Abstract
Mannose trimming is not only essential for N-glycan maturation in mammalian cells but also triggers degradation of misfolded glycoproteins. The crystal structure of the class I α1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2isomer B in the endoplasmic reticulum of Saccharomyces cerevisiae reveals a novel (αα)7-barrel in which an N-glycan from one molecule extends into the barrel of an adjacent molecule, interacting with the essential acidic residues and calcium ion. The observed protein–carbohydrate interactions provide the first insight into the catalytic mechanism and specificity of this eukaryotic enzyme family and may be used to design inhibitors that prevent degradation of misfolded glycoproteins in genetic diseases.
Keywords: class I α1,2-mannosidase/crystallography/drug design/ER quality control/processing glycosidase
Introduction
Class I α1,2-mannosidases (glycosyl hydrolase family 47; Henrissat, 1991) have been conserved throughout eukaryotic evolution for the maturation of N-glycans during glycoprotein biosynthesis (Moremen et al., 1994; Herscovics, 1999a,b,c). N-glycan formation begins with the transfer of a preformed oligosaccharide precursor, usually Glc3Man9GlcNAc2, to nascent polypeptide chains. The oligosaccharide precursor is trimmed immediately by α-glucosidases and α-mannosidases in the endoplasmic reticulum (ER). Glycoproteins that have acquired their native conformation can then be transported to the Golgi apparatus where additional α-mannosidases produce the appropriate substrates for Golgi glycosyltransferases to form the variety of biologically important oligosaccharide structures found on glycoproteins (Varki, 1993). Besides their importance in N-glycan maturation, ER processing glycosidases also play a role in quality control, ensuring that only properly folded glycoproteins are transported to their final destination. Trimming of the oligosaccharide precursor by α-glucosidases I and II controls the interaction of newly formed glycoproteins with the lectin chaperones, calnexin and calreticulin, thus facilitating folding of glycoproteins (Hammond and Helenius, 1995), while trimming of mannose residues in the ER acts as a signal to target misfolded glycoproteins for degradation by the proteasome (Su et al., 1993; Knop et al., 1996; Liu et al., 1997, 1999; Jakob et al., 1998).
In Saccharomyces cerevisiae, there is only one processing α-mannosidase (SwissProt accession No. P32906). This enzyme is a 63 kDa type II ER transmembrane glycoprotein with no significant cytoplasmic tail, an N–terminal transmembrane domain and a large C–terminal catalytic domain (Camirand et al., 1991). The yeast α1,2-mannosidase removes a single mannose residue from Man9GlcNAc2 to form Man8GlcNAc2 isomer B (Byrd et al., 1982; Jelinek-Kelly et al., 1985; Jelinek-Kelly and Herscovics, 1988; Ziegler et al., 1991). An ER α-mannosidase with the same specificity also occurs in mammalian cells (Bischoff and Kornfeld, 1983; Bischoff et al., 1986; Rizzolo and Kornfeld, 1988; Weng and Spiro, 1993, Lal et al., 1998) and has recently been cloned (Gonzalez et al., 1999; Tremblay and Herscovics, 1999). The importance of this enzyme in ER quality control has been demonstrated in both yeast and mammalian cells. In yeast, it was shown that mutant carboxypeptidase Y is stabilized in the mns1 mutant lacking the ER processing α1,2-mannosidase while it is degraded rapidly in wild-type cells (Knop et al., 1996; Jakob et al., 1998). In mammalian cells, the ER degradation of foreign (Su et al., 1993) or abnormal glycoproteins such as mutant α1-antitrypsin (Liu et al., 1999) is prevented by the α1,2-mannosidase inhibitor, 1-deoxymannojirimycin. This processing α1,2-mannosidase may therefore have an important role in genetic diseases characterized by rapid degradation of misfolded glycoproteins, such as cystic fibrosis (Kopito, 1999) and emphysema (Sifers, 1995).
Results and discussion
Structure determination
The crystallization of the yeast α1,2-mannosidase catalytic domain produced in Pichia pastoris as a secreted glycoprotein (beginning at residue 22) was reported earlier (Dole et al., 1997). However, due to the action of trace proteases, these crystals were not reproducible from one batch of protein to the next. N-terminal sequencing revealed heterogeneity and a proteolytic cleavage site between residues 368 and 369 (Camirand et al., 1991). Secondary structure prediction and sequence comparison with other class I α1,2-mannosidases suggested that residues surrounding the proteolytic cleavage site are present in a loop structure that is not conserved in other members of the enzyme family. A new construct was therefore re-engineered in order to eliminate N-terminal heterogeneity by starting at amino acid 34 and, secondly, by eliminating residues 367–371 surrounding the site of proteolytic cleavage. The new construct was expressed in P.pastoris to produce an enzymatically active secreted glycoprotein (Lipari and Herscovics, 1994, 1996; Lipari et al., 1995). Removal of residues 1–33 and 367–371 did not affect the enzyme activity of the protein (data not shown).
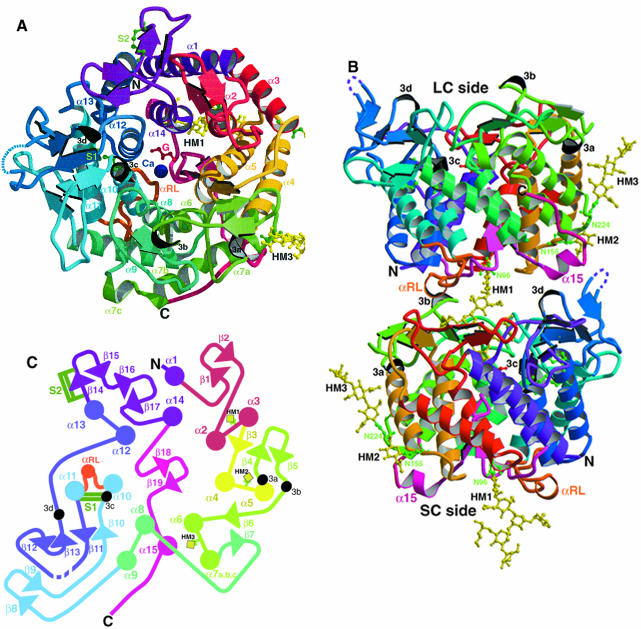
High quality reproducible crystals were obtained following deletion of residues 367–371, and the structure was determined using the single isomorphous replacement with anomalous scattering (SIRAS) technique (see Materials and methods). This is the first processing glycosidase in N-glycan biosynthesis whose three-dimensional structure has been determined. The structure has been refined to an Rcryst of 20.9% and an Rfree of 22.7% for the data between 50 and 1.54 Å resolution (Table I). The final model contains residues 34–366, 372–409 and 411–549 as well as 394 solvent molecules, one glycerol molecule, one calcium ion and three N-glycans. The α1,2-mannosidase catalytic domain is an αα7-helix barrel with overall dimensions of ∼50 × 50 × 50 Å (Figure 1A and B). This is the first example of an αα-helix barrel consisting of seven pairs of helices.
Table I. Data collection, phasing and refinement statistics.
| Native 1 | HgCl2 | Native 2 | Native 3 | |
|---|---|---|---|---|
| Diffraction data | ||||
| X-radiation (λ, Å) | Rigaku | Rigaku | NSLS | NSLS |
| RU200 | RU200 | X8-C | X8-C | |
| (1.54) | (1.54) | (1.00) | (0.975) | |
| Resolution (Å) | 2.71 | 2.71 | 2.00 | 1.54 |
| Unit cell | ||||
| [a, b, c (Å)]a | 90.0, 90.0, 154.9 | 90.1, 90.1, 154.6 | 88.8, 88.8, 153.6 | 88.4, 88.4, 153.3 |
| Temperature (°C) | 20 | 20 | –160 | –160 |
| Measured reflections | 90 034 | 88 356 | 350 472 | 649 016 |
| Unique reflections | 20 423 | 20 049 | 45 749 | 100 985 |
| Redundancy | 4.5 | 4.4 | 7.7 | 6.5 |
| Completenessb | 99 (99) | 99 (99) | 96.4 (98.4) | 99.5 (99.3) |
| Rsymb | 0.071 (0.14) | 0.081 (0.16) | 0.058 (0.27) | 0.062 (0.44) |
| Rderiv | 0.141 | |||
| Sites (n) | 1 | |||
| Rcullis | 0.52 | |||
| Phasing power | 1.63 | |||
| FOM before (after) solvent flattening | 0.42 (0.84) | |||
| Refinement statistics | ||||
| Resolution (Å) | 50–1.54 | |||
| Rcryst | 20.9 | |||
| Rfree | 22.8 | |||
| R.m.s.d. bond length (Å) | 0.005 | |||
| R.m.s.d. bond angles | 1.2 | |||
| R.m.s.d. B-values (Å2) | 2.1 | |||
aSpace group P3121; α = β = 90° and γ = 120°.
bGiven in parentheses are the completeness and Rsym for the last resolution shell.
Rcryst = Σ ||Fo| – |Fc||/Σ |Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. For Rfree, the sum is extended over a subset of reflections (10%) excluded from all stages of refinement (9536 reflections).
Rsym = ΣΣ|Ii – <I>|ΣIi, where <I> is the average of equivalent reflections and the sum is extended over all measured observations for all unique reflections.
Rderiv = Σ|FPH | – |FP||/Σ|FP|
RCullis = Σ||FPH + FP| – |FH(calc)||/Σ|FPH – FP | for centric reflections. Phasing power, root mean square (r.m.s.) FH/r.m.s. ɛ where ɛ is lack of closure and FH is the calculated heavy atom structure factor.
Fig. 1. Yeast α1,2-mannosidase structure. (A) Schematic ribbon representation of the yeast α1,2-mannosidase viewed down the αα7-barrel axis from the LC side. (B) View at 90° to the orientation in (A), showing the protein–carbohydrate and protein–protein interactions. HM1 extends into the barrel of the adjacent molecule. Shown in orange and located at the protein–protein interface is the reconstructed loop (RL). The truncation of this loop was critical for the formation of good quality crystals (see Materials and methods). Due to the orientation of the dimer, the calcium ion is only visible in the protein at the top of the figure. In both (A) and (B), the calcium ion (Ca) is represented as a blue sphere, and the glycerol molecule (G) (red), the three N-glycans (HM1, HM2 and HM3) (yellow), the asparagine residues attached to these N-glycans and the two disulfide bridges (S1: Cys340–Cys385 and S2: Cys468–Cys471) (green) are shown in ball-and-stick representation. (C) Topological two-dimensional representation of the yeast α1,2-mannosidase structure. The figure is roughly oriented as in (A). α-helices, 310 helices and β-strands are represented as colored circles, small black circles and arrows, respectively. The β-hairpin that plugs the barrel is colored in pink. The secondary structure (see below) was assigned with the use of the program PROMOTIF (Hutchinson and Thornton, 1996) and is as follows: α1, residues 40–56; β1, 62–64; β2, 69–71; α2, 84–97; α3, 102–118; β3, 127–126; α4, 130–151; α5, 156–171; 310a, 172–175; β4, 185–187; β5, 193–194; 310b, 199–201; β6, 203–204; α6, 205–222; α7a, 225–232; α7b, 235–242; α7c, 244–248; β7, 255–256; α8, 275–288; α9, 291–307; β8, 309–311; β9, 318–320; β10, 334–336; 310c, 337–341; α10, 342–351; αRL, 356–359; α11, 372–390; β11, 400–403; β12, 414–415; β13, 421–423; 310d, 425–427; α12, 435–447; α13, 451–467; β14, 468–469; β15, 478–479; β16, 482–484; β17, 491–492; α14, 498–511; β18, 521–523; β19, 529–531; α15, 535–540. In all three panels, N and C indicate the N- and C-termini of the molecule. The color scheme for the α-helices, 310 helices and β-strands is conserved in all three panels. The figure was prepared with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merrit and Murphy, 1994).
Overall structure of the molecule
The molecule consists of consecutive helices alternating from outside to inside the barrel. This arrangement results in a topology of seven parallel inner helices (α2, α4, α6, α8, α10, α12 and α14) and a second set of seven parallel outer helices (α1, α3, α5, α7, α9, α11 and α13) concentric to the inner helices and antiparallel to them. The structure is stabilized by a disulfide bond, Cys340–Cys385, which forms a bridge between the 310c helix, located at the beginning of the inner α10 helix, and the outer α11 helix (Figure 1). This disulfide bond occurs between residues conserved in all known members of the family and has previously been shown to be essential for enzyme activity (Lipari and Herscovics, 1996). A second disulfide bond Cys468–Cys471, which links both ends of β-strand 14, is unlikely to be important since these residues are not conserved across the superfamily and mutation of these cysteines does not greatly affect enzyme activity (Lipari and Herscovics, 1996). The two ends of the barrel are structurally distinct (Figure 1B). On one end (short connection or SC side), the pairs of inner and outer helices are connected by short loops of up to four residues, with the exception of the reconstructed loop (RL) that links the α10 and α11 helices. Three N-glycans (HM1, HM2 and HM3) are found at the predicted N-glycosylation sites (Figure 1). These glycans extend away from the surface of the protein on the SC side of the barrel. Only one and three sugar residues of HM2 and HM3, respectively, were found in the electron density due to the flexibility of the oligosaccharides and their lack of stabilization by crystal contacts. In contrast, five mannose and two N-acetyl glucosamine residues were clearly identified in HM1, which behaves as the product of the enzyme, as discussed below (see Figure 2A and B). The other end (long connection or LC side) is structurally more complex and is similar to an ‘open flower’, with β-strands forming the petals of the flower (Figure 1B). The β-strands pack together to form a series of antiparallel β-sheets surrounding the helix barrel. The C-terminus of the protein (residues 512–549) consists of a β-hairpin protruding back into the center of the barrel from the SC side and an additional short helix, α15 (Figure 1A and B). The β-hairpin plugs the inner barrel, preventing an open channel through the protein. The β-hairpin, the inner helices and the β-sheets on the LC side of the barrel result in a cavity of ∼15 Å in depth, parallel to the central axis of the barrel, with a diameter of 25 Å at the level of the β-sheets, decreasing to ∼10 Å at the top of the β-hairpin. This cavity is a consequence of the seven pairs of helices present in the barrel, as no significant cavity is found in αα6-barrel proteins (Aleshin et al., 1992; Alzari et al., 1996; Park et al., 1997; Nagar et al., 1998; Parsiegla et al., 1998). The nine highly conserved acidic residues and the calcium ion, all of which are essential for catalytic activity (Lipari and Herscovics, 1999), are located at the top of this β-hairpin in the center of the αα7-helix barrel, indicating that this region contains the active site of the enzyme.
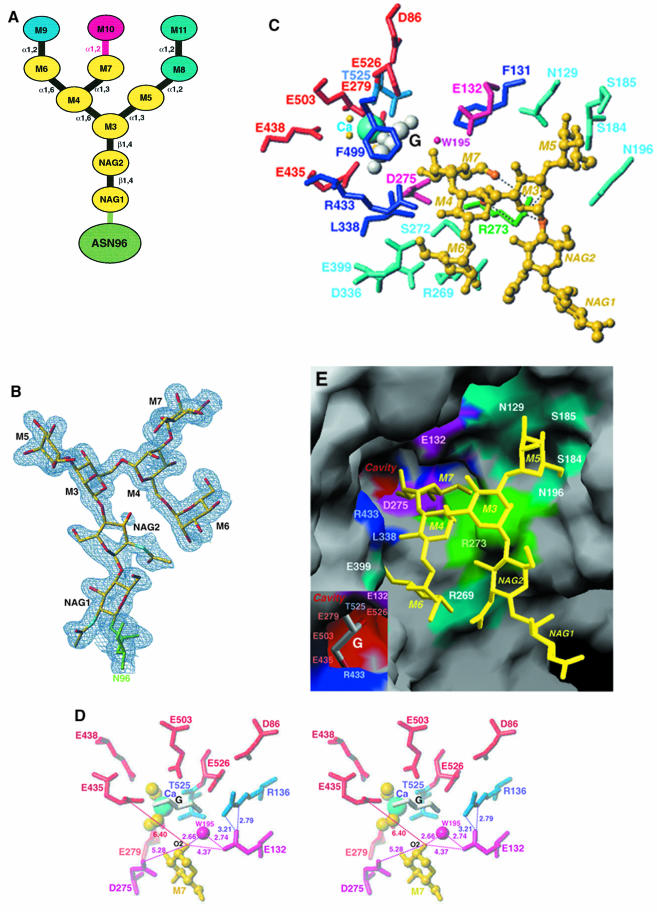
Fig. 2. High-mannose oligosaccharide–enzyme interaction. (A) Schematic representation of the Man9GlcNAc2 high-mannose oligosaccharide. Oligosaccharides of HM1 colored in yellow (M3→M7) are seen in the electron density maps. The pink α1,2-linkage between M7 and M10 is cleaved specifically by the enzyme, yielding Man8GlcNAc2 isomer B, the smallest oligosaccharide found in yeast N-glycans. The color scheme and labeling of the oligosaccharide residues are conserved in (C) and (D) as well as in Figure 3. (B) Final σA-weighted (2Fo– Fc) electron density map of oligosaccharide HM1. The map is contoured at 1.0σ and N96 is colored in green. (C and D) Detailed interactions between (C) HM1 and the protein, and (D) saccharide M7 and the protein. In both (C) and (D), residues strictly conserved in all the class I α1,2-mannosidases are shown, with acidic residues in red and fuschia, and other residues in dark and medium blue. The non-conserved residues are colored in cyan. In (C) R273, which is specific for the yeast and human ER α1,2-mannosidases that form Man8GlcNAc2 isomer B, is colored in green. The HM1 atoms bound to R273 are in orange. Water molecules involved in the catalytic mechanism and calcium binding are represented as small pink and yellow spheres, respectively. (D) A more favorable orientation of the mannose residue M7 and its interactions with E132, D275, E435 and Wat195. Distances are given in Å. (E) van der Waals surface representation of the protein–oligosaccharide interaction. The strictly conserved residues, as well as the glycerol molecule and the calcium ion, are completely buried in the active site. For clarity, not all the residues mentioned in the text are represented in (C), (D) and (E). (B), (C) and (D) were prepared with TURBO-FRODO (Roussel and Cambillau, 1991) and (E) with GRASP (Nicholls et al., 1991).
High-mannose oligosaccharide–protein interactions
A remarkable interaction is observed between adjacent molecules in the crystal, allowing a more detailed characterization of the active site (Figure 1B). The HM1 oligosaccharide (Figure 2A and B) from one molecule extends into the barrel of the adjacent symmetry-related molecule, with the terminal mannose residues located in the cavity containing the essential acidic residues and the calcium ion required for enzyme activity (Figure 2C, D and E). This protein–oligosaccharide interaction corresponds to the enzyme–product complex since the M10 mannose residue of the substrate has been cleaved. The HM1–protein interaction produces ∼47% of the overall intermolecular contacts found in the crystal and it is interesting to note that no crystals were obtained from the N96Q mutant lacking HM1. The presence of the reconstructed loop RL at the interface (Figure 1B) between interacting protein molecules in the crystal and the large contribution of HM1 to the packing stabilization indicate that both protein–protein and protein–oligosaccharide interactions facilitated crystallization. Analysis of the HM1–protein contacts shows that a large proportion of charged residues are involved in carbohydrate stabilization via either H-bonds or van der Waals interactions. At the top of the catalytic cavity, the HM1 core residues, NAG2 and M3, only contact R273 (Figures 2C, D and E, and 3). The α1,6-branch residues M4 and M6 contact R273 and R433, and R269, S272, D336, L338 and E399, respectively, whereas residue M5 of the α1,3-branch contacts residues S184, S185, N129 and N196. When considering the specificity of the yeast α1,2-mannosidase, M7, which would form the target glycosidic bond with M10 in the substrate, is located towards the bottom of the cavity and binds to residues F131, E207, R273 and D275. Of the residues that interact with HM1, the only residues that are conserved in all members of family 47 of the glycosyl hydrolases are D275 and R433. R269, S272 and N196 are specific for the yeast α1,2-mannosidase, while R273 and N129 are only found in the yeast and human class I α1,2-mannosidases known specifically to form the Man8GlcNAc2 B isomer (Figure 2A). The side chain of R273 forms contacts with M3, M4, M7 and NAG2. Arginine R273 may, therefore, together with R269, S272, D336, L338, E399 and R433, stabilize the α1,6-arm of HM1 and thus may dictate the position of the oligosaccharide required for cleavage of the glycosidic linkage between M7 and M10.
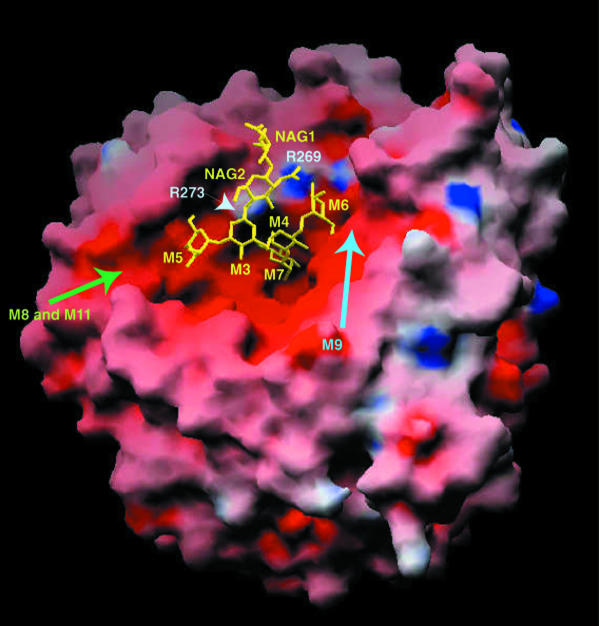
Fig. 3. Electrostatic surface of the yeast α1,2-mannosidase, showing the size of the catalytic groove and the location of the high-mannose oligosaccharide HM1 (in yellow). Acidic and basic residues are colored red and blue, respectively. Labeled in light blue are R269 and R273, two arginine residues that disrupt the overall electronegative surface of the catalytic groove. Although not visible in the electron density maps, the surface clearly indicates where the additional oligosaccharide residues M8, M9 and M11 could be located in the active site (see green and blue arrows). The contour level is at ±20 kT. This figure was prepared with GRASP (Nicholls et al., 1991).
The agreement between the crystal structure described here and the previously reported site-directed mutagenesis experiments (Lipari and Herscovics, 1999) clearly indicates that the protein–carbohydrate interactions observed in the crystal packing are biologically relevant. The close complementarity found between protein and carbohydrate surfaces also supports this hypothesis (Figure 3). Although not visible in the electron density maps, the surface indicates where the additional oligosaccharide residues M8, M9 and M11 could be located in the active site. Additional evidence is provided by the position of a glycerol molecule introduced during crystal freezing. Glycerol has previously been shown to mimic saccharide binding (Schmidt et al., 1998). In the α-mannosidase structure, a glycerol molecule is observed at the bottom of the active site close to M7 and to the calcium ion, suggesting that it partly occupies the putative binding site for M10 (Figure 2C, D and E). The three oxygen atoms of the glycerol molecule hydrogen-bond to glutamic acid (E435, E438, E503) and arginine (R433) residues. Glycerol also forms van der Waals interactions with F499 at the bottom of the catalytic cavity, suggesting that this strictly conserved residue is involved in substrate stabilization.
Catalytic mechanism
The yeast α1,2-mannosidase is an inverting glycosyl hydrolase (Lipari et al., 1995) that specifically cleaves the α1,2-oligosaccharide linkage between the C1 of M10 and O2 of M7, with inversion of the anomeric configuration at C1 of M10 and the formation of a hydroxyl group at O2 of M7. The catalytic mechanism usually involves two carboxylic acids separated by a distance of ∼9.5 Å, one acting as a general base removing a proton from water and the other acting as a general acid donating a proton to the leaving group (Ly and Withers, 1999). Given that M10 is absent from the structure, the observed oligosaccharide–protein interaction most probably represents the enzyme–product complex. The structural data together with the geometrical constraints expected for suitably positioned catalytic residues in inverting enzymes suggest that the only carboxylic acids possibly involved in the catalytic mechanism are E132, D275 and E435. The first two acidic residues are located at the rim of the small cavity containing the glycerol, while E435 is located a little deeper and closer to the glycerol molecule (Figure 2E). Based on the position of M7 in the model, there are two hypotheses that could be considered regarding the catalytic mechanism. In the first hypothesis, E132 could be the catalytic base abstracting a proton from water W195 (Figure 2D). This hypothesis is supported by the fact that the E132Q mutant has residual enzyme activity with a large decrease in kcat(Lipari and Herscovics, 1999). Similar low residual activity has been reported previously for glucoamylase mutated at its catalytic base (Frandsen et al., 1994). The presence of an H-bond between E132 and the strictly conserved residue R136 lends additional support to the role of E132 as the catalytic base by making it more acidic (Figure 2D). Assuming that E132 is the catalytic base, then either D275 or E435 could serve as the acid. These residues are located on the opposite side of M7, 9.5 and 9.6 Å away from E132, respectively. The unambiguous identification of the acid is not possible due to the lack of hydrogen-bond interactions between D275, E435 or any other amino acid and the O2 of M7. In the current model D275 is closer to the O2 of M7 than E435 (5.28 Å versus 6.42 Å), suggesting that D275 is a more attractive candidate for the catalytic acid. This hypothesis is consistent with the complete lack of activity of both the D275N and E435Q mutants (Lipari and Herscovics, 1999).
An alternative, but less favored hypothesis implicates E435 as the catalytic base since it interacts with a water molecule that in turn hydrogen-bonds with the glycerol and the calcium ion. In this instance, E132 would be the most likely candidate for the catalytic acid as D275 is on the same side of the oligosaccharide as E435. Further experimental data are required before the catalytic residues can be identified with certainty.
Calcium has been shown to be essential for activity. The calcium ion binds to the carbonyl oxygen and the Oγ of T525 located at the top of the β-hairpin, and to four water molecules that are in turn hydrogen-bonded to one of the two carboxylate groups of residues E279, E435, E438 and E503, previously demonstrated as crucial for enzymatic activity (Lipari and Herscovics, 1999). Three of these glutamates (E435, E438 and E503) also interact with the glycerol molecule thought to mimic M10 (Figure 2C and D). The lack of direct interaction between the calcium ion and either the carbohydrate moiety or the glycerol molecule seen in the model suggests that the cation does not play a direct role in the catalytic mechanism but is involved in the stabilization of the substrate and/or the enzyme. The latter hypothesis is consistent with the previous observation that calcium protects the enzyme from thermal denaturation (Lipari and Herscovics, 1999).
Orientation of the α-mannosidase in the lumen of the ER
In vivo, the yeast processing α1,2-mannosidase is a resident ER protein with a lumenally oriented catalytic domain and an N-terminal transmembrane domain that specifies its ER localization (Grondin and Herscovics, 1992; Massaad et al., 1999). In the model, the N-terminus of the structure is located on the SC side of the barrel (Figure 1B), indicating that the SC side of the protein probably faces the ER membrane while the LC side projects into the ER lumen in a favorable orientation to interact with newly synthesized glycoprotein substrates. Since there are ∼10 amino acids between the catalytic domain shown in the crystal structure and the transmembrane segment, a certain flexibility in the orientation of the catalytic domain would be possible even when it is anchored to the membrane.
Conclusion
This first structure of a class I processing α1,2- mannosidase provides the framework to understand the mechanism of action, to determine the basis of the differences in specificity of the different family members and to elucidate their respective roles in glycoprotein maturation. In addition, knowledge of the structure of the active site of the yeast enzyme and the mode of substrate binding will aid in the design of specific inhibitors that may be used to prevent the degradation of misfolded glycoproteins in genetic diseases such as emphysema (Sifers, 1995) and cystic fibrosis (Kopito, 1999).
Materials and methods
Protein preparation and crystallization
The α1,2-mannosidase nucleotide sequence encoding amino acids 34–549 was amplified by PCR and ligated into the XhoI and BamHI sites of the pHIL-S1 vector (Invitrogen) producing plasmid YpHΔ33. The nucleotide sequence encoding amino acids 367–371 in the plasmid YpHΔ33 was then deleted using the USE Mutagenesis kit from Pharmacia Biotech Inc. The protein was expressed in P.pastoris and purified as previously described. Crystals were grown by the vapor diffusion method from protein drops (10 mg/ml) equilibrated against well solution (1 ml) containing 17–19% w/v polyethylene glycol MME 2K, 100 mM sodium citrate (pH 5.6) and 250 mM ammonium acetate. Crystals grow within 5 days and exhibit the symmetry of space group P3121 (a = b = 90.04 Å and c = 154.87 Å), with one molecule in the asymmetric unit (59% solvent content).
Data collection, structure determination and model refinement
Native and derivative data were first collected at room temperature using monochromated CuKα X-radiation (Rigaku Rotaflex RU200 rotating anode generator) on a Mar Research (345 mm diameter) imaging plate system. Subsequent native data sets were collected at beamline X8-C at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) using a Quantum4 CCD detector and flash-frozen crystals (cryoprotected in artificial mother liquor containing 25% v/v glycerol). All data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). The structure was determined by the SIRAS phasing method using a single site mercury derivative. The program CNS (Brünger et al., 1998) was used for all stages of the structure determination and refinement. The atomic position of the mercury atom was found using Patterson methods. The mercury coordinates were refined and the calculated phases were subsequently improved by solvent flattening, improving the mean figure of merit (FOM) from 0.42 to 0.84. The initial solvent-flattened SIRAS map showed a monomeric protein well separated from the solvent. Interpretable regions of the map were used to build strands of polyalanine chains with the program TURBO-FRODO (Roussel and Cambillau, 1991). The stepwise re-combination of the phases from the growing polyalanine model with the original SIRAS phases continuously improved the FOM and enhanced the quality of the electron density map. When ∼82% of the protein backbone had been traced, the side chains were inserted using the one disulfide bridge clearly identifiable in the electron density, and the three N-glycosylation sites as starting points for fitting the sequence. The model was refined using the simulated annealing slow-cooling protocol in the resolution range of 50–1.54 Å. The progress of the refinement was monitored by reductions in the Rfactor and Rfree, and the concomitant increase in the FOM of the combined model/SIRAS phases. All residues present in the structure have been modeled, with the exception of residue 410 owing to weak electron density in this region. As detailed previously, residues 367–371 were deleted in the construct. The model comprises 4118 non-hydrogen protein atoms, together with 394 water molecules, 135 carbohydrate atoms, six glycerol atoms plus a Ca2+ ion. In the final structure, none of the non-glycine residues lie in the disallowed region of the Ramachandran plot as analyzed with PROCHECK (Laskowski et al., 1993). The conformation of the oligosaccharides is similar to those observed in other oligosaccharide structures (Petrescu et al., 1999).
Accession number
The coordinates have been deposited in the Protein Data Bank (accession code 1DL2).
Acknowledgments
Acknowledgements
This research is supported by grants from the National Institutes of Health (GM 31265) and the Natural Sciences and Engineering Research Council of Canada (CPG 0181709) to A.H. and P.L.H., a postdoctoral fellowship from the Association pour la Recherche contre le Cancer to F.V. and a scholarship from the Natural Sciences and Engineering Research Council of Canada to F.L. Station X8-C is supported by the US Department of Energy and a Multiuser maintenance grant from the Medical Research Council of Canada and the Natural Science and Engineering Research Council of Canada.
References
- Aleshin A., Golubev, A., Firsov, L.M. and Honzatko, R.B. (1992) Crystal structure of glucoamylase from Aspergillus awamori var. X100 to 2.2Å resolution. J. Biol. Chem., 267, 19291–19298. [DOI] [PubMed] [Google Scholar]
- Alzari P.M., Souchon, H. and Dominguez, R. (1996) The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum.Structure, 4, 265–275. [DOI] [PubMed] [Google Scholar]
- Bischoff J. and Kornfeld, R. (1983) Evidence for an α-mannosidase in endoplasmic reticulum of rat liver. J. Biol. Chem., 258, 7907–7910. [PubMed] [Google Scholar]
- Bischoff J., Liscum, L. and Kornfeld, R. (1986) The use of 1-deoxymannojirimycin to evaluate the role of various α-mannosidases in oligosaccharide processing in intact cells. J. Biol. Chem., 261, 4766–4774. [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998)Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Byrd J.C., Tarentino, A.L., Maley, F., Atkinson, P.H. and Trimble, R.B. (1982) Glycoprotein synthesis in yeast. Identification of Man8GlcNAc2 as an essential intermediate in oligosaccharide processing. J. Biol. Chem., 257, 14657–14666. [PubMed] [Google Scholar]
- Camirand A., Heysen, A., Grondin, B. and Herscovics, A. (1991) Glycoprotein biosynthesis in Saccharomyces cerevisiae.J. Biol. Chem., 266, 15120–15127. [PubMed] [Google Scholar]
- Dole K., Lipari, F., Herscovics, A. and Howell, P.L. (1997) Crystallization and preliminary X-ray analysis of the class I α1,2-mannosidase from Saccharomyces cerevisiae.J. Struct. Biol., 120, 69–72. [DOI] [PubMed] [Google Scholar]
- Frandsen T.P., Dupont, C., Lehmbeck, J., Stoffer, B., Sierks, M.R., Hontzatko, R.B. and Svensson, B. (1994) Site-directed mutagenesis of the catalytic base glutamic acid 400 in glucoamylase from Aspergillus niger and of tyrosine 48 and glutamine 401, both hydrogen-bonded to the γ-carboxylate group of glutamic acid 400. Biochemistry, 33, 13808–13816. [DOI] [PubMed] [Google Scholar]
- Gonzalez D.S., Karaveg, K., Vandersall-Nairn, A.S., Lal, A. and Moremen, K.W. (1999) Identification, expression and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide. J. Biol. Chem., 274, 21375–21386. [DOI] [PubMed] [Google Scholar]
- Grondin B. and Herscovics, A. (1992) Topology of ER processing α-mannosidase of Saccharomyces cerevisiae.Glycobiology, 2, 369–372. [DOI] [PubMed] [Google Scholar]
- Hammond C. and Helenius, A. (1995) Quality control in the secretory pathway. Curr. Opin. Cell Biol., 7, 523–529. [DOI] [PubMed] [Google Scholar]
- Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J., 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A. (1999a) Processing glycosidases of Saccharomyces cerevisiae.Biochim. Biophys. Acta, 1426, 275–285. [DOI] [PubMed] [Google Scholar]
- Herscovics A. (1999b) Importance of glycosidases in mammalian glycoprotein biosynthesis. Biochim. Biophys. Acta, 1473, 96–107. [DOI] [PubMed] [Google Scholar]
- Herscovics A. (1999c) Glycosidases of the asparagine-linked oligosaccharide processing pathway. In Pinto,B.M. (ed.), Comprehensive Natural Products Chemistry. Elsevier, New York, NY, Vol. 3, pp. 13–35. [Google Scholar]
- Hutchinson E.G. and Thornton, J.M. (1996) PROMOTIF: a program to identify and analyze structural motifs in proteins. Protein Sci., 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C.A., Burda, P., Roth, J. and Aebi, M. (1998) Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol., 142, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek-Kelly S. and Herscovics, A. (1988) Glycoprotein biosynthesis in Saccharomyces cerevisiae. Purification of the α-mannosidase which removes one specific mannose residue from Man9GlcNAc2. J. Biol. Chem., 263, 14757–14763. [PubMed] [Google Scholar]
- Jelinek-Kelly S., Akiyama, T., Saunier, B., Tkacz, J.S. and Herscovics, A. (1985) Characterization of a specific α-mannosidase involved in oligosaccharide processing in Saccharomyces cerevisiae.J. Biol. Chem., 260, 2253–2257. [PubMed] [Google Scholar]
- Knop M., Hauser,N. and Wolf,D.H. (1996) N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast, 12, 1229–1238. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. (1999) Biosynthesis and degradation of CFTR. Physiol. Rev., 79, S167–S173. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lal A., Pang, P., Kalelkar, S., Romero, P.A., Herscovics, A. and Moremen, K.W. (1998) Substrate specificities of recombinant murine Golgi α1,2-mannosidases IA and IB and comparison with endoplasmic reticulum and Golgi processing α1,2-mannosidases. Glycobiology, 8, 981–995. [DOI] [PubMed] [Google Scholar]
- Laskowski R., MacArthur, M., Moss, D. and Thornton, J. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 91–97. [Google Scholar]
- Lipari F. and Herscovics, A. (1994) Production, purification and characterization of recombinant yeast processing α1,2-mannosidase. Glycobiology, 4, 697–702. [DOI] [PubMed] [Google Scholar]
- Lipari F. and Herscovics, A. (1996) Role of the cysteine residues in the α1,2-mannosidase involved in N-glycan biosynthesis in Saccharo- myces cerevisiae. The conserved Cys340 and Cys385 residues form an essential disulfide bond. J. Biol. Chem., 271, 27615–27622. [DOI] [PubMed] [Google Scholar]
- Lipari F. and Herscovics, A. (1999) Calcium binding to the class I α1,2-mannosidase from Saccharomyces cerevisiae occurs outside the EF hand motif. Biochemistry, 38, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Lipari F., Gour-Salin, B.J. and Herscovics, A. (1995) The Saccharomyces cerevisiae processing α1,2-mannosidase is an inverting glycosidase. Biochem. Biophys. Res. Commun., 209, 322–326. [DOI] [PubMed] [Google Scholar]
- Liu Y., Choudhury, P., Cabral, C.M. and Sifers, R.N. (1997) Intracellular disposal of incompletely folded human α1-antitrypsin involves release from calnexin and post-translational trimming of asparagine-linked oligosaccharides. J. Biol. Chem., 272, 7946–7951. [DOI] [PubMed] [Google Scholar]
- Liu Y., Choudhury, P., Cabral, C.M. and Sifers, R.N. (1999) Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J. Biol. Chem., 274, 5861–5867. [DOI] [PubMed] [Google Scholar]
- Ly H.D. and Withers, S.G. (1999) Mutagenesis of glycosidases. Annu. Rev. Biochem., 68, 487–522. [DOI] [PubMed] [Google Scholar]
- Massaad M.J., Franzusoff, A. and Herscovics, A. (1999) The processing α1,2-mannosidase of Saccharomyces cerevisiae depends on Rer1p for its localization in the endoplasmic reticulum. Eur. J. Cell Biol., 78, 435–440. [DOI] [PubMed] [Google Scholar]
- Merrit E. and Murphy, M. (1994) RASTER3D: a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Moremen K.W., Trimble, R.B. and Herscovics, A. (1994) Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology, 4, 113–125. [DOI] [PubMed] [Google Scholar]
- Nagar B., Jones, R.G., Diefenbach, R.J., Isenman, D.E. and Rini, J.M. (1998) X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science, 280, 1277–1281. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbon. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Park H.W., Boduluri, S.R., Moomaw, J.F., Casey, P.J. and Beese, L.S. (1997) Crystal structure of protein farnesyltransferase at 2.25Å resolution. Science, 275, 1800–1804. [DOI] [PubMed] [Google Scholar]
- Parsiegla G., Juy, M., Reverbel-Leroy, C., Tardif, C., Belaich, J.-P., Driguez, H. and Haser, R. (1998) The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J., 17, 5551–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu A.J., Petrescu, S.M., Dwek, R.A. and Wormald, M.R. (1999) A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology, 9, 343–352. [DOI] [PubMed] [Google Scholar]
- Rizzolo L.J. and Kornfeld, R. (1988) Post-translational protein modification in the endoplasmic reticulum. Demonstration of fatty acylase and deoxymannojirimycin-sensitive α-mannosidase activities. J. Biol. Chem., 263, 9520–9525. [PubMed] [Google Scholar]
- Roussel A. and Cambillau,C. (1991) The TURBO-FRODO graphics package. In Silicon Graphics Partners Geometry. Silicon Graphics Corporation, p. 81. [Google Scholar]
- Schmidt A., Schlacher, A., Steiner, W., Schwab, H. and Kratky, C. (1998) Structure of the xylanase from Penicillium simplicissimum.Protein Sci., 7, 2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers R.N. (1995) Defective protein folding as a cause of disease. Nature Struct. Biol., 2, 355–357. [DOI] [PubMed] [Google Scholar]
- Su K., Stoller, T., Rocco, J., Zemsky, J. and Green, R. (1993) Pre-Golgi degradation of yeast prepro-α-factor expressed in a mammalian cell. Influence of cell type-specific oligosaccharide processing on intracellular fate. J. Biol. Chem., 268, 14301–14309. [PubMed] [Google Scholar]
- Tremblay L.O. and Herscovics, A. (1999) Cloning and expression of a specific human α1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2isomer B during N-glycan biosynthesis. Glycobiology, 9, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng S. and Spiro, R.G. (1993) Demonstration that a kifunensine-resistant α-mannosidase with a unique processing action on N-linked oligosaccharides occurs in rat liver endoplasmic reticulum and various cultured cells. J. Biol. Chem., 268, 25656–25663. [PubMed] [Google Scholar]
- Ziegler F.D. and Trimble, R.B. (1991) Glycoprotein biosynthesis in yeast: purification and characterization of the endoplasmic reticulum Man9 processing α-mannosidase. Glycobiology, 1, 605–614. [DOI] [PubMed] [Google Scholar]