Abstract
Multiple testing corrections are an active research topic in genetic association studies, especially for genome-wide association studies (GWAS), where tests of association with traits are conducted at millions of imputed SNPs with estimated allelic dosages now. Failure to address multiple comparisons appropriately can introduce excess false positive results and make subsequent studies following up those results inefficient. Permutation tests are considered the gold standard in multiple testing adjustment; however, this procedure is computationally demanding, especially for GWAS. Notably, the permutation thresholds for the huge number of estimated allelic dosages in real data sets have not been reported. Although many researchers have recently developed algorithms to rapidly approximate the permutation thresholds with accuracy similar to the permutation test, these methods have not been verified with estimated allelic dosages. In this study, we compare recently published multiple testing correction methods using 2.5M estimated allelic dosages. We also derive permutation significance levels based on 10,000 GWAS results under the null hypothesis of no association. Our results show that the simpleM method works well with estimated allelic dosages and gives the closest approximation to the permutation threshold while requiring the least computation time.
Keywords: multiple testing, genome-wide association studies, imputed SNPs, allelic dosages
Introduction
An area of interest in statistical methodology for genetic association studies is the optimal genome-wide multiple testing correction threshold [Dudbridge and Gusnanto 2008; Hoggart, et al. 2008; Pe'er, et al. 2008; Risch and Merikangas 1996]. With the ubiquity of genome-wide association studies (GWAS), which test from hundreds of thousands to millions of markers, the issue of multiple comparison adjustment has major implications for the efficiency of GWAS studies and subsequent follow-up studies in large collections of samples. Failure to adjust for multiple testing can mask true signals, or lead to false positive results and cause resources to be expended following up SNPs where the null hypothesis is true. Recently, researchers have begun to use imputed, or estimated, allelic dosages, to conduct GWAS. Imputed allelic dosages are the estimated counts of reference alleles at each SNP. While genotype imputation is now a vital tool in GWAS, the multiple testing issues on imputed allelic dosages have largely been ignored.
The Bonferroni correction is a simple multiple testing correction method. If m tests are conducted and we want to control the experiment-wise error rate (EWER) at a nominal significance level α, we need to adjust the point-wise error rate (PWER) using the Bonferroni formula α/m. However, due to the linkage disequilibrium (LD), this correction is known to be too conservative when large numbers of densely spaced SNPs are evaluated for association with traits. Numerous GWAS and meta-analyses using 2.5M estimated allelic dosages have been reported recently and no study used the Bonferroni correction, 0.05/2.5M = 2×10−8, as the significance level.
In contrast to the Bonferroni correction, permutation tests can give the optimal exact threshold and are considered the gold standard in multiple testing adjustments for genetic association studies. However, this procedure is computationally intensive and this burden can be prohibitive for a large number of random shuffles using millions of SNPs. As a result, researchers have proposed approximation methods to alleviate the computing burden of permutation tests.
To date, there have been three major classes of approximation methods: 1) the effective number of independent tests (Meff); 2) asymptotically multivariate normal distribution (MVN) for the set of all the commonly used association statistics [Lin 2005]; 3) computational optimization. The Meff based methods use dimension reduction methods to filter out the correlation among SNPs so that the denominator in the Bonferroni correction formula can be adjusted correctly. Several Meff methods were designed and tested using limited numbers of genetic markers [Cheverud 2001; Li and Ji 2005; Nyholt 2004]. Recently, Meff methods that target GWAS were also proposed and compared, i.e. simpleM [Gao, et al. 2008] and Keff [Moskvina and Schmidt 2008]. simpleM was reported to perform significantly better than other Meff methods [Gao, et al. 2010; Gao, et al. 2008]. Because the distribution of commonly used association statistics over a set of genetic markers follow multivariate normal (MVN) asymptotically [Lin 2005; Seaman and Müller-Myhsok 2005], some researchers proposed methods to simulate the joint distribution of test statistics in order to avoid permutation [Conneely and Boehnke 2007; Han, et al. 2009; Lin 2005; Seaman and Müller-Myhsok 2005], among which SLIDE showed the best performance on a genome-wide scale [Han, et al. 2009]. There are also methods that optimize the permutation procedure through computational techniques, such as RAT [Kimmel and Shamir 2006] and PRESTO [Browning 2008]. SLIDE was shown to perform better than other MVN methods and RAT [Han, et al. 2009]. A comparison among simpleM, SLIDE and PRESTO using estimated allelic dosages has not been reported yet.
In this work, we compare the performance of simpleM with SLIDE using 2.5M estimated allelic dosages from the NHLBI Family Heart Study. To our knowledge, PRESTO requires both alleles for a SNP to be known, which limits its function only to discrete genotypes. Additionally, other methods have been evaluated in previous publications [Gao, et al. 2010; Gao, et al. 2008; Han, et al. 2009]. simpleM does not require the underlying distribution of joint statistics to be known and is thus a non-parametric method. SLIDE relies on the assumption of MVN and is a parametric method. Therefore, a comparison between simpleM and SLIDE is also a comparison between non-parametric and parametric methods in multiple testing corrections using estimated allelic dosages on a genome-wide scale. We further validate the results using permutation thresholds derived from 10,000 random shuffles × 2.5M GWAS tests of these estimated allelic dosages. To our knowledge, this is also the first report of permutation-based significance level in real data sets for this amount of estimated allelic dosages. The extensive evaluation and permutation can provide guidelines for the study design and the evaluation of GWAS significance using estimated allelic dosages.
Data
We used data from the NHLBI Family Heart Study [Higgins, et al. 1996]. The data was genotyped using the Illumina 550, 610 and 1M chips. We used HapMap Phase II CEU individuals as a reference panel and imputed genotypes to ~2.5 million SNPs. The imputation software, MACH v1.0.16 [Li and Abecasis 2006], was used and imputed genotypes were coded as allelic dosages (fractional counts ranging from 0 to 2). Seven hundred and sixty two unrelated Caucasian individuals were used for evaluating the multiple testing methods. Only SNPs with minor allele frequency (MAF) ≥ 0.01 were utilized and the number of SNPs is shown in Table 1.
Table 1.
Derived significance thresholds for the 2.5M imputed allelic dosages.
| chromosome | Number of SNPs |
Permutation |
simpleM |
SLIDE (w=100) |
||
|---|---|---|---|---|---|---|
| αperm | Meff_G | αG | Meff_S | αS | ||
| 1 | 185487 | 1.07E-06 | 41210 | 1.21E-06 | 61353 | 8.15E-07 |
| 2 | 213518 | 1.15E-06 | 43728 | 1.14E-06 | 59404 | 8.42E-07 |
| 3 | 168156 | 1.38E-06 | 36026 | 1.39E-06 | 55924 | 8.94E-07 |
| 4 | 157187 | 1.35E-06 | 32755 | 1.53E-06 | 47144 | 1.06E-06 |
| 5 | 163016 | 1.38E-06 | 34164 | 1.46E-06 | 51576 | 9.69E-07 |
| 6 | 175230 | 1.35E-06 | 35098 | 1.42E-06 | 46531 | 1.07E-06 |
| 7 | 138470 | 1.50E-06 | 30412 | 1.64E-06 | 42241 | 1.18E-06 |
| 8 | 143112 | 1.49E-06 | 30038 | 1.66E-06 | 40732 | 1.23E-06 |
| 9 | 117448 | 1.79E-06 | 26769 | 1.87E-06 | 35816 | 1.40E-06 |
| 10 | 133125 | 1.75E-06 | 28761 | 1.74E-06 | 39061 | 1.28E-06 |
| 11 | 125417 | 1.64E-06 | 26838 | 1.86E-06 | 36892 | 1.36E-06 |
| 12 | 120016 | 1.78E-06 | 26762 | 1.87E-06 | 37582 | 1.33E-06 |
| 13 | 99766 | 2.40E-06 | 20931 | 2.39E-06 | 29781 | 1.68E-06 |
| 14 | 80985 | 2.50E-06 | 17910 | 2.79E-06 | 26156 | 1.91E-06 |
| 15 | 69391 | 2.57E-06 | 16996 | 2.94E-06 | 23907 | 2.09E-06 |
| 16 | 68561 | 2.45E-06 | 18778 | 2.66E-06 | 25594 | 1.95E-06 |
| 17 | 56130 | 3.19E-06 | 15141 | 3.30E-06 | 20228 | 2.47E-06 |
| 18 | 73988 | 2.71E-06 | 16636 | 3.01E-06 | 23169 | 2.16E-06 |
| 19 | 35517 | 4.33E-06 | 10665 | 4.69E-06 | 13610 | 3.67E-06 |
| 20 | 60777 | 3.10E-06 | 14648 | 3.41E-06 | 21401 | 2.34E-06 |
| 21 | 32880 | 6.21E-06 | 7873 | 6.35E-06 | 10499 | 4.76E-06 |
| 22 | 32153 | 5.25E-06 | 8679 | 5.76E-06 | 10953 | 4.56E-06 |
| genomewide | 2450330 | 8.53E-08 | 540818 | 9.25E-08 | 759554 | 6.58E-08 |
The permutation thresholds were calculated using 10,000 random shuffles. The experiment-wise error rates for each chromosome and the genome-wide adjustment were set to 0.05. Only SNPs with minor allele frequency ≥0.01 were used.
αperm, αG and αS are the point-wise error rates for permutation, simpleM and SLIDE, respectively.
Meff_G and Meff_S are the effective number of independent tests for simpleM and SLIDE, respectively. w=100: window size is set equal to 100.
Methods
simpleM [Gao, et al. 2008] is a principal component analysis (PCA) based approach that calculates the effective number of independent tests, Meff, for a given data set. Then, it uses Meff as the denominator in the Bonferroni correction formula. simpleM uses composite LD to capture the correlation among SNPs and infers the Meff using the number of principal components that jointly contribute to 99.5% of variation in the SNPs. The method has been verified to give multiple correction cut-offs similar to those estimated from the permutation null distribution using data from common SNPs chips [Gao, et al. 2010].
SLIDE [Han, et al. 2009] relies on the assumption that commonly used association statistics for a set of SNPs follow multivariate normal (MVN) distribution asymptotically. SLIDE uses a sliding-window Monte-Carlo approach, which takes into account all the local LD along the genome, to approximate the MVN. One problem, however is that the MVN can be inaccurate in the tails of the true null distribution. SLIDE scales the approximated MVN to overcome the inaccuracy in the tails. A bigger window size may generally yield more accurate results than a smaller window but at the price of increased computing time. In this work, we used a window size of 100 markers (w = 100) and 10K samplings as reported in the original publication.
We also performed permutation tests on the NHLBI Family Heart Study 2.5M estimated allelic dosages. We set the EWERs at 0.05 and derived corresponding PWERs. We conducted permutation tests [Churchill and Doerge 1994; Gao, et al. 2008] with 10,000 random shuffles using the score test for testing the global null hypothesis of beta = 0 in the logistic regression as implemented in SAS v9.2 (SAS Institute, Cary, NC, USA). In each permutation shuffle, half of the samples were randomly assigned as cases and the other half were assigned as controls. We recorded the smallest p-value from each permutation. The smallest p-values were arranged in descending order and the 100α percentile was the permutation-based PWER for the overall significance level of α. We used a nominal value of α = 0.05. Permutation tests were carried out on the computer cluster (800 nodes) of the Division of Statistical Genomics at the Washington University School of Medicine.
Results
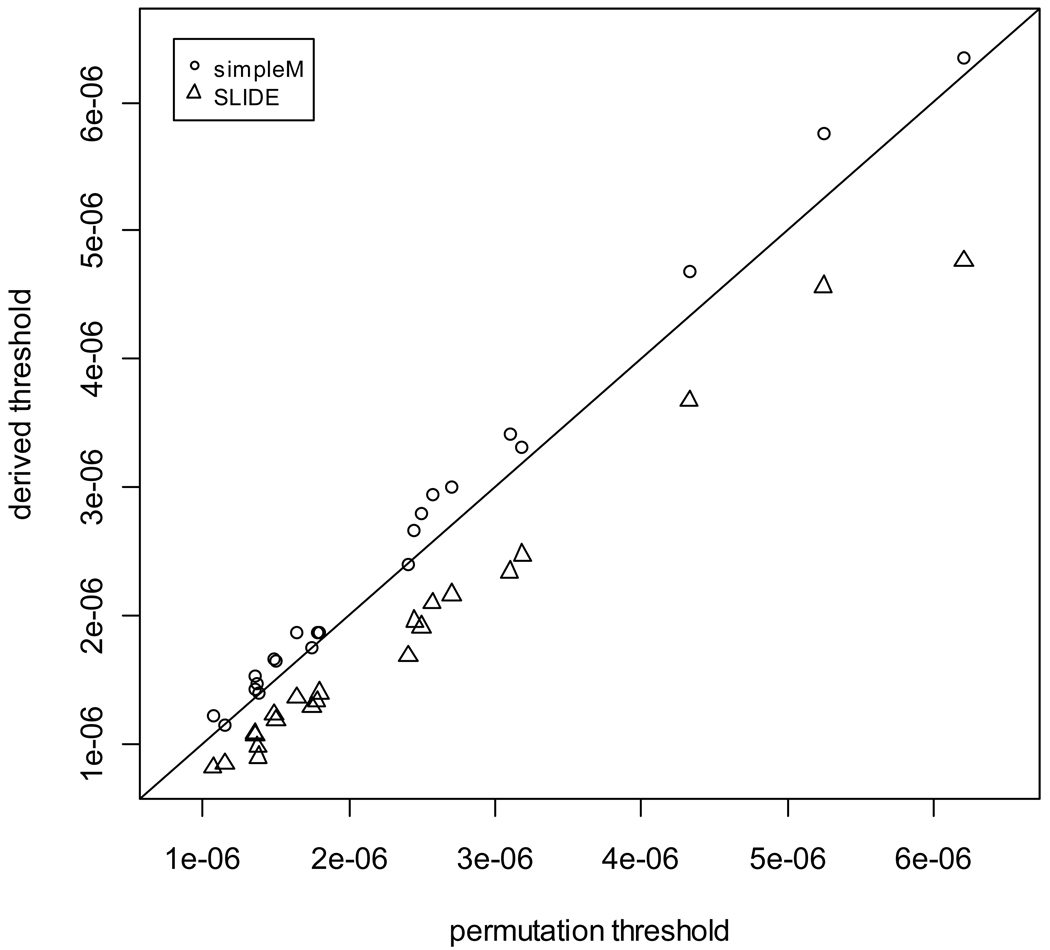
Results for each chromosome (chromosome-wide EWER = 0.05) and at the genome-wide scale (genome-wide EWER = 0.05) are shown in Table 1 for the estimated allelic dosages. The PWERs derived from the simpleM, SLIDE and permutation tests are denoted as αG, αS and αperm, respectively. simpleM gives Meff output. The corresponding αG was derived using the formula, αG = 0.05/Meff. SLIDE gives both Meff and αS output. Comparing αG and αS to αperm, we see that simpleM gives the best approximation to the permutation thresholds while SLIDE is rather conservative. To help visualization, pair-wise plots taking permutation thresholds as the reference for chromosome 1 to 22 are plotted in Figure 1. simpleM × permutation and SLIDE × permutation pair-wise values are indicated as open circles and triangles, respectively. A perfect match with permutation estimates would fall on the diagonal line, y = x. The closer the approximation thresholds to the diagonal line the better. SLIDE thresholds are farther off the diagonal line than simpleM. On a genome-wide scale, the permutation threshold is 8.52×10−8 with 95% confidence interval (CI) as (7.86×10−8, 9.44×10−8), while simpleM and SLIDE give 9.25×10−8 and 6.58×10−8, respectively. It is clear that the simpleM gives the closest approximation to the permutation. The SLIDE estimate, 6.58×10−8, is outside of the 95% CI of the permutation significance level. The corresponding type I error rates for the simpleM and SLIDE thresholds are 0.053 and 0.039, respectively.
Figure 1. Pair-wise plots of the approximation and permutation thresholds derived from the allelic dosage.
The x-axis denotes the permutation significance levels. The y-axis is the derived significance levels from each approximation methods. If the derived thresholds match with the permutation thresholds perfectly, they would fall on the diagonal line, y = x. simpleM × permutation and SLIDE × permutation are represented by open circles and triangles, respectively.
Instead of comparing derived PWERs, we can also compare the Meff estimates. The minimum p-values from permutation tests follow a beta distribution [Gao, et al. 2010]. Using maximum likelihood estimation (MLE), we inferred the permutation genome-wide Meff as 590043. The simpleM Meff estimation, 540818, gave a much closer approximation to the MLE estimate than the SLIDE Meff, 759554.
We also compared the runtime for each approximation method (see Table 2). We ran each method on the chromosome 1 data with 185,487 SNPs using our desktop computer (Intel Core2 2.4G CPU with 3GB memory, Redhat Linux operating system). simpleM and SLIDE took 2 minutes 45 seconds and 9 minutes 30 seconds, respectively. We then compared the runtime on the genome-wide level. simpleM took about 36 minutes and SLIDE took about 126 minutes to finish. simpleM was applied to the allelic dosages directly. In order to run SLIDE on the allelic dosages, the data had to be preprocessed into covariance band matrixes (SLIDE user’s manual), which took a considerable among of time. The runtime for SLIDE was recorded after the data was preprocessed. It took SAS >40 minutes to carry out 10,000 logistic tests on our desktop computer. For permutation tests, we needed to do 10,000×2.5M tests and impractical to carry out on a single PC. All the permutation tests were done using our 800-node computer cluster. Among all the methods tested, simpleM required the least computing time.
Table 2.
Runtime for each multiple testing correction method.
| number of SNPs |
simpleM |
SLIDE |
|
|---|---|---|---|
| chromosome 1 | 185487 | 2 min 45 sec | 9 min 30 sec† |
| genome-wide | 2450330 | ~36 min | ~126 min† |
min: minutes.
sec: seconds.
This is the runtime after the allelic dosages were preprocessed into the covariance band matrix format, which took a considerable amount of time. simpleM does not require this step.
Discussion
With the availability of the HapMap data and the sequence data from the 1000 genomes project, researchers are now using SNP imputation and estimated allelic dosages to conduct GWAS. These innovations pose two main challenges to multiple testing correction methods: 1) do they perform well when SNPs are represented by allelic dosages with uncertainty in SNP calling built in? 2) do they perform well when the number of SNPs are increasing? Here, we have shown that the simpleM method for multiple testing correction gives the closest approximation to the permutation method, and does so in the shortest amount of time.
simpleM and SLIDE are based on two completely different ideas and are implemented in two different languages, i.e. in R and C, respectively. simpleM is platform-independent. SLIDE is currently available only on Linux. simpleM is the fastest and provides the closest approximation to the permutation significance levels. Even though SLIDE is much faster than the permutation tests, it still requires a considerable number of samplings to achieve accuracy. Moreover, in order to run SLIDE on allelic dosages, the data have to be preprocessed into covariance band matrixes.
With the advance of our understanding of the human genome and the endeavor of searching for the missing heretibility, a natural question is how to apply simpleM (essentially the PCA idea) to copy number variants (CNVs) and rare variants. CNVs inferred from microarray intensity data are represented by segments of either discrete copy numbers, e.g. from PennCNV [Wang, et al. 2007], or fractional counts, e.g. from Partek® (http://www.partek.com/). It is not immediately clear how users should do statistical tests and adjust for multiple comparisons with copy number segments. A simple solution is to map the segments back to the microarray matrix with the rows and columns representing locus and individual IDs, and cell values as the inferred copy numbers. Since only a limited number of regions harbor CNVs, all the regions without CNVs (i.e. only show the normal 2 copies for all individuals) can be skipped, which can greatly reduce the multiple comparison burden. Then, simpleM can be applied to this matrix of copy numbers. For rare variants, data analysis usually requires collapsing of neighboring multiple rare mutations into “super” loci [Li and Leal 2008; Madsen and Browning 2009]. simpleM can be easily applied to these “super” loci, which may be represented as fractional counts and weighted by allele frequencies. In the analysis of sequence variations, analyses may be conducted using data from the extremes of the phenotype distribution, e.g. the upper and lower 5% of the phenotype distribution [Cohen, et al. 2004], where small sample sizes are likely to occur and test statistics based on asymptotic theory do not hold. Fisher’s exact and permutation tests can be applied to these situations. Permutation tests may not be formidable in small sample sizes situations as compared to GWAS, which require thousands of individuals.
Multiple testing correction has been a challenging topic in genetic association studies. Though permutation is considered the gold standard, it is computationally intensive, especially for large genetic datasets, and can be impractical for routine analysis using standard statistics software. This problem is likely to grow worse, as the data from the 1000 genomes project becomes available for imputation, and next-generation sequencing data generates more densely spaced genetic variants. Many methods have been proposed to provide approximations of the thresholds for significance from permutation tests. It may be challenging for non-experts to choose among all these methods. Moreover, the permutation thresholds for current commonly used genome-wide imputed SNPs in real data sets have not been reported possibly due to the excessive computing time and resources required. This work can provide end-users with guidelines for choosing the appropriate significance levels and approximation method for addressing multiple testing issues using imputed SNPs and estimated allelic dosages.
Acknowledgments
This research was conducted in part using data and resources from the NHLBI Family Heart Study supported in part by NIH grant 5R01HL08770002. Drs. Todd Edwards and Joshua Starmer gave advice in manuscript preparation. We thank anonymous reviewers for providing constructive feedback for this work.
Footnotes
Web Resources
The URLs for software presented herein are as follows:
simpleM: http://simplem.sourceforge.net/
SLIDE: http://slide.cs.ucla.edu/
References
- Browning BL. PRESTO: rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies. BMC Bioinformatics. 2008;9:309. doi: 10.1186/1471-2105-9-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87(1):52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical Threshold Values for Quantitative Triat Mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. American journal of human genetics. 2007;81(6):1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology. 2008;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34(1):100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genetic Epidemiology. 2008;32(4):361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- Han B, Kang HM, Eskin E. Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet. 2009;5(4):e1000456. doi: 10.1371/journal.pgen.1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, Rao DC, Sprafka JM, Williams R. NHLBI Family Heart Study: objectives and design. Am J Epidemiol. 1996;143(12):1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32(2):179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- Kimmel G, Shamir R. A fast method for computing high-significance disease association in large population-based studies. Am J Hum Genet. 2006;79(3):481–492. doi: 10.1086/507317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79 [Google Scholar]
- Lin DY. An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics. 2005;21(6):781–787. doi: 10.1093/bioinformatics/bti053. [DOI] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V, Schmidt KM. On multiple-testing correction in genome-wide association studies. Genetic Epidemiology. 2008;32(6):567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A Simple Correction for Multiple Testing for Single-Nucleotide Polymorphisms in Linkage Disequilibrium with Each Other. American journal of human genetics. 2004;74(4):765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Seaman SR, Müller-Myhsok B. Rapid Simulation of P Values for Product Methods and Multiple-Testing Adjustment in Association Studies. American journal of human genetics. 2005;76(3):399–408. doi: 10.1086/428140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]