Abstract
Sp3 is a ubiquitously expressed transcription factor closely related to Sp1 (specificity protein 1). We have disrupted the mouse Sp3 gene by homologous recombination. Sp3-deficient embryos are growth retarded and invariably die at birth of respiratory failure. The cause for the observed breathing defect remains obscure since only minor morphological alterations were observed in the lung, and surfactant protein expression is indistinguishable from that in wild-type mice. Histological examinations of individual organs in Sp3–/– mice show a pronounced defect in late tooth formation. In Sp3 null mice, the dentin/enamel layer of the developing teeth is impaired due to the lack of ameloblast-specific gene products. Comparison of the Sp1 and Sp3 knockout phenotype shows that Sp1 and Sp3 have distinct functions in vivo, but also suggests a degree of functional redundancy.
Keywords: knockout/post-natal death/Sp1/Sp3/tooth development
Introduction
Many housekeeping, tissue-specific and viral genes contain functionally important GC and related GT/CACC boxes. It has been known for some time that the general transcription factor Sp1 (specificity protein 1) (Kadonaga et al., 1987) can bind to and act through these elements, and it was generally accepted that Sp1 is involved in the expression of many different genes. More recently, however, it became clear that Sp1 is not the only protein acting through ‘Sp1-binding sites’ but represents the first identified and cloned protein of a large, still growing family of transcription factors united by the presence of a highly conserved DNA-binding domain consisting of three zinc fingers. This Sp/XKLF superfamily of proteins includes the Sp transcription factors, their close relatives BTEB1, TIEG1 and TIEG2, and the Krüppel-like factors (reviewed in Philipsen and Suske, 1999).
The Sp subfamily of transcription factors is composed of four proteins (Sp1, Sp2, Sp3 and Sp4) characterized by very similar structural features (Suske, 1999). In addition to the highly conserved DNA-binding domain, all four proteins contain glutamine-rich activation domains adjacent to serine/threonine-rich stretches in their N–terminal region. The linkage to the four human Hox gene clusters also documents their close evolutionary relationship (Kalff-Suske et al., 1995, 1996; Scohy et al., 1998). Sp1, Sp3 and Sp4 are more closely related to each other than to Sp2 (Philipsen and Suske, 1999; Suske, 1999). Consistently, Sp1, Sp3 and Sp4 recognize the classical GC box and the related GT/CACC box with identical affinity (Hagen et al., 1992, 1994). Sp3 and Sp1 are ubiquitously expressed, unlike Sp4, which shows a complex expression pattern but is most abundant in neuronal tissues (Supp et al., 1996).
A large variety of biological functions have been assigned to Sp1-binding sites. This raises the question of which of the tasks are performed by which protein in vivo. This question is particularly interesting for Sp1 and Sp3 because both proteins are ubiquitously expressed. Mice lacking Sp1 and Sp4 have been reported (Supp et al., 1996; Marin et al., 1997). The most interesting aspect of the Sp4 phenotype is the complete absence of mating behavior in Sp4 null males. Since their reproductive organs are fully developed and apparently normal, the most likely cause of this behavioral abnormality is a neurological defect.
Sp1 null embryos are severely growth retarded, and die after day 10 of embryonic development (E10). They display a wide range of abnormalities, but all characteristic hallmarks up to this developmental stage are present. Blastocyst injections of Sp1 null embryonic stem (ES) cells showed that these cells contribute efficiently to early chimeric embryos, but after embryonic day 11 (E11) this declines very rapidly with no detectable contribution to any tissue of newborn animals. Thus, Sp1 deficiency causes a cell-autonomous defect, and it appears that Sp1 function is generally required for cellular survival after E10 (Marin et al., 1997).
It was surprising that the Sp1 null embryos survive until the post-implantation stage. More than 3000 publications have implicated this factor in the activation of a very large number of genes and in cellular processes such as cell cycle regulation, chromatin remodeling and the propagation of methylation-free CpG islands. Thus, a cell lacking Sp1 would be predicted to stand little chance of surviving, but surprisingly Sp1 null ES cells have normal growth characteristics and survival rates. It has been suggested that this might be due to the presence of Sp3, the most closely related Sp family member (Marin et al., 1997).
Sp3 has been shown to act as a transcriptional activator on many promoters similarly to Sp1 (e.g. Udvadia et al., 1995; Liang et al., 1996; Ihn and Trojanowska, 1997; Zhao and Chang, 1997). In other promoter settings, however, Sp3 remained inactive or acted only as a weak activator (e.g. Hagen et al., 1994; Majello et al., 1994; Dennig et al., 1995; Kumar and Butler, 1997). Biochemically, the most obvious differences between Sp3 and Sp1 are the presence of a potent inhibitory domain in Sp3 (Dennig et al., 1996) and the existence of three Sp3 isoforms (Kennett et al., 1997; Suske, 1999). However, the significance of these differences for in vivo functions remains an open question.
Here we describe the targeted disruption of the mouse Sp3 gene by homologous recombination and analysis of Sp3 null mice. We find that the absence of Sp3 has consequences for mouse development that are very different from those observed in Sp1 knockout mice. Sp3 null mice develop until birth with no obvious gross abnormalities other than a reduction in body weight. After natural birth or Caesarian section, the knockout mice invariably die within 10 min, apparently of respiratory failure. Histological examination revealed minor structural abnormalities in the lungs. Furthermore, the mice show a pronounced defect in dentin/enamel layer formation of the developing teeth. We conclude that Sp1 and Sp3 may have similar, and therefore redundant, functions during early development but exert distinct and highly specific functions in later developmental stages.
Results
Isolation of mouse Sp3 gene fragments
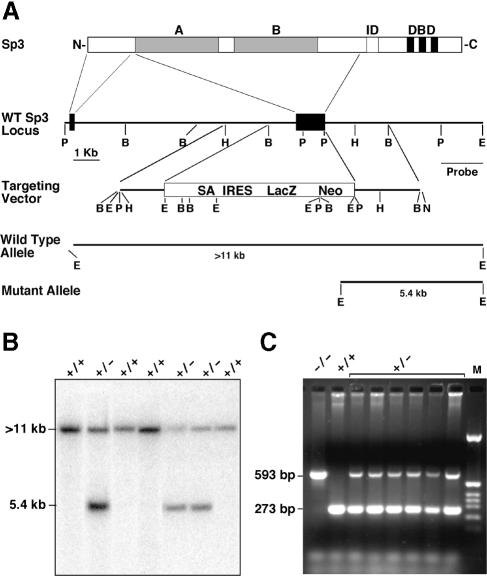
Primers derived from the human Sp3 cDNA were used to amplify a fragment of the mouse Sp3 gene by PCR. The primers A3/K (5′–CAGATCATTCCTGGCTCT–3′) and A3/L (5′–TCTAGATCGACACTATTGAT–3′) that produced a unique 210 bp fragment with human and mouse genomic DNA were used to screen a genomic mouse ES cell P1 phage library (Genome Systems Inc.). A single P1 clone of ∼80 kbp was mapped and several fragments were subcloned into the Bluescript KS vector. Sequence analyses revealed that both activation domains of Sp3 are encoded by a single large exon (Figure 1A).
Fig. 1. Targeted disruption of the mouse Sp3 gene. (A) Schematic representation of the Sp3 protein structure. The glutamine-rich activation domains A and B, the inhibitory domain (ID) and the zinc fingers (black bars) of the DNA-binding domain (DBD) are indicated. Connecting lines with the corresponding murine Sp3 gene regions indicate the derivation of the N–terminal part of the Sp3 protein. Both glutamine-rich activation domains (A and B) are encoded by a single large exon. In the targeting vector, this exon was replaced by a cassette containing a splice acceptor site (SA), an internal ribosomal entry site (IRES) and a lacZ–neo fusion gene (LacZ Neo) (Mountford et al., 1994). The positions of relevant restriction sites (B, BamHI; E, EcoRI; H, HindIII; P, PstI; N, NotI) and the probe used for Southern blotting are indicated. Restriction of genomic DNA with EcoRI and hybridization with the indicated probe detects a >11 kbp fragment of the wild-type allele and a 5.4 kbp fragment of the mutated allele. (B) Southern blot analysis of targeted ES cells. (C) PCR analysis of mouse embryos. The primers produce a 273 bp DNA fragment from the wild-type and a 593 bp fragment from the targeted allele.
Targeted disruption of the mouse Sp3 gene
The target vector pBS-P-B-Δ-lacZ-neo–5′–Sp3 (Materials and methods) was designed to substitute exon 2 [amino acids 38–490 according to Yajima et al. (1998)] of the murine Sp3 gene for IRES-LacZ-neo-polyA sequences (Figure 1A). The lacZ–neo fusion gene is expressed under the control of the endogenous Sp3 promoter. The deleted sequences encode both glutamine-rich activation domains of Sp3 (Dennig et al., 1996). Thus, the disruption should result in a null allele.
Plasmid pBS-P-B-Δ-lacZ-neo–5′–Sp3 was linearized at a unique NotI site present in the vector for transfection into E14 ES cells. Cells were maintained subsequently under G418 selection. A total of 36 G418-resistant colonies were analyzed by Southern blotting for the homologous recombination event. Hybridization of EcoRI-restricted DNA from individual clones with an EcoRI–PstI intron fragment was predicted to show a >11 kbp fragment from the wild-type locus and a 5.4 kbp fragment from a correctly targeted locus (Figure 1A). Fourteen clones showed the predicted mutant fragment of 5.4 kbp (Figure 1B). In addition, PCR analysis with a set of primers specific for the wild-type and the targeted Sp3 gene (Figure 1C) confirmed the targeted disruption of the mouse Sp3 gene.
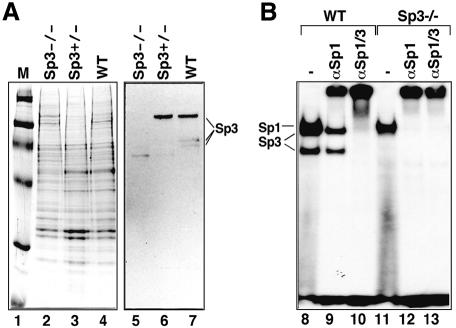
Two of the targeted ES clones with the correct karyotype were injected into C57BL/6 blastocysts. Breeding of the chimeras revealed germline transmission of the mutated Sp3 allele. Mating of heterozygous animals resulted in embryos deficient in all three Sp3 isoforms (Figure 2).
Fig. 2. Sp3 protein expression in wild-type and Sp3 mutant mice. (A) Western blot analysis of wild-type, Sp3+/– and Sp3–/– animals. Nuclear extracts (6 μg of protein) from brains of wild-type (WT), heterozygous (Sp3+/–) and Sp3-deficient (Sp3–/–) mice were fractionated through 7.5% SDS–polyacrylamide gels, stained with Coomassie Blue (lanes 1–4; M, marker lane) or blotted on nitrocellulose filter and incubated with Sp3-specific antibodies (lanes 5–7). (B) Electrophoretic mobility shift assay of GC box binding activity in wild-type (WT) and Sp3-deficient (Sp3–/–) mouse embryonic fibroblasts. Crude nuclear extracts (1.25 μg of protein) were incubated with 32P-labeled GC box oligonucleotide in the absence (lanes 8 and 11) or presence of antisera against Sp1 (lanes 9 and 12, αSp1) or a mixture of antisera against Sp1 and Sp3 (lanes 10 and 13, αSp1/3).
Mice lacking Sp3 die immediately after birth due to respiratory failure
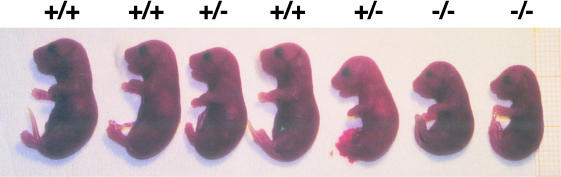
Heterozygous Sp3+/– mice derived from male germline chimeras exhibited no discernible phenotype. Sp3-deficient mice derived from heterozygous matings were not viable (Table I). Genotyping of embryos obtained by Caesarian section shortly before the parturition date (E18.5) showed no statistically significant loss of Sp3–/– mice up to birth (Table I). The weight of these pups was ∼25% lower than that of their wild-type littermates (Figure 3; Table II).
Table I. Genotype distribution of Sp3 heterozygous crossings.
| Total | +/+ | +/– | –/– | |
|---|---|---|---|---|
| E18.5 | 242 | 70 (28.8%) | 115 (47.1%) | 57 (23.4%) |
| Day 10 | 68 | 24 (35.3%) | 44 (64.7%) | 0 (0%) |
The genotype was determined by PCR analysis as described in Materials and methods.
Fig. 3. Sp3 mutant E18.5 embryos. Genotyped E18.5 embryos of one litter that are wild-type (+/+), heterozygous (+/–) or homozygous (–/–) for the targeted Sp3 gene locus. Homozygous mutants are ∼25% smaller than wild-type and heterozygous embryos.
Table II. Weight of embryos (g) of Sp3 heterozygous crossings.
| +/+ | +/– | –/– | |
|---|---|---|---|
| E13.5 | 0.16, SD = 0.02, n = 19 | 0.15, SD = 0.02, n = 31 | 0.12, SD = 0.02, n = 15 |
| E18.5 | 1.10, SD = 0.16, n = 18 | 1.05, SD = 0.14, n = 39 | 0.72, SD = 0.08, n = 17 |
Close monitoring of E18.5 embryos after Caesarian section and of litters after birth revealed that Sp3–/– mice were born alive and made visible efforts to breathe. In contrast to their wild-type and heterozygous littermates, the Sp3 null mice died within 10 min post-partum. The duration of the pregnancy was prolonged by up to 2 days with progesterone injections in an attempt to promote the maturation of the Sp3 null fetuses. However, this did not rescue the neonatal lethality of the knockout mice.
Expression of lung-specific genes is not affected in Sp3–/– mice
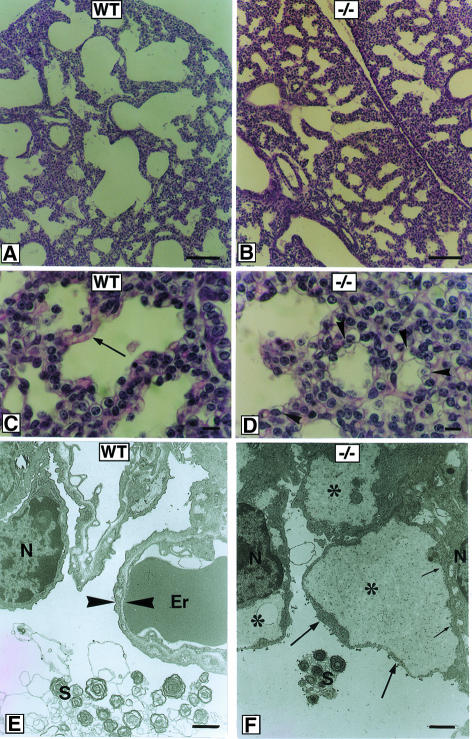
Lung tissue dissected from several mutant neonates obtained from independent litters failed to float on water, indicating that the alveoli were never filled with air. Histopathological examination of the lung (Figure 4) revealed that lung tissue of Sp3 null mice is more compact, featuring a smaller mean alveolar space diameter and thicker septa between the alveoli. In addition, fewer capillaries are observed in Sp3–/– lung tissue, but instead abnormal cubical pale cells are present at the inner surface of the alveoli. Ultrastructural analysis of these cells shows disruption of the apical membrane along with a small rim of cytoplasm leading to an artificial intracellular space filled with slightly electron-dense material (Figure 4).
Fig. 4. Histological and ultrastructural analyses of lung tissue in wild-type and Sp3–/– mice. Lung tissue at E18.5 of wild-type (WT) (A, C and E) and Sp3 knockout mice (–/–) (B, D and F) is shown in histological sections stained with hematoxylin and eosin (A–D), or in ultrastructural sections (E and F). At low magnification (A and B), lung tissue from Sp3–/– mice is more compact, with a smaller mean diameter of alveolar spaces and a thicker septum between individual alveoli. At higher magnification (C and D), capillaries are regularly seen at the inner surface of the alveoli in wild-type mice (C, arrow). Lung tissue from Sp3-deficient mice exhibits small alveoli with the inner surface lined with cuboid pale cells (D, arrowheads). Ultrastructural analysis revealed that the apical membranes of these cells together with a small rim of cytoplasm (F, large arrows) are disrupted from their original location near the nucleus (N, small arrows in F). These lead to artificial intracellular spaces (asterisks) filled with slightly electron-dense material. In wild-type lung tissue, a regular air–blood boundary (between arrowheads in E) consisting of an endothelial cell, a basement membrane and a pneumocyte type I is observed. An erythrocyte (‘Er’ in E) filling the space of a capillary and surfactant (‘S’ in E and F) are indicated. Bar in (A) and (B), 100 μm; in (C) and (D), 10 μm; in (E) and (F), 0.5 μm.
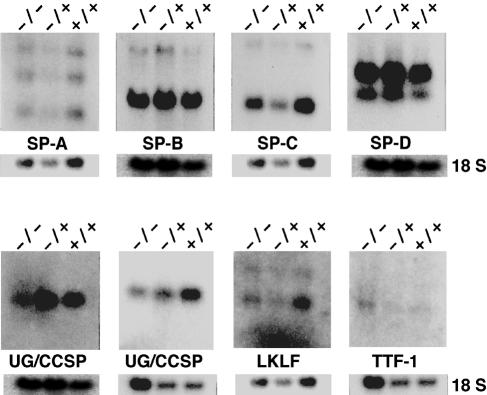
The histopathological examination suggests that the inability of the Sp3–/– mice to breathe might be attributed to an intrinsic inability of the lungs to expand. We have examined the presence of transcripts encoding surfactant and other lung-specific proteins involved in lung development and function. Northern blot analyses of RNA from E18.5 mice revealed that expression of surfactant proteins A, B, C and D, and the transcription factors, thyroid transcription factor 1 (TTF-1), which is highly expressed in lung Clara cells, and lung Krüppel-like factor (LKLF), was not affected in lungs of Sp3–/– embryos (Figure 5). In addition, elastin and p21 mRNAs were expressed at normal levels (data not shown). The only transcript that was slightly reduced (2–fold) was uteroglobin/Clara cell secretory protein (UG/CCSP). However, uteroglobin/CCSP-deficient mice develop normally and are apparently healthy (Stripp et al., 1996). Thus, the 2–fold lower expression of uteroglobin/CCSP cannot account for the breathing failure. In conclusion, the molecular cause of the respiratory defect remains unknown at present.
Fig. 5. Expression of putative Sp3 target genes in the lung. RNA was extracted from lungs of wild-type (+/+), Sp3+/– (+/–) and Sp3-deficient (–/–) E18.5 embryos, subjected to electrophoresis through 1.2 or 0.8% formaldehyde–agarose gels and transferred to nylon membranes. The filters were hybridized with cDNA fragments of surfactant proteins A (SP-A), B (SP-B), C (SP-C) and D (SP-D), uteroglobin/Clara cell secretory protein (UG/CCSP; two different membranes), lung Krüppel-like factor (LKLF) and thyroid transcription factor (TTF–1). As a control, the filters were probed with an 18S rRNA-specific oligonucleotide.
Impact on tooth development
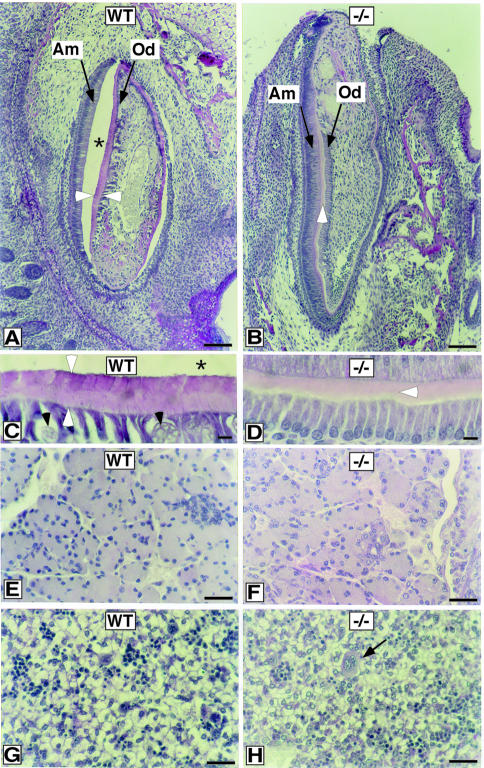
In the course of analyzing histological sections of mouse E18.5 embryos, we observed a morphological deviation in the developing teeth of Sp3–/– mice. In our sections, the dentin/enamel layer of the teeth is generally disrupted from the ameloblast cells in wild-type mice, leading to the creation of an artificial empty space (Figure 6). This was not observed in Sp3 null mice. At late embryonic stages, the processes of dentinogenesis (dentine formation) and amelogenesis (enamel formation) take place. Close inspection of the sections revealed that the developing teeth of wild-type mice contain a continuous row of ameloblasts and a row of odontoblasts interrupted by small vessels (Figure 6A). Two layers of differentially stained pink and purple material, consisting of predentin, dentin and enamel, are visible between the ameloblasts and odontoblasts (Figure 6C). In Sp3–/– mice, only a few vessels are observed and the odontoblasts are arranged in a continuous row. Most significantly, teeth of Sp3–/– mice contain only the pink layer and lack the purple layer (Figure 6D).
Fig. 6. Histological analysis of developing teeth in wild-type and Sp3–/– mice. Teeth at E18.5 of wild-type (A and C) and Sp3-deficient mice (B and D) are shown in histological sections stained with PAS. At low power magnification, the developing tooth of wild-type mice shows a continuous row of ameloblasts (‘Am’, arrow) and odontoblasts (‘Od’, arrow) interrupted by small vessels interspersed between the odontoblasts (black arrowheads in C). Between the ameloblasts and odontoblasts, a sheet of pink and purple stained material that contains dentin and enamel is visible (marked in A and C by two white arrowheads). This layer of secretory products has been disrupted from the ameloblasts, leaving an artificial space (asterisks in A and C) that is regularly observed in wild-type mice of this age, but not in Sp3 null mice. In contrast, Sp3 knockout mice show only a pink layer (white arrowheads in B and D). In addition, only a few vessels are observed in the dental papilla, leaving the odontoblasts in a continuous row (‘Od’ with arrow in B and bottom cell layer in D). (E–H) PAS-stained histological sections of E18.5 tissues with no obvious morphological alterations in knockout mice. (E and F) Pancreas. (G and H) Liver; the arrow in (H) points to a megakaryocyte. Liver cells have a pale cytoplasm and are interspersed by small clusters of hematopoetic cells. Bar in (A) and (B), 100 μm; in (C) and (D), 10 μm; in (E–H), 30 μm.
Lack of ameloblast-specific transcripts in Sp3–/– mice
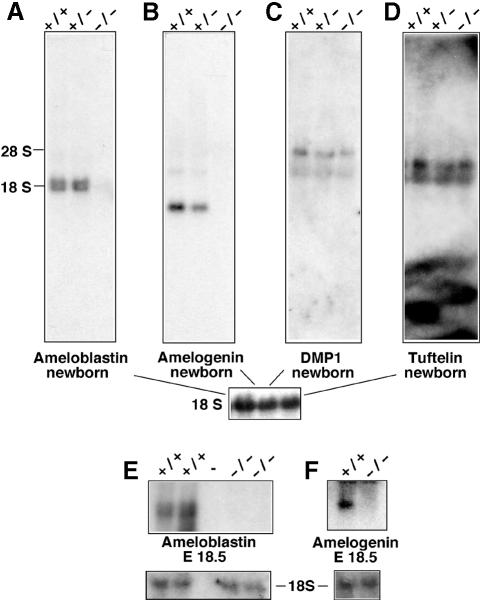
Odontoblasts and ameloblasts express genes that are required to form the dentin and the enamel extracellular matrix, respectively. We asked whether the altered dentin/enamel layers in sections of teeth from Sp3–/– mice might reflect the lack of odontoblast- and/or ameloblastspecific gene products. Amelogenin (Snead et al., 1983; Couwenhoven and Snead, 1994) and ameloblastin (Krebsbach et al., 1996; Lee et al., 1996) are ameloblast-specific gene products involved in enamel formation. The dentin-matrix protein 1 (DMP1) is expressed specifically in odontoblasts (MacDougall et al., 1998a). Tuftelin, yet another specialized protein secreted into the developing enamel matrix, is also synthesized in odontoblasts at this stage of development (Diekwisch et al., 1997). Northern blot analyses were performed with RNA extracted from jaws or heads of newborn mice and of E18.5 embryos (Figure 7). Amelogenin and ameloblastin mRNA was detectable at birth (Figure 7A and B) and at E18.5 (Figure 7E and F) in wild-type and heterozygous mice but not in Sp3–/– mice. In accordance with published data, hybridization with the ameloblastin-specific probe detected two distinct transcripts that were both expressed only in teeth (Krebsbach et al., 1996). The odontoblast-specific transcript encoding DMP1 (MacDougall et al., 1998a) as well as transcripts encoding tuftelin (Diekwisch et al., 1997; Zeichner-David et al., 1997; MacDougall et al., 1998b) were detectable at similar intensities in wild-type, Sp3+/– and Sp3–/– mice (Figure 7C and D). Thus, ameloblast-specific but not odontoblast-specific gene products are missing in Sp3-deficient mice. These results strongly suggest that the absence of ameloblast-specific proteins contributes to the observed histological tooth phenotype in Sp3–/– mice.
Fig. 7. Expression of putative Sp3 target genes in teeth. RNA was extracted from head or jaws of wild-type (+/+), Sp3+/– (+/–) and Sp3-deficient (–/–) newborn or E18.5 embryos, subjected to electrophoresis through 1.2% formaldehyde–agarose gels and transferred to nylon membranes. The filters were hybridized with cDNA fragments encoding ameloblastin (A and E), amelogenin (B and F), DMP1 (C) and tuftelin (D). As a control, the filters were probed with an 18S rRNA-specific oligonucleotide.
Discussion
Sp3-deficient mice are not viable
Our results demonstrate that the ubiquitous transcription factor Sp3 is essential for immediate post-natal survival of mice. Sp3–/– mice were found in expected Mendelian ratios until birth, but they all died within a few minutes post-partum due to respiratory failure. The molecular cause for the observed breathing defect of the Sp3–/– mutants remains obscure. Only slight morphological alterations were observed in the lung that we attributed initially to developmental delay. However, a prolonged pregnancy did not rescue the lethal phenotype. In addition, surfactant protein gene expression in lung of Sp3 mutant mice was apparently indistinguishable from that in wild-type mice. Thus, undiscovered physiological alterations, such as in the enervation of respiratory muscles, are more likely to be responsible for the respiratory failure of Sp3 null mice.
Tooth development is impaired in Sp3-deficient mice
Our analyses of Sp3-deficient embryos provide evidence that Sp3 plays an essential role in late tooth development. Newborn Sp3–/– embryos show only partial formation of the dentin/enamel layer, indicating that dentinogenesis or amelogenesis (dentin and enamel formation) is impaired. A consequence of epithelial cell differentiation into ameloblasts is the expression of proteins that form the enamel extracellular matrix, a scaffold required to nucleate calcium phosphate salts to form calcium hydroxylapatite. Although the ameloblast cell layer is present in Sp3–/– mice, the ameloblast-specific products amelogenin and ameloblastin are absent. In contrast, the odontoblast-specific gene products DMP1 and tuftelin are present in Sp3 null embryos. At this stage, we do not know whether Sp3 functions as a direct regulator of ameloblast-specific genes. Recently, the cloning and preliminary characterization of the murine ameloblastin (Dhamija et al., 1999) and the amelogenin promoter (Chen et al., 1998) have been reported. The ameloblastin promoter contains a potential Sp3-binding site (CACCC box) in a promoter region that contributes to its cell type-specific expression. Thus, the amelogenin and ameloblastin genes might be direct targets of Sp3.
Physiological functions of Sp1 and Sp3
Sp3 is a ubiquitously expressed transcription factor with structural features similar to Sp1. Sp1 and Sp3 recognize the same DNA elements and, in many reports, Sp3 has been shown to act as a potent transcriptional activator similar to Sp1 (Udvadia et al., 1995; Liang et al., 1996; Ihn and Trojanowska, 1997; Zhao and Chang, 1997). The physiological function of Sp1 and Sp3, however, appears to be significantly different. In contrast to Sp3 null mice, Sp1-deficient embryos are already severely retarded at early embryonic stages and die around day 10–11 of gestation (Marin et al., 1997). Thus, the obvious structural similarity, the common DNA recognition sites and the ubiquitous expression of Sp1 and Sp3 do not reflect identical physiological functions. Nevertheless, there still might be many overlapping functions of Sp1 and Sp3 in vivo. The Sp1 and Sp3 knockouts demonstrate that each protein on its own is not responsible for the expression of a very large number of essential genes, such as housekeeping, tissue-specific and cell cycle-regulated genes. This suggests that Sp1 and Sp3 have a wide range of redundant functions that can compensate for each other in Sp1 and Sp3 knockout mice.
Another, tissue-restricted member of the Sp family of transcription factors, Sp4, might also contribute to redundancy in vivo. Sp4, although expressed predominantly in the brain, is also detectable in epithelial tissues, testis and developing teeth (Supp et al., 1996). Disruption of the mouse Sp4 gene revealed that Sp4 is also important for early post-natal survival since approximately two-thirds of the Sp4–/– mice die within a few days after birth for unknown reasons (S.Philipsen and G.Suske, unpublished data). So far, essential target genes for Sp4 have not been identified.
On many reporter constructs containing multiple Sp-binding sites, Sp3, unlike Sp1, is inactive or acts only as a weak activator. The molecular basis for the inactivity of Sp3 under these conditions has been mapped to an inhibitory domain located between the second glutamine-rich activation domain and the zinc finger region (Dennig et al., 1996). The integrity of a charged amino acid triplet (KEE) within this domain is absolutely essential for inhibitor function. Mutation of these amino acids converts Sp3 to a strong activator (Dennig et al., 1996) that is almost indistinguishable from Sp1. A most intriguing possibility would be that this molecular difference between Sp1 and Sp3 reflects the differences in their physiological functions. Can a mutant of Sp3 lacking the inhibitory domain rescue the Sp1 knockout? The generation of transgenic lines expressing different isoforms and mutants of Sp3 and intercrossings with heterozygous Sp1, Sp3 and Sp4 mice could provide new insight into the physiological and biochemical relationships between these three transcription factors.
The early embryonic lethality of Sp1 knockout mice and the post-natal lethality of Sp3 knockout mice preclude the analysis of later developmental stages. In order to achieve this, conditional disruption of the Sp1 and Sp3 genes in specific tissues at any given stage of development, as well as the generation of compound knockouts of Sp1, Sp3 and Sp4 will be an important step to unravel further the physiological roles of these proteins in vivo.
Materials and methods
Generation of the Sp3 homologous recombination construct
As starting plasmid, we chose the Bluescript derivative pBS4.5BamHI that contains a 4.5 kb BamHI fragment encoding both glutamine-rich activation domains on a single exon and ∼1 kbp upstream and 2.5 kbp downstream intron sequences of the Sp3 gene locus (see Figure 1A). We removed the exon and the 5′ intron sequences by restriction with PstI and re-ligation, leading to pBS-P-B. β–galactosidase sequences within the Bluescript vector that may interfere with the lacZ gene (see below) were removed by NaeI and KspI restriction, Klenow filling and re-ligation (plasmid name: pBS-P-B-Δ). Next, we introduced an IRES-LacZ-neo cassette obtained as a 7.3 kbp SalI fragment from the plasmid pGT1.8Iresβgeo (Mountford et al., 1994) into the SalI polylinker site of pBS-PstI–BamHIΔlacZ, leading to pBS-P-B-Δ-lacZ-neo. In a final step, we cloned 5′ intron sequences of the Sp3 gene as a 1.7 kbp HindIII–BamHI fragment into the XhoI polylinker site of pBS-P-B-Δ-lacZ-neo using XhoI linkers, leading to the knockout construct pBS-P-B-Δ-lacZ-neo–5′–Sp3. For transfection into ES cells, the plasmid was linearized with NotI.
Transfection and selection of ES cells
E14 ES cells were electroporated with 15 μg of NotI-linearized targeting vector pBS-P-B-Δ-lacZ-neo–5′–Sp3. Clones were selected with G418 (200 μg/ml) and homologous recombination was analyzed by Southern blotting of EcoRI-restricted genomic DNA with the probe indicated in Figure 1. Unwanted random integrations were detected by hybridizing these blots with a Bluescript vector-specific probe.
Generation of chimeric and Sp3-deficient mice
Sp3+/– ES cell clones were karyotyped, and two clones (#10 and #38) were microinjected in C57BL/6 host blastocysts. Chimeric males were mated to C57BL/6 females. Germline transmission was obtained with both clones. The F1offspring were interbred to expand the stocks. No viable Sp3 null animals were obtained with mice derived from clones #10 and #38. Preliminary in utero analysis of Sp3 null fetuses revealed no obvious differences between the two lines. For the analyses presented herein, we have used line #10. To extend the duration of pregnancy, pregnant females received daily subcutaneous injections of progesterone (5 mg/kg body weight) from day 16 after discovery of the vaginal plug. Pregnancies extended 2 days beyond their normal duration were delivered by Caesarian section.
Genotyping of embryos by PCR
DNA was prepared from tail snips and analyzed for the presence of the wild-type and knockout Sp3 allele by PCR. Three primers were used: a sense primer in the Sp3 gene amplifying the wild-type allele (5′–ACTACTAGTGGGCAAGTCCA–3′), a sense primer in the neo gene amplifying the knockout allele (5′–AGCGCATCGCCTTCTATCG–3′) and an antisense primer in the Sp3 gene (5′–TACCATTGCACA– TTTAATGA–3′). PCR conditions were 94°C, 1 min; 60°C, 1 min; 72°C, 1 min; for 30 cycles.
Nuclear extracts and gel retardation assays
Nuclear extracts were prepared according to Andrew and Faller (1991). Electrophoretic mobility shift assays (EMSAs) were performed by pre-incubating 1–3 μl of nuclear extract with 1.5 μg of unspecific competitor poly(dI–dC) in a buffer containing 10 mM HEPES pH 7.9, 150 mM KCl, 1 mM dithiothreitol (DTT), 0.5 mM MgCl2, 0.1 mM EDTA, 8.5% glycerol for 10 min on ice. Subsequently, 0.1 ng of 32P-labeled double-stranded GC box oligonucleotide was added to a final volume of 20 μl, and samples were incubated for another 20 min on ice. Antisera against Sp1 and Sp3 used for supershift assays were described previously (Hagen et al., 1994). Usually, 1 μl of the appropriate antiserum was added to the binding reaction, and incubation was continued at room temperature for another 20 min. Samples were analyzed on 4% native polyacrylamide gels in 45 mM Tris, 45 mM boric acid, 1.6 mM EDTA. Gels were transferred to Whatman 3MM paper, dried under heat and vacuum, and exposed to X-ray films overnight.
Western blotting
Nuclear extracts (6 μg of protein) were prepared from mouse embryonic E18.5 brains according to Gorski et al. (1986), separated on 10% SDS–polyacrylamide gels, blotted on nitrocellulose or PVDF membranes, and probed with a rabbit αSp3 serum (Hagen et al., 1994). Primary antibodies were visualized using the Amersham ECL kit.
Northern blot analyses
Total RNA from embryonic mouse tissues and cell lines was extracted by the guanidinium/isothiocyanate procedure using the Qiagen kit. RNA was separated through 0.8 and 1.2% agarose gels containing 2.2 M formaldehyde and blotted to nylon membranes. Pre-hybridization and hybridization were carried out as described (Braun and Suske, 1998). Gene-specific probes were obtained from appropriate plasmids or primer sets. Detailed information is available upon request.
Morphological analysis
Embryos were dissected at day 18.5 post-coitum, and a small tail snip was removed for genotyping. Fetuses were cut in half longitudinally and fixed in Carnoy's solution (60% ethanol, 30% chloroform, 10% acetic acid) at 4°C overnight and embedded in paraffin according to standard procedures. Sections were stained with hemalaun and either eosin (HE) or periodic acid–Schiff (PAS) reagent. For electron microscopy, lung tissue samples were cut into small pieces of ∼1 mm3 and immersion fixed with 2.5% paraformaldehyde, 2.5% glutaraldehyde and 0.05% picrinic acid in 0.067 M cacodylate buffer pH 7.4 for 2 h at 4°C. Standard procedures for dehydration and embedding in Epon were employed. Thin sections were stained with uranyl acetate and lead citrate, and examined in an EM 109 electron microscope (Zeiss).
Acknowledgments
Acknowledgements
We thank Iris Rohner and Waltraud Sperling for excellent technical assistance. Drs Martha Kalff-Suske and Jörg Klug are gratefully acknowledged for critically reading the manuscript. This work was supported by grants of the Deutsche Forschungsgemeinschaft to G.S. and the NOW to F.G.
References
- Andrew N.C. and Faller, D.V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res., 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun H. and Suske, G. (1998) Combinatorial action of HNF3 and Sp family transcription factors in the activation of the rabbit uteroglobin/CC10 promoter. J. Biol. Chem., 273, 9821–9828. [DOI] [PubMed] [Google Scholar]
- Chen E., Yuan, Z.A., Collier, P.M., Greene, S.R., Abrams, W.R. and Gibson, C.W. (1998) Comparison of upstream regions of X- and Y-chromosomal amelogenin genes. Gene, 216, 131–137. [DOI] [PubMed] [Google Scholar]
- Couwenhoven R.I. and Snead, M.L. (1994) Early determination and permissive expression of amelogenin transcription during mouse mandibular first molar development. Dev. Biol., 164, 290–299. [DOI] [PubMed] [Google Scholar]
- Dennig J., Hagen, G., Beato, M. and Suske, G. (1995) Members of the Sp transcription factor family control transcription from the uteroglobin promoter. J. Biol. Chem., 270, 12737–12744. [DOI] [PubMed] [Google Scholar]
- Dennig J., Beato, M. and Suske, G. (1996) An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J., 15, 5659–5667. [PMC free article] [PubMed] [Google Scholar]
- Dhamija S., Liu, Y., Yamada, Y., Snead, M.L. and Krebsbach, P.H. (1999) Cloning and characterization of the murine ameloblastin promoter. J. Biol. Chem., 274, 20738–20743. [DOI] [PubMed] [Google Scholar]
- Diekwisch T.G., Ware, J., Fincham, A.G. and Zeichner-David, M. (1997) Immunohistochemical similarities and differences between amelogenin and tuftelin gene products during tooth development. J. Histochem. Cytochem., 45, 859–866. [DOI] [PubMed] [Google Scholar]
- Gorski K., Carneiro, M. and Schibler, U. (1986) Tissue-specific in vitro transcription from the mouse albumin promoter. Cell, 47, 767–776. [DOI] [PubMed] [Google Scholar]
- Hagen G., Müller, S., Beato, M. and Suske, G. (1992) Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res., 20, 5519–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Müller, S., Beato, M. and Suske, G. (1994) Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J., 13, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H. and Trojanowska, M. (1997) Sp3 is a transcriptional activator of the human α2(I) collagen gene. Nucleic Acids Res., 25, 3712–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T., Carner, K.R., Masiarz, F.R. and Tjian, R. (1987) Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell, 51, 1079–1090. [DOI] [PubMed] [Google Scholar]
- Kalff-Suske M., Kunz, J., Grzeschik, K.-H. and Suske, G. (1995) Human Sp4 transcription factor gene (SP4) maps to chromosome 7p15. Genomics, 26, 631–633. [DOI] [PubMed] [Google Scholar]
- Kalff-Suske M., Kunz, J., Grzeschik, K.-H. and Suske, G. (1996) Human Sp3 transcriptional regulator gene (SP3) maps to chromosome 2q31. Genomics, 37, 410–412. [DOI] [PubMed] [Google Scholar]
- Kennett S.B., Udvadia, A.J. and Horowitz, J.M. (1997) Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res., 25, 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach P.H., Lee, S.K., Matsuki, Y., Kozak, C.A., Yamada, K.M. and Yamada, Y. (1996) Full-length sequence, localization and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J. Biol. Chem., 271, 4431–4435. [DOI] [PubMed] [Google Scholar]
- Kumar A.P. and Butler, A.P. (1997) Transcription factor Sp3 antagonizes activation of the ornithine decarboxylase promoter by Sp1. Nucleic Acids Res., 25, 2012–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Krebsbach, P.H., Matsuki, Y., Nanci, A., Yamada, K.M. and Yamada, Y. (1996) Ameloblastin expression in rat incisors and human tooth germs. Int. J. Dev. Biol., 40, 1141–1150. [PubMed] [Google Scholar]
- Liang Y., Robinson, D.F., Dennig, J., Suske, G. and Fahl, W.E. (1996) Transcriptional regulation of the SIS/PDGF-B gene in human osteosarcoma cells by the Sp family of transcription factors. J. Biol. Chem., 271, 11792–11797. [DOI] [PubMed] [Google Scholar]
- MacDougall M., Gu, T.T., Luan, X., Simmons, D. and Chen, J. (1998a) Identification of a novel isoform of mouse dentin matrix protein 1: spatial expression in mineralized tissues. J. Bone Miner. Res., 13, 422–431. [DOI] [PubMed] [Google Scholar]
- MacDougall M., et al. (1998b)Cloning, characterization and tissue expression pattern of mouse tuftelin cDNA. J. Dent. Res., 77, 1970–1978. [DOI] [PubMed] [Google Scholar]
- Majello B., De Luca, P., Hagen, G., Suske, G. and Lania, L. (1994) Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res., 22, 4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M., Karis, A., Visser, P., Grosveld, F. and Philipsen, S. (1997) Transcription factor Sp1 is essential for early development but dispensable for cell growth and differentiation. Cell, 89, 619–628. [DOI] [PubMed] [Google Scholar]
- Mountford P., Zevnik, B., Duwel, A., Nichols, J., Li, M., Dani, C., Robertson, M., Chambers, I. and Smith, A. (1994) Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl Acad. Sci. USA, 91, 4303–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S. and Suske, G. (1999) A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res., 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy S., Van Vooren, P., Szpirer, C. and Szpirer, J. (1998) Assignment of Sp genes to rat chromosome bands 7q36 (Sp1), 10q31→q32.1 (Sp2), 3q24→q31 (Sp3) and 6q33 (Sp4) and of the SP2 gene to human chromosome bands 17q21.3→q22 by in situ hybridization. Cytogenet. Cell Genet., 81, 273–274. [DOI] [PubMed] [Google Scholar]
- Snead M.L., Zeichner-David, M., Chandra, T., Robson, K.J., Woo, S.L. and Slavkin, H.C. (1983) Construction and identification of mouse amelogenin cDNA clones. Proc. Natl Acad. Sci. USA, 80, 7254–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripp B.R., Lund, J., Mango, G.W., Doyen, K.C., Johnston, C., Hultenby, K., Nord, M. and Whitsett, J.A. (1996) Clara cell secretory protein: a determinant of PCB bioaccumulation in mammals. Am. J. Physiol., 271, L656–L664. [DOI] [PubMed] [Google Scholar]
- Supp D.M., Witte, D.P., Branford, W.W., Smith, E.P. and Potter, S.S. (1996) Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability and male fertility. Dev. Biol., 176, 284–299. [DOI] [PubMed] [Google Scholar]
- Suske G. (1999) The Sp-family of transcription factors. Gene, 238, 291–300. [DOI] [PubMed] [Google Scholar]
- Udvadia A.J., Templeton, D.J. and Horowitz, J.M. (1995) Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl Acad. Sci. USA, 92, 3953–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima S., Lee, S.H., Minowa, T. and Mouradian, M.M. (1998) Sp family transcription factors regulate expression of rat D2 dopamine receptor gene. DNA Cell Biol., 17, 471–479. [DOI] [PubMed] [Google Scholar]
- Zeichner-David M., et al. (1997)Timing of the expression of enamel gene products during mouse tooth development. Int. J. Dev. Biol., 41, 27–38. [PubMed] [Google Scholar]
- Zhao L. and Chang, L.S. (1997) The human POLD1 gene. Identification of an upstream activator sequence, activation by Sp1 and Sp3 and cell cycle regulation. J. Biol. Chem., 272, 4869–4882. [PubMed] [Google Scholar]