 prc1-1) cells were radiolabeled at 24°C and chased at 37°C. Aliquots were taken at specific time points, the cells lysed and Vph1p immunoprecipitated from solubilized extracts (A). Vph1p-depleted extracts were then used to immunoprecipitate CPY* (B). ACY76 (vma22Δ ssa1-ts ssa2 ssa3 ssa4) cells carrying either pRS313 or pAC427 (SSA1) were radiolabeled at 24°C and chased at either 24°C (C) or 37°C (D). Vph1p was then immunoprecipitated as described.
Misfolded/unassembled integral membrane proteins in the ER also expose domains to the cytosol where they may be identified as ERQC substrates by cytosolic molecular chaperones. In yeast, the SSA sub-family of hsp70 cytosolic molecular chaperones, encoded by SSA1–SSA4, are required for the folding of certain nascent cytosolic proteins (Kim et al., 1998) in addition to assisting the translocation of some proteins into the ER (Becker et al., 1996). To determine if the SSA family of chaperones contribute to the turnover of Vph1p, the strain ACY76 (vma22Δ ssa1-ts ssa2Δ ssa3Δ ssa4Δ) was constructed which lacks Ssa2p, Ssa3p and Ssa4p and contains a temperature-sensitive mutation in the Ssa1p peptide-binding domain (Becker et al., 1996). ACY76 was transformed with either a vector plasmid (pRS313) or a plasmid containing the wild-type SSA1 gene (pAC427) and radiolabeled at 24°C. As it is not known whether Ssa1p function is required for Vph1p import into the ER membrane, the chase period proceeded for 8 min at 24°C to ensure the complete translocation of the radiolabeled Vph1p before either heated medium was added to raise the temperature immediately to the non-permissive temperature of 37°C or the chase was continued at 24°C. Aliquots were taken at specified time points, the cells were lysed and solubilized Vph1p was immunoprecipitated and quantified (Figures 9C and D). In contrast to the Kar2p results, Ssa1p activity was found to be required for Vph1p turnover as shown by the significant increase in Vph1p half-life when Ssa1p activity was inactivated at 37°C.
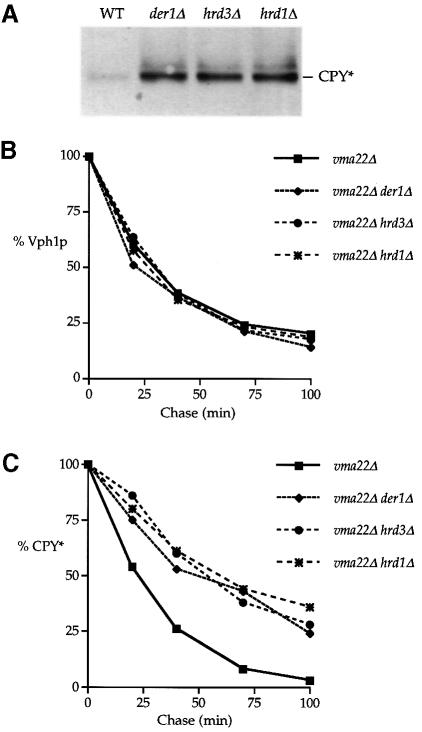
Fig. 9. Der1p, Hrd3p and Hrd1p/Der3p do not participate in the ERQC of Vph1p. (A) Protein extracts prepared from vma22Δ (KHY125), vma22Δ der1Δ (KHY127), vma22Δ hrd3Δ (KHY138)and vma22Δ hrd1Δ (KHY140)cells were resolved by Western blotting and probed with anti-CPY antibodies. (B) KHY125, KHY127, KHY138 and KHY140 cells were radiolabeled, and Vph1p was immuno- precipitated from solubilized extracts and analyzed as outlined in Figure 4. (C) Vph1p-depleted extracts were then used to immuno- precipitate CPY* and analyzed as above.
Vph1p degradation occurs independently of the previously identified ERQC components Der1p, Hrd1p/Der3p and Hrd3p
Genetic screens in yeast employing CPY* and Hmg2p as ERQC substrates have identified three genes DER1, HRD1/DER3 and HRD3 required for their degradation. The potential involvement of these ER membrane proteins in the turnover of Vph1p was examined by disrupting the DER and HRD genes individually in a vma22Δ vph1Δ strain that also carried the CPY*-encoding prc1-1 allele. Strains carrying plasmid-borne VPH1 (pMM322) were grown and protein extracts prepared as described by Knop et al. (1996). Samples were resolved by SDS–PAGE and the resulting immunoblots probed with anti-CPY antibodies. As expected for these mutations, the steady-state level of CPY* from the der1Δ (KHY127), hrd3Δ (KHY138) and hrd1Δ (KHY140) cells was significantly elevated relative to that of wild-type (KHY125) cells (Figure 9A). To determine whether these mutations had any effect on the ERQC of unassembled Vph1p, a kinetic analysis was performed on radiolabeled KHY125, KHY127, KHY138 and KHY140 cells. Our experiments indicated that the absence of Hrd1p, Hrd3p and Der1p had no effect on the degradation of Vph1p as compared with that in KHY125 cells. This surprising result led us to confirm by a radiolabeling kinetic analysis that our derΔ/hrdΔ strains displayed a reduced turnover rate of CPY*. The radiolabeling experiment was therefore repeated, sequentially immunoprecipitating first Vph1p (Figure 9B) and then CPY* (Figure 9C) from the same cell extracts, allowing a direct comparison of the data from our kinetic analysis. CPY* was found to be stabilized in radiolabeled der1Δ, hrd3Δ and hrd1Δ cells as expected, but again these mutations had no effect on the degradation of Vph1p.
Discussion
Vma22p is required for assembling Vph1p into the V–ATPase
Vma22p has been identified as an ER resident protein required for the formation of the yeast V–ATPase. In the absence of Vma22p, the 100 kDa subunit of the V–ATPase, Vph1p, is degraded rapidly and specifically, requiring neither vacuolar proteases nor transport to the Golgi (Hill and Stevens, 1995). In this work, we characterize the fate of unassembled Vph1p in vma22Δ cells.
Fractionation experiments found the steady-state level of Vph1p in vma22Δ cells to be localized to the ER membrane and suggest two possible models for Vma22p function: one in which Vma22p is an assembly protein, required in the ER to assist Vph1p assembly into the V–ATPase, and another where Vma22p functions to aid in the insertion of Vph1p into the ER membrane. If Vma22p functioned in the insertion of Vph1p into the ER membrane, we may expect that its absence would result in a Vph1p molecule that was only peripherally associated with the membrane. However, data from carbonate extraction and membrane floatation experiments (Figures 2 and 7) indicate that the Vph1p remaining in vma22Δ cells behaved as an integral membrane protein similar to wild-type Vph1p. Furthermore, the similar profile of cleavage products seen in protease shaving assays indicates that Vph1p attained the same topology in both vma22Δ and wild-type cells. It is therefore most likely that Vma22p acts to assist in the assembly of Vph1p into the Vo sector of the V–ATPase after the protein has translocated successfully into the ER membrane. This role is very similar to that proposed for Vma12p (Jackson and Stevens, 1997), while Vma21p may play a different role in the V–ATPase assembly (Graham et al., 1998).
Vph1p degradation involves ubiquitylation and the 26S proteasome
Inhibition of Vph1p turnover by mutations in the 26S proteasome and UBC genes indicates that ubiquitylation contributes to the ERQC–mediated degradation of Vph1p. Other ERQC substrates have been found to be ubiquitylated during the course of their degradation, and it is likely that Vph1p is also ubiquitylated. Although approaches using tagged forms of ubiquitin so far have been unsuccessful in demonstrating that unassembled Vph1p is ubiquitylated, we have identified in vma22Δ cells a Vph1p-related species that migrates ∼7 kDa larger than Vph1p (ubiquitin Mr = 8.6 kDa) which is not seen in VMA22 cells (K.Hill and A.A.Cooper, unpublished results).
Our characterization of the effects of UBC genes found that no single ubc mutation inhibited Vph1p turnover but that Ubc4p played a central role, with a ubc1Δ ubc4Δ double mutation causing the greatest inhibition of degradation amongst those ubc combinations tested. Ubc4p shares 92% identity with Ubc5p, and together they contribute to the selective degradation of many short-lived and abnormal proteins. Together, Ubc1p, Ubc4p and Ubc5p are considered to constitute a UBC subfamily providing an essential function as judged by the lethality of a ubc1Δ ubc4Δ ubc5Δ mutation (Seufert et al., 1990). Our evidence suggests that Ubc1p, Ubc4p and Ubc5p would contribute the majority of the Vph1p-ubiquitylating activity and predicts that a ubc4Δ ubc5Δ double mutant would severely inhibit the degradation of unassembled Vph1p. To investigate this, we have repeatedly attempted to construct a vma22Δ ubc4Δ ubc5Δ strain by both gene disruption and genetic crossing, but so far have been unsuccessful, suggesting that the disruption of VMA22 ina ubc4Δ ubc5Δ strain is lethal.
In contrast to the yeast ERQC substrates characterized to date, the ER-associated ubiquitin-conjugating enzymes Ubc6p and Ubc7p are not involved in the degradation of Vph1p. The subcellular localization of Ubc6p and Ubc7p at the ER membrane made them attractive candidates for the ubiquitylation of ERQC substrates, but this does not disqualify other Ubc proteins from participating in the process. Although novel, it is perhaps not surprising that Ubc proteins other than Ubc6p and Ubc7p contribute to the turnover of ERQC substrates, as it is unlikely that two Ubc proteins can recognize all possible ERQC substrates. Even within the pair of Ubc6p and Ubc7p enzymes, there is a difference in their contributions to substrate ubiquitylation as shown by the varying effects of their absence on ERQC substrate degradation. The deletion of UBC6 has no effect on HMG–R degradation (Hampton and Bhakta, 1997) yet significantly reduces the turnover of CPY* (Hiller et al., 1996) and Sec61-2p (Biederer et al., 1996).
The molecular basis for being classified as an ERQC substrate is at present unknown, and it is possible that the process that identifies unassembled proteins such as Vph1p as ERQC substrates may differ from that used to identify misfolded ERQC substrates such as CPY*, and this process may therefore involve different Ubc proteins. Furthermore, the above discussion centers on the UBC/E2 class of enzymes dictating substrate selection for ubiquitylation and it may be that E3 or ubiquitin–protein ligases alone, or in conjunction with E2s, determine substrate specificity.
The impairment of Vph1p degradation in cim5 or doa4 cells demonstrates that the 26S proteasome participates in the degradation of unassembled Vph1p. The lack of proteolytic intermediates observed during Vph1p turnover and the absence of full-length Vph1p extracted into a soluble fraction prior to degradation suggest that Vph1p is simultaneously extracted from the membrane and degraded by the proteasome in a concerted manner, as has been suggested for other ERQC substrates (Mayer et al., 1998; Plemper et al., 1998; Xiong et al., 1999). Such an event would presumably involve the recruitment of the 26S proteasome to the ER membrane. A significant portion of 26S proteasomes have been localized to the ER and nuclear envelope network in Saccharomyces cerevisiae (Enenkel et al., 1998), and such positioning of the proteasomes, especially if bound in the vicinity of the cytosolic face of the translocon, would simplify delivery of an ERQC substrate for degradation. In addition, the six ATPase subunits within the 19S cap of the proteasome might contribute to the driving force required to retro-translocate a ubiquitylated substrate (Mayer et al., 1998; Plemper et al., 1998; Brodsky et al., 1999). However, the attractive possibility that the proteasome solely provides the required extraction force presents an apparent conflict with the findings that certain ERQC substrates, both soluble and membrane-spanning, are released as soluble proteins into the cytosol when proteasome activity is inhibited (McCracken and Brodsky, 1996; Wiertz et al., 1996a, b).
Molecular chaperone involvement in Vph1p degradation
Kar2p, the yeast BiP homolog, is required for the degradation of soluble ERQC substrates including CPY* (Plemper et al., 1997; this study), pro-α-factor and the PiZ variant of α1-antitrypsin (Brodsky et al., 1999). BiP is also implicated in the degradation of soluble proteins associated with the mammalian ER in that a strong correlation has been found between the extent of BiP interaction with unassembled immunoglobulin light chains and the rate of light chain degradation (Skowronek et al., 1998). Vph1p is the first ERQC substrate whose turnover has been examined for the role of both ER lumenal molecular chaperones such as BiP and cytosolic molecular chaperones including Ssa1p. We have found that peptide-binding activity of Kar2p is not required for the degradation of unassembled Vph1p. The turnover of another ERQC substrate, the membrane-spanning Pdr5p* (a mutant yeast ABC transporter), is unaffected in a different kar2 mutant strain (Plemper et al., 1998). This BiP-independent degradation of membrane-spanning ERQC substrates is also suggested in mammalian cells where truncated fragments of the influenza type I membrane protein HA do not associate with BiP during degradation (Zhang et al., 1997).
We have found that the SSA family of cytosolic hsp70s is required for the degradation of Vph1p. Similarly in mammalian cells, cytosolic hsp70 maintains a prolonged interaction with the misfolded membrane-spanning protein CFTR-ΔF508 prior to its degradation (Yang et al., 1993). Furthermore, apolipoprotein B interacts with cytosolic hsp70 when lipid availability is reduced in the ER, and undergoes rapid degradation that is enhanced upon overexpression of hsp70 (Fisher et al., 1997). However, the SSA family of hsp70s are not needed for the turnover of yeast soluble ERQC substrates including pro-α-factor and the PiZ variant of α1-antitrypsin (Brodsky et al., 1999). Soluble ERQC substrates appear to require lumenal but not cytosolic chaperones for their degradation and, conversely, membrane-spanning substrates require cytosolic but not lumenal chaperones to effect their turnover. Different components of the ERQC system may participate in the degradation of soluble and membrane-spanning substrates. Soluble ERQC substrates are probably released from the translocon into the ER lumen (Plemper et al., 1999) where BiP/Kar2p, having identified and retained the protein as a misfolded protein/ERQC substrate, may also act to return it to the translocon for retro-translocation into the cytosol. Kar2p's ability to interact with the translocon via Sec63p may contribute to this step (Brodsky et al., 1999). Der1p might also participate in this step, as to date it has been found to be required only for the turnover of soluble ERQC substrates (Knop et al., 1996). Membrane-spanning ERQC substrates may either enter the ER membrane via a lateral gating of the translocon (Singer, 1990) or remain associated with the translocon. Re-entry of the membrane-spanning substrate into the translocon might occur by a reversal of the lateral gating mechanism. In either situation, membrane-spanning substrates could remain associated or re-associate with the translocon in a BiP-independent manner.
The functions of Ssa1p in the ERQC-mediated degradation of Vph1p might include: (i) acting as a motor or ratchet to extract Vph1p from the membrane into the cytosol in much the same manner as Kar2p is thought to provide directionality to the import of proteins into the ER (Brodsky et al., 1999); (ii) to maintain the unassembled Vph1p in a non-aggregated state accessible to the proteasome; or (iii) Ssa1p may act to partially unfold the cytosolic domain of Vph1p either to expose a ubiquitylation site for UBCenzymes in a manner similar to that proposed for hsc70 (Bercovich et al., 1997) or to assist in unfolding as a prelude for entry into the proteasome. If Ssa1p acts to prevent aggregation of membrane-spanning substrates, then why would this function not be required for retro-translocated soluble substrates? It is possible that soluble substrates exiting the translocon are ubiquitylated rapidly and immediately enter a proteasome bound at the ER membrane, which would eliminate the substrate's opportunity to aggregate in the cytosol.
Any role proposed for the participation of molecular chaperones in ERQC must account for a difference in the way that chaperones, both cytosolic and ER lumenal, facilitate the folding of temporarily misfolded nascent proteins that ultimately will attain the correct mature structure in contrast to effecting the degradation of terminally misfolded proteins (Gottesman et al., 1997). It may be that misfolded proteins that never attain the correct mature structure, due to a mutation or the lack of subunit assembly, might either remain bound to, or continually be rebound by, a molecular chaperone. If the molecular chaperone remained bound to an ‘unfoldable’ protein, the increased time of association might result in entry into the ERQC pathway.
Vph1p turnover occurs independently of DER/HRD gene products
Three ER membrane proteins (Der1p, Hrd1p/Der3p and Hrd3p) were characterized previously as components of the yeast ERQC system. Der1phas so far been found to contribute only to the degradation of soluble ERQC substrates (Knop et al., 1996), whereas Hrd1p/Der3p plays a significant role in the turnover of HMG–R (Hampton et al., 1996), CPY* and Sec61–2p (Bordallo et al., 1998). Hrd3p is required for the degradation of HMG–R (Hampton et al., 1996), and our work has extended its involvement to the turnover of CPY*. Given the participation of Hrd1p/Der3p and Hrd3p in the degradation of both soluble and membrane-spanning ERQC substrates, it was surprising that the absence of either of these gene products had no effect on Vph1p turnover. Vph1p appears to possess the traits of a ‘classical’ ERQC substrate in that it is an unassembled protein in the ER that is degraded rapidly in a process involving ubiquitylation and the proteasome. However, the ability to degrade Vph1p independently of the known components of the ERQC system identified to date (Ubc6p, Ubc7p, Der1p, Hrd1p/Der3p and Hrd3p) suggests that either (i) the ERQC system can bypass the normal requirements for these proteins to degrade Vph1p, perhaps because Vph1p is unassembled rather than misfolded, or (ii) that other ERQC components, perhaps comprising an alternative parallel pathway, exist to effect the degradation of Vph1p. The lack of complete stabilization of Vph1p in proteasome mutants supports the possibility of an alternative degradative system contributing to Vph1p turnover. An alternative pathway might employ the proteolytic activity recently identified in mammalian cells treated with proteasome inhibitors (Glas et al., 1998). Vacuolar proteases can be eliminated as candidates for this activity as the disruption of PEP4 ina cim5-1 strain did not stabilize Vph1p further (data not shown). Components of such a pathway should be identified by the genetic screen presently underway to isolate mutants that stabilize Vph1p. This plate-based assay utilizes a Vph1p-based reporter substrate which, as expected, is not stabilized by disruptions of DER1, HRD1/DER3 or HRD3 (K.Hill and A.A.Cooper, unpublished data).
Materials and methods
Materials
Restriction enzymes were purchased from New England Biolabs. Chemicals were purchased from either Fisher or Sigma.
Strains, plasmids, media and microbiological techniques
Media were prepared as described by Hill and Stevens (1994). Yeast strains used in this study are listed in Table II and the plasmids used are described in Table III.
Table II. Yeast strains.
| Strain | Genotype |
|---|---|
| SNY28 | MATα leu2-3,112 ura3-52 his4-519 ade6a |
| KHY38 | vma22Δ-1::URA3 in SNY28b |
| RH448 | MATa his4 leu2 ura3 lys2 bar1c |
| KHY70 | vma22Δ-2::URA3 in RH448 |
| MHY501 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1d |
| KHY172 | vma22Δ-2::URA3 in MHY501 |
| RH3136 | MATa ubc1::HIS3 ura3 leu2 his3 trp1 lys2 bar1c |
| KHY17 | vma22Δ-2::URA3 in RH3136 |
| RH3134 | MATa ubc2::HIS3 ubc4::TRP1 ura3 leu2 his3 trp1 lys2 bar1c |
| KHY154 | vma22Δ-2::URA3 in RH3134 |
| RH3130 | MATa ubc2::HIS3 his3 ura3 leu2 trp1 bar1c |
| KHY180 | vma22Δ-1::URA3 in RH3130 |
| RH3132 | MATa ubc4::TRP1 ura3 leu2 his3 trp1 lys2 ade2 bar1c |
| KHY67 | vma22Δ-1::URA3 in RH3132 |
| RH3142 | MATa ubc5::LEU2 ura3 leu2 his3 his4 lys2 trp1 bar1c |
| KHY58 | vma22Δ-2::URA3 in RH3142 |
| MHY552 | MATα ubc6::HIS3 ubc7::LEU2 his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1d |
| KHY173 | vma22Δ-2::URA3 in MHY552 |
| RH3145 | MATa ubc8::URA3 ura3 leu2 his3 trp1 ade2 bar1c |
| KHY13 | MATα vma22Δ-4::LEU2 leu2-3,112 his4-519 ade6 ura3-52b |
| KHY183 | vma22Δ-4::LEU2 |
| KHY184 | vma22Δ-4::LEU2 ubc8::URA3 |
| RH3147 | MATa ubc1::HIS3 ubc4::TRP1 ura3 leu2 his3 lys2 trp1 bar1c |
| KHY177 | vma22Δ-2::URA3 in RH3147 |
| MHY623 | MATα doa4Δ1::LEU2 his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1d |
| KHY51 | vma22Δ-1::URA3 in MHY623 |
| YPH499 | MATa ura3-52 leu2-Δ1 his3-Δ200 trp1Δ63 lys2-801 ade2-101e |
| KHY78 | vma22Δ-6::KAN in YPH499 |
| CM765 | MATα cim5-1 ura3-52 leu2-Δ1 his3-Δ200 e |
| KHY77 | vma22Δ-6::KANin CM765 |
| SEY6211 | MATa leu2-3,112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9a |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9a |
| ACY77 | MATα vph1Δ::leu2::KANvma22Δ-3::TRP1 lys2-801 his3-200 ura3-52 |
| KHY94 | vma22Δ-5::STOP in ACY77 |
| ACY73 | vph1Δ::LEU2 vma22Δ-3::TRP1 in SEY6211 |
| ACY74 | vph1Δ::leu2::KAN vma22Δ-3::TRP1 in ACY73 |
| KHY92 | vma22Δ-5::STOP in ACY74 |
| KHY102 | prc1-1 in KHY92 |
| KHY125 | vph1Δ::kan::LEU2 in KHY102 |
| KHY127 | der1Δ::KAN in KHY125 |
| KHY138 | hrd3Δ::KAN in KHY125 |
| KHY140 | hrd1Δ::LEU2 in KHY102 |
| MY767 | MATα kar2-1 ura3-52 leu2-3,112 ade2 his4g |
| KHY50 | vma22Δ-4::LEU2 in SEY6211a |
| KHY85 | KAR2 vma22Δ-4::LEU2 |
| KHY86 | kar2-1 vma22Δ-4::LEU2 |
| KHY181 | prc1-1 in KHY85 |
| KHY190 | prc1-1 in KHY86 |
| a1-45ΔU | MATα ssa1-ts ssa2-1 ssa3-1 ssa4-2 his3-11,3–15 leu2-3,112 ura3-52 trp1-Δ1 lys2f |
| ACY76 | MATα vma22Δ-2::URA3 ssa1-ts ssa2-1 ssa3-1 ssa4-2 |
aHill and Stevens (1994); bHill and Stevens (1995); cDrs L.Hicke and H.Riezman; dDr M.Hochstrasser; eDr R.Haguenauer-Tsapis; fDr E.Craig; gDr M.Rose. Strains KHY183 and KHY184 are spores from a cross between RH3145 and KHY13. KHY85 and KHY86 are spores from a cross between KHY50 and MY767.
Table III. Plasmids.
| Plasmid | Description |
|---|---|
| pKH2212 | Hill and Stevens (1995) |
| pKH2213 | Hill and Stevens (1995) |
| pMM322 | Manolson et al. (1992) |
| pvph1Δ::LEU2 | Manolson et al. (1992) |
| YCp50-SSA1 | A gift from Dr E.Craig |
| pDJ65C | From Dr D.Jackson |
| pAC334 | A 1.1 kb URA3 fragment from pJJ252 inserted into StyI-digested (and blunted) pKH2212. pAC334 was cut by SpeI–XhoI prior to transformation |
| pAC335 | pvph1Δ::LEU2 was digested with NheI–EcoRV, and a SmaI–SpeI fragment from pF6-KanMX6 inserted. pAC335 was digested with BamHI–ApaI prior to transformation |
| pAC336 | pKH2212 was digested with StyI, blunt ended and treated with alkaline phosphatase. A SmaI–EcoRV kanr-containing DNA fragment from pF6-MX4 was then inserted. pAC336 was digested with SpeI–XhoI prior to transformation |
| pAC338 | DER1 was amplified by PCR from genomic DNA prepared from SEY6211 cells to yield an 842 bp fragment which was digested with BamHI–ClaI and inserted into BamHI–ClaI-digested pBluescript (Stratagene) |
| pAC341 | pAC338 was digested with BsaBI, and a SmaI–Ecl136II fragment from pF6-KanMX4 inserted. pAC341 was digested with BamHI–SalI prior to transformation |
| pAC356 | The prc1-1 allele was amplified from genomic DNA prepared from SF648-5Da cells using PCR. The resulting 3 kb fragment was cloned into the SrfI site of pCR-Script |
| pAC357 | pAC356 was digested with HindIII, and a 1.1 kb HindIII DNA fragment containing URA3 from pJJ242 inserted. pAC357 was digested with BglII prior to transformation |
| pAC359 | pKH2212 was digested with HincII, and an XbaI linker (Stratagene) inserted |
| pAC361 | pAC359 was digested with XhoI–SpeI and inserted into the corresponding sites in pRS306. pAC361 was digested with PflMI prior to transformation |
| pAC367 | pAC370 was digested with BglII–NsiI to release an 825 bp fragment which was then replaced with a BamHI–PstI LEU2 fragment from pJJ250. pAC367 was digested with DraI prior to transformation |
| pAC368 | A 2.5 kb fragment containing HRD3 was amplified by PCR from genomic DNA isolated from SEY6211 cells and inserted into the ScfI site of pCR-Script |
| pAC370 | HRD1 was amplified by PCR as described above to yield a 1.8 kb fragment which was inserted into the ScfI site of pCR–Script (Stratagene) |
| pAC372 | pAC368 was digested with BglII–HpaI, and a BamHI–EcoRV fragment from pF6-KanMX4 inserted. pAC372 was digested with BamHI–NheI prior to transformation |
| pAC427 | YCp50-SSA1 was digested with BamHI–HpaI and the SSA1-containing fragment inserted into EcoRV–BamHI-digested pRS313 to create pAC427 |
In order to introduce the prc1-1 allele, pAC357 was transformed into KHY92 cells and Ura+ prototrophs placed on 5-fluoro-orotic acid (5-FOA). Ura– colonies were screened for replacement of the wild-type PRC1 gene by Western blot to detect CPY*. DER1 was disrupted in KHY125 cells by transformation with pAC341 and selection on YEPD pH 5.0 medium containing 200 μg/ml G418 (Sigma). G418R candidates were screened by Western blotting to detect stabilized CPY*. The structure of the der1Δ disruption in KHY127 was confirmed by PCR analysis on genomic DNA utilizing oligonucleotides specific for the DER1 locus. HRD3 was disrupted in KHY125 cells using pAC372 and selection on G418-containing YEPD pH 5.0. The structure of the disruption in KHY138 was confirmed by PCR analysis as described. HRD1 wasdeleted from KHY102 cells using pAC367 and selecting Leu+ prototrophs. Prototrophs were screened for stabilized CPY* by Western blotting, and the structure of the disruption in KHY140 confirmed by PCR analysis. KHY125, KHY127, KHY138 and KHY140 cells were transformed with pMM322 before radiolabeling/immunoprecipitation to detect Vph1p and CPY*. The vph1Δ::LEU2 allele contained in ACY73 and ACY77 was introduced as described (Manolson et al., 1992). Leu+ prototrophs were examined by Western blotting to confirm the absence of Vph1p. The vma22Δ-1::URA3 allele contained in KHY38, KHY51, KHY67 and KHY180 was created by PCR amplification as described (Hill and Stevens, 1995). The vma22Δ-2::URA3 allele contained in KHY58, KHY70, KHY104, KHY154, KHY172, KHY173, KHY175 and KHY177 cells was introduced by transformation with pAC334, and Ura+ transformants were tested for pH sensitivity. ACY73 and ACY77 contain vma22Δ-3::TRP1 introduced via PCR amplification as described (Hill and Stevens, 1995) except that pRS314 was used as a template. KHY13 contains vma22Δ-4::LEU2 introduced by transformation with pKH2213 (Hill and Stevens, 1995). The vma22Δ-5::STOP allele contained in KHY92 and KHY94 was introduced by transformation with pAC361. The vma22Δ-3::TRP1 allele was then excised via selection on 5-FOA, and pH-sensitive Trp– colonies selected. The vma22Δ-6::KAN allele contained in KHY77 and KHY78 was created using pAC336 and selection on YEPD pH 5.0 + G418. Resistant colonies were screened for pH sensitivity. The vph1Δ::leu2::KAN allele contained in ACY74, KHY102 and KHY125 cells was created by transformation with pAC335 and selection on YEPD pH 5.0 + G418. G418R colonies were then tested for a Leu– phenotype and the absence of Vph1p was confirmed by Western blot.
Protein preparation, antibodies and Western blotting
Whole-cell protein extracts were prepared as described (Hill and Stevens, 1994). Secondary antibodies were purchased from Bio-Rad. Immunoblots were detected using chemifluorescent or chemiluminescent detection (Amersham). Polyclonal antibodies were generated by HTI Bio-Products Inc. Monoclonal antibodies were purchased from Molecular Probes. Protease shaving experiments were performed as outlined by Jackson and Stevens (1997).
Radiolabeling and immunoprecipitation
Pulse–chase immunoprecipitations of Vph1p were performed essentially as described (Hill and Stevens, 1994). Samples were removed at set times during the chase period, processed for immunoprecipitation and resolved by SDS–PAGE. Gels were fixed, dried and exposed either to a phosphor cassette prior to quantitation using a phosphorimager (Molecular Dynamics) or to X-ray film. Immunoprecipitation of CPY was carried out as described previously (Hill and Stevens, 1994). Sodium carbonate fractionation was performed as described previously (Hill and Stevens, 1994).
Vesicle pelleting and floatation
Microsomal pellets were prepared as described above except that microsomes were collected at 100 000 g for 30 min. Microsomal floatation experiments were performed as described by Xiong et al. (1999).
Membrane fractionation
Membrane fractionation was performed as described by Roberg et al. (1997) except the gradients were centrifuged at 50 000 r.p.m. in an SW55.1 rotor for 14 h before samples were collected from the bottom of the tube.
Acknowledgments
Acknowledgements
We thank Till Bartke and Sabrina Caldwell for assistance in strain construction, Dr Douglas Crawford for critical reading of this manuscript, and Drs M.Hochstrasser, H.Riezman, M.Manolson, R.Haguenauer-Tsapis, T.Stevens, E.Craig, M.Bard and M.Rose for generously providing strains and plasmids used in this work. We thank Helen Yu and Eric Epping for sucrose gradient suggestions, and Dr D.Jackson for advice on protease shaving assays. This work was supported by the National Institutes of Health (NIH) grant GM55848 and American Heart Association (Kansas Affiliate) grant (KS-96-GB-47).
References
- Becker J., Walter, W., Yan, W. and Craig, E. (1996) Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo.Mol. Biol. Cell, 16, 4378–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich B., Stancovski, I., Mayer, A., Blumenfeld, N., Laszlo, A., Schwartz, A.L. and Ciechanover, A. (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem., 272, 9002–9010. [DOI] [PubMed] [Google Scholar]
- Biederer T., Volkwein, C. and Sommer, T. (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin–proteasome pathway. EMBO J., 15, 2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman, A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J., Plemper, R.K., Finger, A. and Wolf, D.H. (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell, 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L., Werner, E.D., Dubas, M.E., Goeckeler, J.L., Kruse, K.B. and McCracken, A.A. (1999) The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem., 274, 3453–3460. [DOI] [PubMed] [Google Scholar]
- Enenkel C., Lehmann, A. and Kloetzel, P.M. (1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope–ER network in yeast. EMBO J., 17, 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A., Knop, M. and Wolf, D.H. (1993) Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem., 218, 565–574. [DOI] [PubMed] [Google Scholar]
- Fisher E.A., Zhou, M., Mitchell, D.M., Wu, X., Omura, S., Wang, H., Goldberg, A.L. and Ginsberg, H.N. (1997) The degradation of apolipoprotein B100 is mediated by the ubiquitin–proteasome pathway and involves heat shock protein 70. J. Biol. Chem., 272, 20427–20434. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard, A.L., Fowler, S. and Lazarow, P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol., 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.M., Cantegrit, B., Garnier, C., Namy, O. and Haguenauer-Tsapis, R. (1998) ‘ER degradation’ of a mutant yeast plasma membrane protein by the ubiquitin–proteasome pathway. FASEB J., 12, 315–323. [DOI] [PubMed] [Google Scholar]
- Glas R., Bogyo, M., McMaster, J.S., Gaczynska, M. and Ploegh, H.L. (1998) A proteolytic system that compensates for loss of proteasome function. Nature, 392, 618–622. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy, A. and Mann, C. (1993) S.cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature, 366, 358–362. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Wickner, S. and Maurizi, M.R. (1997) Protein quality control: triage by chaperones and proteases. Genes Dev., 11, 815–823. [DOI] [PubMed] [Google Scholar]
- Graham L.A. and Stevens, T.H. (1999) Assembly of the yeast vacuolar proton-translocating ATPase. J. Bioenerg. Biomembr., 31, 39–47. [DOI] [PubMed] [Google Scholar]
- Graham L.A., Hill, K.J. and Stevens, T.H. (1998) Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol., 142, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y. and Bhakta, H. (1997) Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl Acad. Sci. USA, 94, 12944–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., Gardner, R.G. and Rine, J. (1996) Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell, 7, 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S.A. and Dice, J.F. (1996) Roles of molecular chaperones in protein degradation. J. Cell Biol., 132, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.J. and Stevens, T.H. (1994) Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H+-ATPase complex. Mol. Biol. Cell, 5, 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.J. and Stevens, T.H. (1995) Vma22p is a novel endoplasmic reticulum-associated protein required for assembly of the yeast vacuolar H+-ATPase complex. J. Biol. Chem., 270, 22329–22336. [DOI] [PubMed] [Google Scholar]
- Hiller M.M., Finger, A., Schweiger, M. and Wolf, D.H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science, 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Jackson D.D. and Stevens, T.H. (1997) VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J. Biol. Chem., 272, 25928–25934. [DOI] [PubMed] [Google Scholar]
- Kim S., Schilke, B., Craig, E.A. and Horwich, A.L. (1998) Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc. Natl Acad. Sci. USA, 95, 12860–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Finger, A., Braun, T., Hellmuth, K. and Wolf, D.H. (1996) Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J., 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Loayza D., Tam, A., Schmidt, W.K. and Michaelis, S. (1998) Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in S.cerevisiae.Mol. Biol. Cell, 9, 2767–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson M.F., et al. (1992)The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J. Biol. Chem., 267, 14294–14303. [PubMed] [Google Scholar]
- Mayer T.U., Braun, T. and Jentsch, S. (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J., 17, 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A.A. and Brodsky, J.L. (1996) Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin and ATP. J. Cell Biol., 132, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F.R., Amerik, A.Y. and Hochstrasser, M. (1999) Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell, 10, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M., Schekman, R. and Romisch, K. (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J., 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R.K., Bohmler, S., Bordallo, J., Sommer, T. and Wolf, D.H. (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature, 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Egner, R., Kuchler, K. and Wolf, D.H. (1998) Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem., 273, 32848–32856. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Deak, P.M., Otto, R.T. and Wolf, D.H. (1999) Re-entering the translocon from the lumenal side of the endoplasmic reticulum. Studies on mutated carboxypeptidase yscY species. FEBS Lett., 443, 241–245. [DOI] [PubMed] [Google Scholar]
- Roberg K.J., Rowley, N. and Kaiser, C.A. (1997) Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae.J. Cell Biol., 137, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., McGrath, J.P. and Jentsch, S. (1990) UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J., 9, 4535–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons J.F., Ferro-Novick, S., Rose, M.D. and Helenius, A. (1995) BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol., 130, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S.J. (1990) The structure and insertion of integral proteins in membranes. Annu. Rev. Cell Biol., 6, 247–296. [DOI] [PubMed] [Google Scholar]
- Skowronek M.H., Hendershot, L.M. and Haas, I.G. (1998) The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc. Natl Acad. Sci. USA, 95, 1574–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., Misra, L.M. and Rose, M.D. (1990) Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol., 110, 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Omura, S. and Kopito, R.R. (1995) Degradation of CFTR by the ubiquitin–proteasome pathway. Cell, 83, 121–127. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Jones, T.R., Sun, L., Bogyo, M., Geuze, H.J. and Ploegh, H.L. (1996a) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell, 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J.H.J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T.R., Rapoport, T.A. and Ploegh, H.L. (1996b) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature, 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Xiong X., Chong, E. and Skach, W.R. (1999) Evidence that ER-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J. Biol. Chem., 274, 2616–2624. [DOI] [PubMed] [Google Scholar]
- Yang Y., Janich, S., Cohn, J.A. and Wilson, J.M. (1993) The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl Acad. Sci. USA, 90, 9480–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.X., Braakman, I., Matlack, K.E. and Helenius, A. (1997) Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C–terminal truncations. Mol. Biol. Cell, 8, 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]