Abstract
Pseudomonas putida is a gram-negative rod-shaped gammaproteobacterium that is found throughout various environments. Members of the species P. putida show a diverse spectrum of metabolic activities, which is indicative of their adaptation to various niches, which includes the ability to live in soils and sediments contaminated with high concentrations of heavy metals and organic contaminants. Pseudomonas putida strains are also found as plant growth-promoting rhizospheric and endophytic bacteria. The genome sequences of several P. putida species have become available and provide a unique tool to study the specific niche adaptation of the various P. putida strains. In this review, we compare the genomes of four P. putida strains: the rhizospheric strain KT2440, the endophytic strain W619, the aromatic hydrocarbon-degrading strain F1 and the manganese-oxidizing strain GB-1. Comparative genomics provided a powerful tool to gain new insights into the adaptation of P. putida to specific lifestyles and environmental niches, and clearly demonstrated that horizontal gene transfer played a key role in this adaptation process, as many of the niche-specific functions were found to be encoded on clearly defined genomic islands.
Keywords: Pseudomonas putida, comparative genomics, W619, KT2440, F1, GB-1
Introduction
The group of the pseudomonads comprises a heterogeneous set of microorganisms that can be isolated from many different niches (Clarke, 1982). They belong to the class Gammaproteobacteria and nearly 100 different strains have been described. Within this group of microorganisms, a number of strains have been associated to the species Pseudomonas putida, and in this review, we analyzed some of the most characteristic features derived from the analysis of their genome sequences. Bacteria of this species are known for their ability to colonize soils and for their capacity to degrade a wide variety of chemicals, including many natural and man-made compounds.
The rhizospheric bacterium P. putida KT2440 was isolated from garden soil in Japan based on its ability to use 3-methylbenzoate (Nakazawa, 2002). Since the publication of its complete 6.18 Mbp genome sequence in 2002, which represented the first sequenced Pseudomonas genome and that allowed a comparative analysis of the metabolically versatile P. putida KT2440 as a function of this strain's ability to survive in marginal and polluted soils (Nelson et al., 2002), our knowledge of this strain has increased significantly. As a result, P. putida KT2440 represents the best-characterized strain from this species, and is considered as the workhorse for Pseudomonas research (Timmis, 2002; Dos Santos et al., 2004;). Pseudomonas putida KT2440 is a microorganism generally recognized as safe (GRAS certified) for the cloning and expression of foreign genes, and although there is a high level of genome conservation with the pathogenic species Pseudomonas aeruginosa (85% of the predicted coding regions are shared), key virulence factors including exotoxin A and type III secretion systems were found to be lacking from its genome (Nelson et al., 2002). Its unusual wealth of determinants for high-affinity nutrient acquisition systems, mono- and di-oxygenases, oxido-reductases, ferredoxins and cytochromes, dehydrogenases, sulfur metabolism proteins, for efflux pumps and glutathione-S transfereases and for the extensive array of extracytoplasmatic function sigma factors, regulators and stress response systems constitutes the genomic basis for the exceptional nutritional versatility and opportunism of P. putida KT2440. This metabolic diversity was successfully exploited for the experimental design of novel catabolic pathways as well as for its application in the production of high-added value compounds whose synthesis is not coupled to cell survival (Dos Santos et al., 2004). These include the hyperproduction of polymers (such as polyhydroxyalkanoates) (Huijberts et al., 1992; Huijberts & Eggink, 1996;), industrially relevant enzymes, the production of epoxides, substituted catechols, enantiopure alcohols and heterocyclic compounds (Schmid et al., 2001). Biotechnology applications of P. putida KT2440 were further facilitated by the genome-scale reconstruction and analysis of metabolic network models (Nogales et al., 2008; Puchalka et al., 2008;). Both models were used successfully to improve the production of polyhydroxyalkanoates from various carbon sources as examples of how central metabolic precursors of a compound of interest not directly coupled to the organism's growth function might be increased via the modification of global flux patterns. Because the species P. putida encompasses many strains with a wide range of metabolic features and numerous isolates with unique phenotypes, these models provide an important basic scaffold upon which future models of other P. putida strains, specifically addressing important properties for their niche-specific adaptation such as the growth of P. putida F1 on aromatic hydrocarbons, can be tailored as a function of strain-specific metabolic pathways.
Pseudomonas putida W619 is a plant growth-promoting endophytic bacterium, which was isolated from Populus trichocarpa×deltoides cv. ‘Hoogvorst,’ and represents one of the most commonly found species among the poplar endophytes isolated so far (Taghavi et al., 2005, 2009). Very similar bacteria were isolated from the poplar rhizosphere, and as endophytes from root and stem material. Like P. putida KT2440, strain W619 is an excellent host for the expression of foreign genes. This property was exploited to successfully introduce and express the ability to degrade toluene and trichloroethylene (TCE). In situ bioaugmentation with P. putida W619-TCE reduced TCE evapotranspiration by 90% under field conditions. This encouraging result, which represents one of the few examples of successful bioaugmentation, was achieved after the establishment and enrichment of P. putida W619-TCE as a poplar root endophyte and by further horizontal gene transfer of TCE metabolic activity to members of the poplars endogenous endophytic population (Weyens et al., 2009). Because P. putida W619-TCE was engineered via horizontal gene transfer, its deliberate release is not restricted under European genetically modified organisms regulations.
Pseudomonas putida F1 was isolated from a polluted creek in Urbana, IL, by enrichment culture with ethylbenzene as the sole source of carbon and energy. F1 is considered a reference strain for the degradation and growth of P. putida on aromatic hydrocarbons, including benzene, toluene, ethylbenzene and p-cymene. Mutants of strain F1 that are capable of growing on n-propylbenzene, n-butylbenzene, isopropylbenzene and biphenyl as the sole carbon sources are easily obtained (Choi et al., 2003). In addition to aromatic hydrocarbons, the broad substrate toluene dioxygenase of strain F1 can oxidize TCE, indole, nitrotoluenes, chlorobenzenes, chlorophenols and many other substituted aromatic compounds (Spain & Gibson, 1988; Wackett & Gibson, 1988; Spain et al., 1989;). Although P. putida F1 cannot use TCE as a source of carbon and energy, it is capable of degrading and detoxifying TCE in the presence of an additional carbon source. The ability of P. putida F1 to degrade benzene, toluene and ethylbenzene has a direct bearing on the development of strategies for dealing with environmental pollution.
Pseudomonas putida GB-1 was isolated from fresh water and characterized as a robust manganese (Mn2+) oxidizer (Rosson & Nealson, 1982). When supplied with Mn2+, the cells deposit manganese oxide outside the outer membrane during the early stationary growth phase, a process that is enzymatically catalyzed (Okazaki et al., 1997). This process was further studied at both the genetic and the physiological level, making GB-1, along with the closely related strain MnB1, the model organism for studies of Mn(II) oxidization (Okazaki et al., 1997; Caspi et al., 1998; de Vrind et al., 1998; Brouwers et al., 1999; Villalobos et al., 2003;).
This review focuses on the diversity and adaptation of four P. putida strains (KT2440, W619, F1 and GB-1) in terms of their respective environments from a genome point of view, and provides further insights into their potential applications for the bioremediation of contaminated environments or as plant growth-promoting bacteria.
Comparative genomics of P. putida
Structure and general features of the P. putida genomes
A whole-genome comparison using megan (Huson et al., 2007) between P. putida W619 and the publicly available genome sequences of members of the genus Pseudomonas reveals the presence of 3708 shared coding sequences (CDS) (Fig. 1). An additional 684 CDS are shared between W619 and the genomes of the other three P. putida strains, plus 82, 47 and 108 CDS are uniquely shared between W619 and strains KT2440, GB-1 and F1, respectively. Among the Pseudomonas sp. outside the species P. putida, Pseudomonas entomophila L48 is the closest relative, sharing 110 additional genes with W619. Because this review focuses on the genomic analyses of representative strains of P. putida, P. entomophila L48 has not been included in this comparison. It should also be noticed that W619 contains 170 CDS that have no hit (at E value of <10−5) to any CDS present among the sequenced Pseudomonas genomes, which could indicate that these genes potentially originate from organisms outside of the genus Pseudomonas.
Fig. 1.
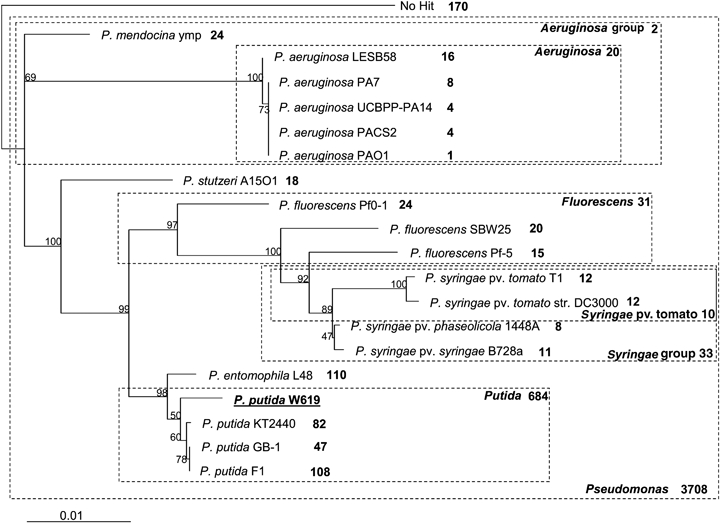
Phylogenetic relationship between members of the genus Pseudomonas. 16S rRNA gene comparison was used to build the phylogenetic tree for those members of the genus Pseudomonas with publicly available genome sequences. The numbers at nodes represent the bootstrap values (1000 replicates), and the numbers in bold correspond to the number of coding sequences (CDS) preferentially shared by W619 and the corresponding organisms (with E value of 10−5). The numbers of CDS equally found in two or more organisms are indicated for this subset of organisms (boxed with dotted lines) that correspond to a group of taxa/phylum according to the NCBI taxonomy. Whole-genome comparison for shared CDS was based on megan (Huson et al., 2007).
The P. putida W619 genome was manually annotated using the MaGe annotation system (Vallenet et al., 2006) (http://www.genoscope.cns.fr/agc/website/), and compared with the well-analyzed KT2440 and automatically annotated GB-1 and F1 genomes. The general genome features of the four P. putida strains are summarized in Table 1. Strains W619, F1 and GB-1 are predicted to encode 5471 CDSs with a coding density of 89%, 5300 CDSs with a coding density of 90% and 5417 CDSs with a coding density of 90%, respectively. These findings are comparable to the 5420 CDSs predicted for KT2440, with a coding percentage of 86%. The four strains share many general genome features. For example, all possess a single circular chromosome that displays a clear GC skew transition. Their chromosomal replication origin has the typical organization for P. putida, which is distinct from the enteric-type origins (Yee & Smith, 1990; Weigel et al., 1997;). The oriC site locates between the rpmH and the dnaA genes and contains conserved DnaA-binding boxes (TTATCCACA). Figure 2 shows the Genome Atlas of P. putida W619 (Hallin et al., 2009; Ussery et al., 2009;) with customized blast lanes added for F1, KT2440 and GB-1. The outer three lanes show genes of W619 shared with the other three P. putida strains. The numbers around the chromosomal atlas indicate the locations of the predicted genomic islands in W619 (see details in Supporting Information, Table S1). These regions are generally characterized by lack of synteny, along with a high intrinsic curvature and stacking energy, which is indicative of their higher recombination activity as compared with other chromosomal regions that seem to constitute the P. putida core genome. The putative orthologous relations and synteny group arrangements of the four genomes are illustrated in the comparative syntheny line plots (Fig. 3) and by the syntheny statistics (Table S2). Based on the comparative synteny line plots (Fig. 3), P. putida W619 has considerable inverted alignments on both sides of the chromosomal replication origin as compared with the other three P. putida strains. This indicates that the W619 genome might have undergone a major DNA rearrangement that involved swapping two DNA segments that are flanking the chromosomal replication origin. Members of the genus Pseudomonas are characterized by their ability to grow on defined minimal media using a huge variety of organic compounds as energy and carbon sources. The biosynthetic pathways for proteinogenic amino acids and vitamins have been solved recently for P. putida KT2440, and this information is well conserved in the other analyzed strains of this species (Molina-Henares et al., 2009, 2010). Using the COGs functional categories in the NCBI database, the P. putida strains were compared with the Gammaproteobacteria and total bacteria, and were found to share a similar distribution for most COG classes (Table S3) (Tatusov et al., 2000). However, all four P. putida strains displayed a higher percentage of genes putatively coding for signal transduction mechanisms (T), pointing to the presence of sophisticated regulatory networks to control gene expression in function of a highly variable environment, and inorganic ion transport and metabolism (P). On the other hand, the P. putida genomes were less dense in genes putatively involved in carbohydrate transport and metabolism (G) and replication, recombination and repair (L).
Table 1.
Comparison of the general genome features of Pseudomonas putida strains W619, F1, GB-1 and KT2440
| W619 | F1 | GB-1 | KT2440 | |
|---|---|---|---|---|
| Number of bases | 5 774 330 | 5 959 964 | 6 078 430 | 6 181 860 |
| Number of CDSs | 471 | 5300 | 5417 | 5420 |
| CDSs with predicted function (%) | 70.0 | 73.5 | 74.9 | 76.9 |
| CDSs without function with a homolog (%) | 25.6 | 23.6 | 22.7 | 19.1 |
| CDSs without function without a homolog (%) | 4.4 | 0.65 | 0.7 | 4.0 |
| Pseudogenes | 26 | 49 | 8 | ND |
| Coding % | 89 | 90 | 90 | 86 |
| %GC | 61 | 61 | 62 | 62 |
| tRNAs | 75 | 76 | 74 | 73 |
| rRNA genes (clusters) | 22 (7) | 20 (6) | 22 (7) | 22 (7) |
| Putative orthologous relations (%) | ||||
| W619 | 100 | 77 | 75 | 75 |
| F1 | – | 100 | 80 | 82 |
| GB-1 | – | – | 100 | 77 |
| KT2440 | – | – | – | 100 |
The numbers of genome features have been retrieved from the NCBI site (http://www.ncbi.nlm.nih.gov/sites/genome/) and the JGI site (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). ND, not determined; the absence of pseudogenes in KT2440 is based on information provided by NCBI, which might be out of date. Orthologous relations are predicted using the RefSeq synteny statistic in MaGe platform (Vallenet et al., 2006).
Fig. 2.
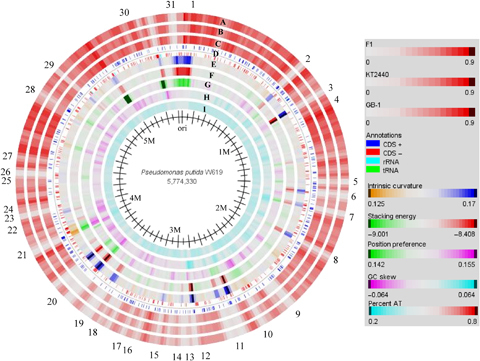
Genome atlas for the chromosome of Pseudomonas putida W619. The numbers outside the atlas indicate the locations of the predicted genomic islands on the W619 chromosome. Details of these genomic islands are provided in Table S1. From the outside to the inside the circles represent the three blast atlases for chromosome comparisons with P. putida strains F1 (a), KT2440 (b) and GB-1 (c), respectively, followed by an overview of specific W619 chromosome properties: (d) annotations for coding sequences on the +and – strand, rRNA and tRNA genes; (e) intrinsic curvature; (f) stacking energy; (g) position preference; (h) GC skew; and (i) percent AT. The explanation of the color codes is presented in the figure legend. The atlas was generated using the GeneWiz browser 0.91 (http://www.cbs.dtu.dk/services/gwBrowser/).
Fig. 3.
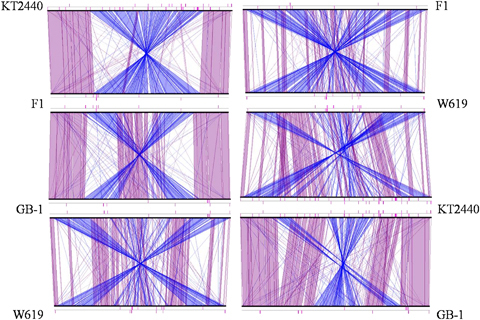
Comparative syntheny line plots showing the orthologous relations among Pseudomonas putida W619, F1, GB-1 and KT2440. The synteny regions are displayed with strand conservation (in purple) and stand inversion (in blue). The pink bars represent the positions of transposable elements. The ‘Conserved Synteny LinePlot’ tool in MaGe was used to generate this figure (Vallenet et al., 2006). The corresponding RefSeq synteny statistics are provided in Table S2.
Mobile elements in P. putida
Conserved IS elements between the P. putida strains were identified using the IS Finder (http://www-is.biotoul.fr/) database and blast searches with a stringent E value threshold (below e−100). Complete putative IS elements identified in W619, KT2440, F1 and GB-1 are listed in Table 2A. As can be seen in the table, the KT2440 genome contains 36 IS elements, a very high number especially in comparison with strain F1 (only 2 IS elements of the IS3 and IS5 families, respectively), with the majority of the IS elements in KT2440 being present in multicopies. In comparison, strains W619 and GB-1 contain eight and nine IS elements, respectively. The high number of IS elements may be related to the versatile catabolic ability of KT2440, which also requires an increased level of genome plasticity in order to adapt to various environments, this in comparison with the other strains that thrive in more specialized niches. When applying a higher threshold E score, several incomplete or truncated remnants of IS elements were identified among the various genomes (Table S4).
Table 2.
Mobile elements' comparison for the Pseudomonas putida strains W619, KT2440, F1 and GB-1
| Family/Group | Name | Origin | Length | IR | DR | W619ORF ID (PputW619) | KT2440ORF ID (PP_) | F1ORF ID (Pput_) | GB-1ORF ID (PputGB1_) |
|---|---|---|---|---|---|---|---|---|---|
| (A) IS elements | |||||||||
| IS3/IS51 | ISPpu22 | P. putida GB-1 | 1232 | 29/45 | 3 | – | – | 0419-0415 | 1613-16142972-29713805-38065424-5425 |
| IS4/IS4 | ISPpu8 | P. putida KT2440 | 1419 | 18 | 11 | – | 1865 19902114 22182522 4318 | – | – |
| IS5/IS5 | ISPa16 | P. aeruginosa | 1192 | 12 | 4 | 3354 | – | – | – |
| IS5/IS5 | IS1246 | P. putida mt-2 (pWWO) | 1275 | 15/16 | ND | – | 2971 | – | – |
| IS5/IS5 | IS2000 | P. aeruginosa JES | 1186 | 14/17 | 4 | – | – | 1356 | – |
| IS5/IS427 | ISPs1 | P. syringae | >203 (800-1000) | ND | ND | 2346-2347 | – | – | 4818-48174824-4825 |
| IS30 | IS1382 | P. putida H | 1093 | 35 | 3 | 1990 | – | – | - |
| IS66 | ISPpu14 | P. putida KT2440 | 2383 | 27/33 | 8 | 0037-0039 5173-5175 | 3501-34993964-39663979-39814443-44414439-44375398-5396 | – | – |
| IS66 | ISPpu15 | P. putida KT2440 | 2041 | 22/28 | 8 | – | 0638-06374024-40254092-40914746-4745 | – | 0521-05221718-17194791-4792 |
| IS66 | ISPpu13 | P. putida KT2440 | 2370 | 19/22 | 8 | – | 3986-39843113-3115 | – | – |
| IS110 | ISPpu9 | P. putida KT2440 | 2043 | 6/11 | 2 | – | 1133 1260 2570 3381 3586 4603 4791 | – | – |
| IS110 | ISPpu10 | P. putida KT2440 | 1314 | 0 | 0 | – | 0526 1653 2134 3502 4599 50505290 | – | – |
| IS110/IS1111 | ISPpu11 | P. putida KT2440 | 1348 | 12 | 0 | – | 0334 3498 | – | – |
| IS256 | ISPa27 | P. aeruginosa | 1361 | 23/29 | 7 | 3315 | – | – | – |
| IS1182 | ISPpu16 | P. putida KT2440 | 1677 | 15/16 | 2 | 3317 | 1931 | – | – |
| ISL3 | ISPpu12 | P. putida pWW0 | 3372 | 21/24 | ND | 2325-2328 | – | – | – |
| IS elements were identified using IS Finder: http://www-is.biotoul.fr/ | |||||||||
| IR, the length(s) of the terminal IR(s) in base pairs. A single number refers to two IRs with the same length; DR, the number of target base pairs duplicated on insertion; ND, not determined. | |||||||||
| W619* | KT2440 | F1 | GB-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (B) Phage | |||||||||
| PputW6191301-1357 | PP1536-1584 | Pput_3359-3401 | PputGB1_1181-1221a | ||||||
| PputW6193919-3971b | PP2266-2297 | Pput_4096-4150a | PputGB1_1720-1762 | ||||||
| PputW6194030-4047a | PP3026-3066 | PputGB1_3388-3473 | |||||||
| PP3859-3920 | PputGB1_4147-4177b | ||||||||
| *Phages with the same label inserted in the same position on the genomes of respective strains. The presence of phages was predicted using the prophinder (Lima-Mendez et al., 2008) tool. | |||||||||
| Putative product (gene)* | W6 19 ORF ID (PputW619) | F1 ORF ID (Pput_) | KT2440 ORF ID (PP_) | GB-1ORF ID (PputGB1_) | |||||
|---|---|---|---|---|---|---|---|---|---|
| (C) Other transposable elements | |||||||||
| Tn7 family protein (tnsA) | 5195a | – | 5406g | – | |||||
| Tn7 family protein (tnsB) | 5194a | – | 5405g | – | |||||
| Tn7 family protein (tnsC) | 5193a0036 | – | 5404g | – | |||||
| Tn7 family protein (tnsD) | 5192a | – | 5407g | – | |||||
| Tn4652 cointegrate resolution protein T (tnpT) | 2672b2275c | 2522f | 32392982h | 2650i | |||||
| Tn4652 cointegrate resolution protein S (tnpS) | 2680b2276c | 2535f | 31812981h | 2669i | |||||
| Tn4652 transposase subunit AB | – | – | 2976–2977 | – | |||||
| Tn4652 transposase (tnpA) | – | – | 2964 | – | |||||
| Tn4652 tnpA regulatory protein (tnpC) | – | – | 2965 | – | |||||
| Tn3 transposase(tnpA) | 2321d | – | – | – | |||||
| Tn3 resolvase (tnpR) | 2322d4533 | – | – | – | |||||
| Resolvase (tniR) | 2332e | – | – | – | |||||
| Truncated transposase (tniA) | 2334e | – | – | – | |||||
| Tyrosine recombinase (xerC) | 0242 | 5139 | 5230 | 5291 | |||||
| Tyrosine recombinase (xerD) | 4154 | 4253 | 1468 | 1073 | |||||
*ORFs with the same label are located in the same region on the chromosome and are supposedly part of the same composite transposable element. IS elements were identified using IS Finder: http://www-is.biotoul.fr/
It should also be noted that several of the IS elements were unique to a P. putida genome, including seven copies of both ISPpu9 and ISPpu10, six copies of ISPpu8, two copies of both ISPpu13 and ISPpu11 and a copy of IS1246 found in KT2440, copies of ISPa16, ISPa27 and IS1382 in W619 and a copy of IS2000 in F1. None of the identified IS elements was present on more than two of the genomes.
IS elements are often associated with resistance and accessory functions that bacteria have acquired via horizontal gene transfer, or with DNA rearrangements in order to affect the expression or the stability of the newly acquired functions (Mahillon & Chandler, 1998). For example in P. putida W619, the unique mhp aromatic degradation operon was found between one incomplete copy of IS1182 (PputW619_1976-1977) and the copy of IS1382 (PputW619_1990), indicating that this operon was acquired via horizontal gene transfer. Also, a mercury resistance operon was found adjacent to the unique copy of ISPpu12 (PputW619_2325-2328) (Williams et al., 2002), while in its proximity, other transposable elements were identified, including a putative Tn3 family transposon (PputW619_2321-2322), a recombinase and a truncated transposase. Therefore, this gene segment might represent a composite mercury resistance transposon in W619, acquired via horizontal gene transfer and absent in P. putida F1, KT2440 and GB-1. Additional transposable elements are summarized in Table 2C. Among them, tnsABCD from the Tn7 family was found in association with a gene cluster encoding heavy metal resistance in W619 and KT2440.
The prophinder (Lima-Mendez et al., 2008) tool was used for prophage prediction in the P. putida genomes. Prophages were identified in all four genomes (Table 2B), with their numbers ranging between 2 and 4. Several prophages were found to be inserted at the same location among different strains. For example, of the three prophages found in strain W619, one was found at the same location in strain GB-1, while another prophage (PputW619_4030-4047) was found inserted adjacent to a ferredoxin-related gene in both GB-1 (PputGB1_1181-1221) and F1 (Pput_4096-4150). This points to the existence of an insertion hotspot for this particular prophage in the P. putida core genome (Manna et al., 2004).
Genomic islands in P. putida
We predicted specific genomic regions for each of the four P. putida strains in comparison with the other three strains using the MaGe annotation system (Vallenet et al., 2006). Based on the automatic prediction algorithm, 61, 54 and 66 putative regions were identified in P. putida KT2440, F1 and GB-1, respectively; along with the manual annotation of W619, 31 putative genomic islands were identified for P. putida W619 (Table S1). Different from 105 predicted genomic islands of KT2440 as described by Weinel et al. (2002), a number that was based on the compositional bias of the GC, di- and tetranucleotides of the chromosome itself, the 61 putative genomic islands in KT2440 that we identified in our analysis were predicted based on the lack of synteny against the genomes of the other three P. putida strains, GC bias and the existence of mobile genetic elements. For example, genomic island 19 (coordinates 922000–945000) predicted by Weinel et al. (2002) was not included in our analysis because this region is conserved in the other three P. putida strains. On the other hand, region 31 (coordinates 3121963–3224467) of KT2440, which was not considered as a genomic island by Weinel et al. (2002), lacks synteny to the genomes of W619 and GB-1, and carries genes that are involved in sugar transport (PP_2757-PP_2761).
Among the various putative genomic islands, many are suggested to have been acquired via horizontal gene transfer due to the presence of integrases or mobile genetic elements (transposons, IS elements) and an alternative matrix of codon usage compared with the rest of the chromosome. As shown in the following sections, these putative genomic islands have conferred the P. putida species with many accessory capabilities, including heavy metal resistance, aromatic compound degradation and stress responses, all of which are highly relevant for the specific niche adaption of P. putida.
Adaption of P. putida to a polluted soil environment
Heavy metal resistances in P. putida
Based on its genome sequence, P. putida KT2440 was predicted to tolerate various heavy metals (Canovas et al., 2003), and this strain's metal resistance properties were experimentally confirmed. Pseudomonas putida W619 strain was originally isolated from the roots of poplar cuttings that were obtained from trees growing on a field site aimed to remediate groundwater contaminated with various compounds, including BTEX compounds (benzene, toluene, ethylbenzene and xylene), TCE and Ni (Taghavi et al., 2005). It was therefore expected that strain W619 also possesses the capacity to deal with heavy metals. Based on the strain's genome sequence, 78 genes were identified that encode proteins putatively involved in heavy metal resistance and homeostasis, a number that exceeds the number of genes identified on the KT2440 genome (Canovas et al., 2003). Furthermore, when comparing the MIC values of strains W619 and KT2440 for various heavy metals, we found that W619 displayed an increased resistance to Cd(II) (MIC values of 0.5 and 0.25 mM for W619 and KT2440, respectively), Cu and Ni (see details below) and similar resistance to Zn(II) (MIC 0.5 mM) and Co(II) (MIC 0.1 mM). We therefore used the W619 genome as the reference to compare the different P. putida genomes for functions putatively involved in heavy metal resistance.
The majority of the P. putida W619 heavy metal resistance genes are found on two genomic regions, referred to as region 1 and 31 that are located on opposite sites of the chromosomal origin of replication. A similar organization is observed for F1 and GB-1 and in some way for KT2440. Most of the heavy metal resistance genes located in region 1 (coordinates 7447–75941) are conserved among all four P. putida strains (Table 3 and Fig. 4a). On the 5′-end, this region is delimitated by an integrase gene (PputW619_0006). This integrase is also found in the equivalent regions of F1 and GB-1, but is absent in KT2440. Interestingly, region 1 contains three different systems for putative copper resistance: copRSABMG (PputW619_0018-11) for periplasmic detoxification, with CopA being a multicopper oxidase (MCO) that is thought to oxidize Cu(I) to Cu(II) in the periplasm (Rensing & Grass, 2003), and CopB being an outer membrane protein (Cha & Cooksey, 1991); a P-type ATPase gene copF (PputW619_0029) for the cytoplasmic detoxification of monovalent Cu(I); and the copper/silver resistance operon cusFcusABC (PputW619_0023-0020) for the periplasmic detoxification of Cu(I) and Ag(I) via expulsion by the three-component efflux system (cusABC) across the outer membrane. The accessory protein CusF is believed to be a periplasmic copper chaperon delivering Cu(I) to the CusABC complex (Franke et al., 2003). Region 1 also seems to encode for resistance to cadmium, zinc and cobalt based on the presence of the czcABC (a three-component efflux system for the periplasmic detoxification of Cd, Zn and Co) and the czcDRS gene [including the cation diffusion facilitator (CDF) protein CzcD] clusters with some rearrangement (PputW619_0060-62, 0043, 0046-0047). In addition, a cadA gene (PputW619_0058), encoding a CadA P-type ATPase, is present as well as a gtrABM (PputW619_0052-0050) locus putatively involved in heavy metal resistance by repairing the membrane, whose integrity may have been altered by the precipitation of heavy metal on the bacterial surface. The clustering of the czc efflux system, the membrane maintenance genes gtrABM and a putative porin (PputW619_0063) (Fig. 4a) are features that resemble those described for the Cd, Zn and Co resistance cluster (ORF82-106) of the Cupriavidus metallidurans CH34 plasmid pMOL30, but with some rearrangements (Monchy et al., 2007). It should also be noted that a czcN homolog, described to play a role in the regulation of the czc operon (Dong & Mergeay, 1994; Grosse et al., 1999;), is located 18 kb upstream of the czc cluster in between the putative cusF and copF genes.
Table 3.
Summary and comparison of genes found in Pseudomonas putida that might be involved in metal resistance and homeostasis based on genomic analysis
| Gene | Metal | W619 (region)*ORF ID PputW619_ | F1 ORF ID Pput_ | KT2440 ORF ID PP_ | GB-1 ORF ID PputGB1_ | Product |
|---|---|---|---|---|---|---|
| copG | Cu | 0011 (1) | 0011 | 5377 | 0013 | Involved in survival in the presence of high bioavailable Cu(II) |
| copM | Cu | 0012 (1) | 0012 | 5378 | 0014 | Cytochrome c family protein |
| copB′ | Cu | 0013 (1) | 0013 | 5379 | 0015 | Copper resistance protein B |
| copB″ | Cu | 0014 (1) | 0014 | – | 0016 | Putative CopB (frameshifted) |
| copA | Cu | 0015 (1) | 0015 | 5380 | 0017 | Copper resistance protein A |
| copR | Cu | 0017 (1) | 0017 | 5383 | 0019 | Transcriptional activator CopR |
| copS | Cu | 0018 (1) | 0018 | 5384 | 0020 | Sensor protein CopS |
| cusC | Cu/Ag | 0020 (1) | 0020 | 5385 | 0022 | Cu/Ag tricomponent efflux outer membrane porin |
| cusB | Cu/Ag | 0021 (1) | 0021 | 5386 | 0023 | Cu/Ag tricomponent efflux membrane fusion protein |
| cusA | Cu/Ag | 0022 (1) | 0022 | 5387 | 0024 | Copper transporter, RND family |
| cusF | Cu/Ag | 0023 (1) | 0023 | 5388 | 0025 | Periplasmic copper-binding protein |
| - | ? | 0024 (1) | 0024 | 5389 | 0026 | S-isoprenylcysteine methyltransferase (czcN homolog) |
| copF | Cu | 0029 (1) | 0030 | – | 0031 | Copper P-type ATPase |
| czcD | Cd/Zn/Co | 0043 (1) | 0040 | 0026 | 0040 | Cation Diffusion Facilitator (CDF) |
| czcR | Cd/Zn/Co | 0046 (1) | 0043 | 0029 | 0043 | DNA-binding response regulator |
| czcS | Cd/Zn/Co | 0047 (1) | 0044 | 0030 | 0044 | Sensory histidine kinase |
| gtrM | – | 0050 (1) | 0047 | 0033 | 0047 | Glycosyltransferase (protein o-glycosylation) |
| gtrB | – | 0051 (1) | 0048 | 0034 | 0048 | Glycosyltransferase (bactoprenol) |
| gtrA | – | 0052 (1) | 0049 | 0035 | 0049 | Bactoprenol-linked glucose translocase (Flippase) |
| cadA | Cd/Zn | 0058 (1) | 0055 | 0041 | 0055 | Cadmium translocating P-type ATPase |
| czcA | Cd/Zn/Co | 0060 (1) | 0057 | 0043 | 0057 | Cobalt/zinc/cadmium efflux RND transporter |
| czcB | Cd/Zn/Co | 0061 (1) | 0058 | 0044 | 0058 | Cobalt/zinc/cadmium efflux RND transporter |
| czcC | Cd/Zn/Co | 0062 (1) | 0059 | 0045 | 0059 | Cobalt/zinc/cadmium resistance protein |
| - | Cd/Zn/Co | 0063 (1) | 0060 | 0046 | 0060 | Putative porin, OprD family |
| czcR | Cd/Zn/Co | 0064 (1) | 0061 | 0047 | 0061 | DNA-binding heavy metal response regulator |
| cadR | Cd/Zn | 0325 | 5013 | 5140 | 5193 | Transcriptional regulator |
| cadA | Cd/Zn | 0326 | 5012 | 5139 | 5192 | Cadmium translocating P-type ATPase |
| arsC | As | 1207 | 4072 | 1645 | 1247 | Arsenate reductase |
| cinA | Cu | 1676 | 3583 | 2159 | 1700 | Copper-containing azurin-like protein |
| cinQ | Cu | 1677 | 3584 | 2160 | 1701 | Pre-Q0 reductase |
| copB | Cu | 1712 (8) | – | – | – | Copper resistance protein B |
| copA | Cu | 1713 (8) | – | – | – | Copper resistance protein A |
| - | Zn | 1714 (8) | – | – | – | Cation efflux protein (Putative Zinc transporter ZitB) |
| arsB | As | 1715 (8) | – | – | – | Arsenite efflux pump |
| merR | Hg | 2323 (11) | – | – | – | Mercuric resistance operon regulatory protein |
| merB | Hg | 2324 (11) | – | – | – | Alkylmercury lyase (organomercurial lyase) |
| merR | Hg | 2325 (11) | – | – | – | Transcriptional regulator, MerR-family |
| - | ? | 2326 (11) | – | – | – | Heavy metal/H+ antiporter, CDF family |
| merE | Hg | 2336 (11) | – | – | – | Mercuric resistance protein |
| merD | Hg | 2337 (11) | – | – | – | HTH-type transcriptional regulator |
| merA | Hg | 2338 (11) | – | – | – | Mercuric Hg(II) reductase |
| merP | Hg | 2339 (11) | – | – | – | Mercuric transport protein periplasmic component |
| merT | Hg | 2340 (11) | – | – | – | Mercuric transport protein |
| merR | Hg | 2341 (11) | – | – | – | Mercuric resistance operon regulatory protein |
| nikR | Ni | 3004 (18) | – | 3341 | – | Putative nickel-responsive regulator |
| nikA | Ni | 3005 (18) | – | 3342 | – | Nickel ABC transporter, periplasmic nickel-binding protein |
| nikB | Ni | 3006 (18) | – | 3343 | – | Nickel transporter permease NikB |
| nikC | Ni | 3007 (18) | – | 3344 | – | Nickel transporter permease NikC |
| nikD | Ni | 3008 (18) | – | 3345 | – | Nickel import ATP-binding protein NikD |
| nikE | Ni | 3009 (18) | – | 3346 | – | Nickel import ATP-binding protein NikE |
| chrA | Cr | 3017 (18) | – | – | – | Chromate transporter |
| modA | Mo | 3197 | 1942 | 3828 | 3543 | Mo ABC transporter, periplasmic Mo-binding protein |
| modB | Mo | 3198 | 1941 | 3829 | 3544 | Mo ABC transporter, permease protein |
| modC | Mo | 3199 | 1940 | 3830 | 3545 | Mo ABC transporter, ATP-binding protein |
| arsC | As | 4088 | 4193 | 1531 | 1140 | Arsenate reductase (glutaredoxin family) |
| - | Cu | 4576 | 0627 | 0588 | 0633 | Putative copper-binding protein |
| - | Cu | 4578 | 0625 | 0586 | 0631 | Putative copper-translocating P-type ATPase |
| merE | Cu | 4579 | 0624 | 0585 | 0630 | Transcription regulator heavy metal-dependent MerE family |
| znuA | Zn | 5108 | 0137 | 0120 | 0135 | Zinc uptake ABC transporter, periplasmic binding protein |
| zur | Zn | 5109 | 0136 | 0119 | 0134 | Transcriptional repressor of Zn transport system |
| znuC | Zn | 5110 | 0135 | 0118 | 0133 | Zinc ABC transporter, ATP-binding protein |
| znuB | Zn | 5111 | 0134 | 0117 | 0132 | Zinc ABC transporter, permease protein |
| arsR | As | 5146 (31) | 3034 | 2718/1930 | 3077 | Arsenical resistance operon repressor |
| arsC | As | 5147 (31) | 3036 | 2716/1928 | 3079 | Arsenate reductase |
| arsB | As | 5148 (31) | 3035 | 2717/1929 | 3078 | Arsenite efflux transporter |
| arsH | As | 5149 (31) | 3037 | 2715/1927 | 3080 | Arsenical resistance protein |
| chrF | Cr | 5156 (31) | – | – | – | Chromate resistance regulator |
| chrA | Cr | 5157 (31) | 3159 | 2556 | 3384 | Chromate transporter |
| chrB | Cr | 5158 (31) | – | – | 3383 | Chromate resistance protein |
| nreB | Ni | 5159 (31) | – | – | – | Major facilitator superfamily (nickel efflux family) |
| - | ? | 5161 (31) | – | – | – | Putative cation diffusion facilitator (CDF) |
| copS | Cu | 5177 (31) | – | – | – | Sensor protein CopS |
| copR | Cu | 5178 (31) | – | – | – | Transcriptional activator CopR |
| copA | Cu | 5180 (31) | 0574 | 2205 | 1828 | Copper resistance protein A |
| copB″ | Cu | 5181 (31) | – | – | – | Putative CopB (frameshifted) |
| copB′ | Cu | 5182 (31) | 0573 | 2204 | 1827 | Copper resistance protein B |
| copM | Cu | 5183 (31) | 0572 | – | – | Cytochrome c family protein |
| copG | Cu | 5184 (31) | – | 2203 | 1826 | Involved in survival in the presence of high bioavailable Cu(II) |
| czcC | Cd/Zn/Co | – | 3287 | 2408 | 2042 | Cobalt/zinc/cadmium resistance protein |
| czcB | Cd/Zn/Co | – | 3286 | 2409 | 2043 | Cobalt/zinc/cadmium efflux RND transporter |
| czcA | Cd/Zn/Co | – | 3285 | 2410 | 2044 | Cobalt/zinc/cadmium efflux RND transporter |
| mrdH | Ni/Cd/Zn | – | – | 2968 | – | Ni/Cd/Zn resistance-associated protein |
| mreA | ? | – | – | 2969 | – | Metal resistance-associated cytoplasmic protein |
For the reference genome W619, the numbers in brackets indicate the genomic region of the gene, which was omitted if the gene was not located in a genomic region.
?, the metal specificity remains to be determined. The gene annotations and comparisons were obtained using the MaGe annotation platform (Vallenet et al., 2006).
Fig. 4.
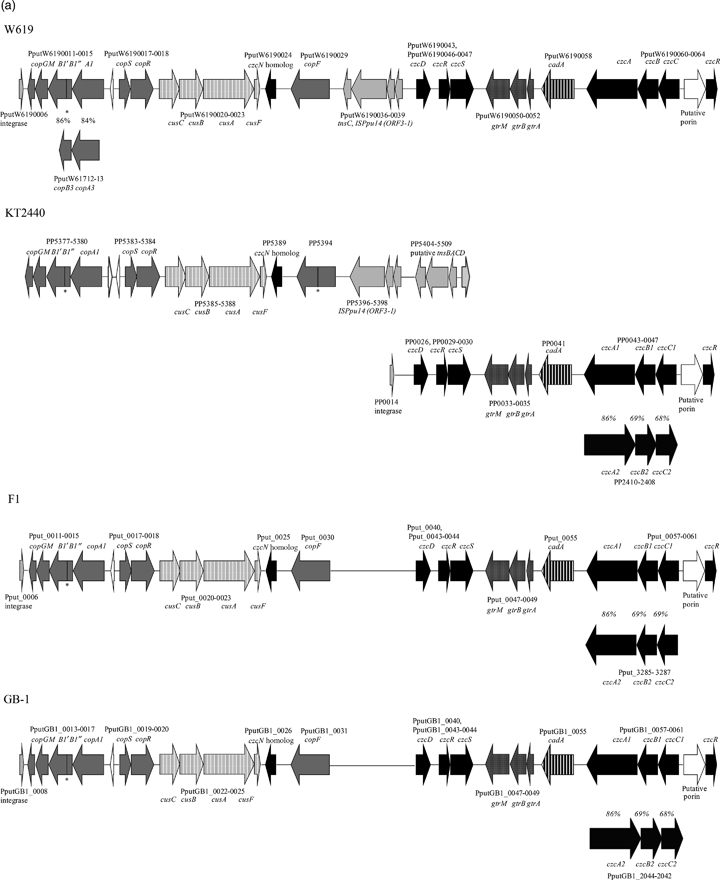
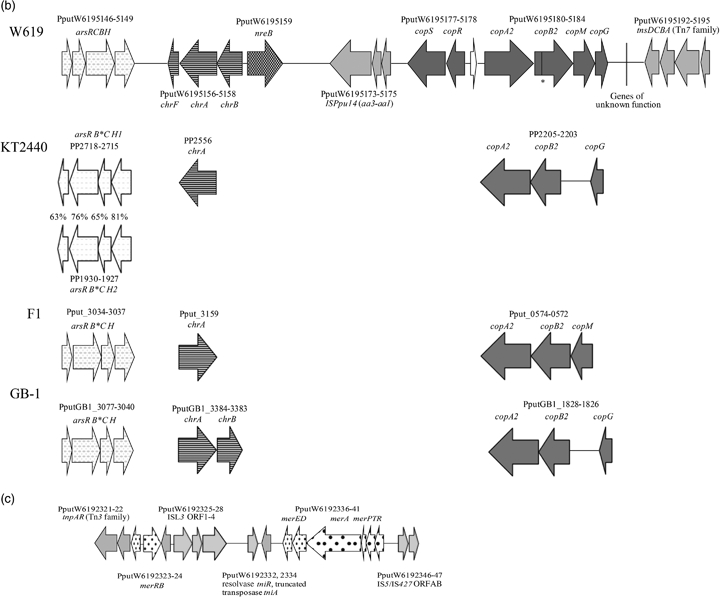
Genetic organization of heavy metal resistance determinants located on the chromosomes of Pseudomonas putida W619, KT2440, F1 and GB-1. (a) Genetic organization of heavy metal resistance determinants on the putative genomic island region 1 of P. putida W619, and its comparison with the corresponding regions located on the chromosomes of P. putida KT2440, F1 and GB-1. PP5394, located on the P. putida KT2440 chromosomes, encodes a P-type ATPase (Ag/Cu efflux), which was reported to have a frameshift mutation (http://www.tigr.org). Identities are provided as percentages between two copies of homologous genes. * The position of a frame shift mutation in comparison with the full-length ORF. (b) Genetic organization of heavy metal resistance determinants on putative genomic island region 31 of P. putida W619, and its comparison with the corresponding regions located on the chromosomes of P. putida KT2440, F1 and GB-1. The arsB gene in P. putida W619 and the arsB* genes in P. putida KT2440, F1 and GB-1 are from different sources. (c) The mercury resistance region and its flanking mobile genetic elements on the chromosome of the P. putida W619 genome. This region is absent in P. putida KT2440, F1 and GB-1.
In between copF and czcDRS, a copy of ISPpu14 (PputW619_0037-0039) was found, separating the copper/silver resistance cluster from the cadmium/zinc/cobalt resistance cluster. The region 1 counterparts of F1 and GB-1 seem to be identical in organization with very close synteny to W619, the only difference being the absence of ISPpu14. In the case of KT2440, region 1 is divided into two sections that are separated by the chromosomal replication origin: section 1, containing the copper/silver resistance cluster that ends with a copy of ISPpu14, and section 2, which starts with a putative integrase and contains the cadmium/zinc/cobalt resistance cluster.
Region 31 (coordinates: 5706900–5753604) of P. putida W619 is less conserved among the other three P. putida strains, and contains genes involved in arsenite/arsenate, chromium, nickel and copper resistances (Fig. 4b). The arsenite/arsenate resistance operon (arsRCBH; PputW619_5146-5149) has homologs on the KT2440, F1 and GB-1 genomes, except that their arsB genes are different: the gene from W619 is closely related to the arsB gene from Acinetobacter baumannii AYE, while the arsB genes from the other P. putida strains are closer to the Escherichia coli K12 arsB. Still, P. putida W619 and KT2440 showed similar growth and resistance to As(III) and As(V) in the range of concentration we tested (both viable in the presence of NaAsO2 (0.01 to ∼2 mM) and Na2HAsO4 (0.001 to ∼5 mM). Besides this arsRCBH operon, two additional arsC genes encoding the arsenate reductase were present on the P. putida chromosomes, as well as an additional copy of arsRCBH in KT2440 (PP_1927-1930).
The chromate resistance genes chrFAB (PputW619_5156-5158) and the nickel resistance gene nreB (PputW619_5159) are located downstream of the ars operon, and are incomplete or absent in KT2440, F1 and GB-1. They are followed by a second copy of the copper resistance genes copSRABMG (PputW619_5177-5178, 5180-5184), which is flanked by a copy of ISPpu14 (PputW619_5173-5175) and by a copy of a Tn7-like transposon (tnsABCD; PputW619_5195-5192). Remnants of the cop operon are found in KT2440, F1 and GB-1. As for region 1, the region 31 cop operon seems to be part of a composite mobile element. Although the organization of both cop operons is very similar, they showed only 80% protein identity, which rules out a recent duplication.
The cop determinant in Pseudomonas syringae pv. tomato contains six genes (copABCDRS) (Mellano & Cooksey, 1988). However, the presence of copAB was sufficient to render the cells copper resistant (Mellano & Cooksey, 1988; Cha & Cooksey, 1993;). CopAB are known to be involved in the detoxification and efflux of copper ions from the periplasm (Rensing et al., 2000; Monchy et al., 2006;). In P. syringae, the CopA and CopB proteins contain the MXXMXHXXM (MDH) motif repeated several times throughout the sequence. This motif also appears in CopA1 (PputW619_0015), where it is repeated 12 times. However, this repeat is truncated for CopA2 (PputW619_5180) and is only present five times. A similar organization was described for the two copA genes in KT2440 (Canovas et al., 2003), having this motif repeated 14 and five times in CopA1KT2440 and CopA2KT2440, respectively, and was also observed for F1 and GB-1.
Both copies of CopB in W619 contained a frame shift mutation. For instance, in the case of CopB1, if this frame shift was ignored, the resulting complete protein would contain six copies of the MDH motif and 21 histidine residues, which are known to bind copper efficiently. The truncated CopB1 protein, however, only contains three MDH motifs and 13 histidine residues. A similar frame shift was also found in KT2440, F1 and GB-1, but only in one of the two CopB copies. In addition to the two copies of copSRAB, an additional copAB locus was identified on the W619 genome (PputW619_1712-1713, located in region 8; coordinates: 1875105–1910820), which might make up for the frame shift mutations. In addition, W619 has a copF gene putatively encoding a P1-type ATPase, which is located in the inner membrane and involved in Cu(I) efflux from the cytoplasm to the periplasm (Rensing et al., 2000; Mergeay et al., 2003;). The copF gene is absent in KT2440, which might explain the higher level of copper resistance for W619 compared with KT2440, with MIC values of 0.5 and 0.1 mM Cu, respectively.
Because P. putida W619 was isolated from a nickel-contaminated site, it was no surprise that it showed elevated levels of nickel resistance compared to KT2440, with MIC values of 1 and 0.1 mM, respectively. The only putative nickel resistance protein identified in W619 was a copy of nreB (PputW619_5159), located in region 31, which is absent in KT2440, F1 and GB-1. The presence of nreB is consistent with the observation that in A. xylosoxidans and E. coli, nreB expression was sufficient for nickel resistance (Grass et al., 2001). In KT2440, a novel metal resistance determinant mrdH (PP_2968) was identified recently, which encodes a protein with a chimeric domain organization from RcnA and CzcB (Haritha et al., 2009). MrdH was found to be involved in nickel, cadmium and zinc resistance. The mrdH gene and mreA (PP_2969), the latter showing similarity to NreA-like proteins, lack homologs in P. putida W619, GB-1 and F1, and are located on genomic island 34 (GI 55 according to Weinel et al. 2002) of KT2440. The presumably higher specificity for nickel resistance of nreB compared with mrdH may explain strain W619's much higher MIC value for Ni(II) as compared to KT2440. It should also be noted that the activities of the mobile genetic elements Tn4652 (PP_2964-65) and IS1246 (PP_2971), which are uniquely found on the KT2440 chromosome flanking mrdH and mreA, were found to be induced by the presence of Cd, Ni and Zn.
Additional putative heavy metal-responsive genes, many of which are absent in KT2440, F1 and GB-1 (Table 3), can be found on putative genomic islands on the W619 chromosome. A gene coding for a CDF putatively involved in zinc transport (PputW619_1714) and an additional copy of arsB (PputW619_1715) are located on region 8 (coordinates: 1875105–1910820). This region is located next to genes encoding proteins putatively involved in iron uptake (PputW619_1716-1719). On region 11 (coordinates: 2520458–2644620), two clusters of genes involved in mercury resistance (Fig. 4c) were found: a merRB (PputW619_2323-2324) locus, which contains a merB organomercurial lyase required for cleavage of mercury-alkyl bounds, and that is flanked by a copy of Tn3 (PputW619_2321-2322) and an IS element from the ISL3 family (ISPpu12; PputW619_2325-2328); mercury resistance is completed by a merRTPADE operon (PputW619_2341-2336). This operon is flanked by a truncated transposase and a complete recombinase gene, and a copy of ISPs1. This organization suggests the acquisition of mercury resistance via two events of horizontal gene transfer, followed by DNA rearrangements. On the putative genomic island region 18 (coordinates 3322597–3400760), a copy of nikABCDE (PputW619_3005-3009) encoding an ABC nickel uptake transporter, which was also found in KT2440, and a copy of chrA (PputW619_3017) involved in the transport of chromate were found. Additional heavy metal resistance or homeostasis genes were also identified outside putative genomic islands and include cadAR (PputW619_0325-0326), modABC (PputW619_3197-3199), znuABC (PputW619_5108-5111) and cinAQ (PputW619_1676-1677). These genes are common for all four P. putida strains. Among them, the copper-inducible genes cinAQ encode a copper-containing azurin-like protein and a pre-Q0 reductase, respectively, and are located adjacent to their two-component regulatory system cinRS. Gene disruptions of cinA and cinQ did not lead to a significant increase in the copper sensitivity of P. putida KT2440, which might result from the redundancy of copper resistance systems (Quaranta et al., 2007). In addition, KT2440, F1 and GB-1 have a duplicated Cd/Zn/Co resistance system, czcABC, which is lacking in W619.
Degradation of organic solvents and aromatic compounds by P. putida
Previous genome analysis of P. putida KT2440 revealed many metabolic pathways for the transformation of aromatic compounds (Jimenez et al., 2002; Nelson et al., 2002;). Putative genes coding for the degradation of aromatic compounds were compared among the four P. putida strains (see Table 4 and Table S5). Some of these aromatic compounds (ferulate, coumaryl alcohols, aldehydes and acids, p-hydroxybenzoate) may arise from the decomposition of plant materials, as can be found in the rhizosphere.
Table 4.
Aromatic compound degradation pathway in Pseudomonas putida F1, and comparison with KT2440, W619 and GB-1
| Gene | Product | F1 ORF ID Pput_ | KT2440 ORF ID PP_ | GB-1 ORF ID PputGB1_ | W619 ORF ID PputW619_ |
|---|---|---|---|---|---|
| sepA/ttgA | RND family efflux transporter | 2867 | 1386 | 4427 | 1026 |
| sepB/ttgB | Hydrophobe/amphiphile efflux | 2868 | 1385 | 4428 | 1025 |
| sepC/ttgC | RND efflux outer membrane lipoprotein | 2869 | 1384 | 4429 | 1024 |
| todT | Response regulator | 2871 | – | – | – |
| todS | Signal transduction histidine kinase | 2872 | – | – | – |
| todE | 3-methylcatechol 2,3-dioxygenase | 2876 | – | – | – |
| todD | cis-Toluene dihydrodiol dehydrogenase | 2877 | – | – | – |
| todA | Aromatic-ring-hydroxylating dioxygenase | 2878 | – | – | – |
| todB | Ferredoxin | 2879 | – | – | – |
| todC2 | Toluene dioxygenase | 2880 | – | – | – |
| todC1 | Toluene dioxygenase | 2881 | – | – | – |
| todF | 2-Hydroxy-6-oxo-2,4-heptadienoate hydrolase | 2882 | – | – | – |
| todX | Membrane protein | 2883 | – | – | – |
| - | Enoly-coenzyme A hydratase | 2887 | – | – | – |
| mhpT | 3-Hydroxyphenylpropionic acid transporter | – | – | – | 1985 |
| cmtG (mhpE) | 4-Hydroxy-2-oxovalerate aldolase | 2888 | – | – | 19842007 |
| cmtH (mhpF) | Acetaldehyde dehydrogenase | 2889 | – | – | 19832008 |
| cmtF (mhpD) | 2-Hydroxypenta-2,4-dienoate hydratase | 2890 | – | – | 19822011 |
| mhpC | 2-Hydroxy-6-ketonona-2,4-dienedioic acid hydrolase | – | – | – | 1981 |
| mhpB | 2,3-Dihydroxyphenylpropionate 1,2-dioxygenase | – | – | – | 1980 |
| mhpA | 3-(3-Hydroxy-phenyl)propionate hydroxylase | – | – | – | 1979 |
| mhpR | Mhp operon transcriptional activator | – | – | – | 1978 |
| cmtE | HOMODA hydrolase | 2891 | – | – | – |
| cmtI | Protein of unknown function | 2892 | – | – | – |
| cmtD | HCOMODA decarboxylase | 2893 | – | – | – |
| cmtAd | p-cumate dioxygenase ferredoxin subunit | 2894 | – | – | – |
| cmtB | 2,3-dihydroxy-2,3-dihydro-p-cumate dehydrogenase | 2895 | – | – | – |
| cmtC | 2,3-dihydroxy-p-cumate-3,4-dioxygenase | 2896 | – | – | – |
| cmtAc | p-Cumate dioxygenase small subunit | 2897 | – | – | – |
| cmtAb | p-Cumate dioxygenase large subunit | 2898 | – | – | – |
| cmtAa | p-Cumate dioxygenase ferredoxin reductase subunit | 2899 | – | – | – |
| cymE | Acetyl-coenzyme A synthetase | 2900 | – | – | – |
| cymD | Outer membrane protein | 2901 | – | – | – |
| cymAb | p-Cymene monooxygenase reductase subunit | 2902 | – | – | – |
| cymAa | p-Cymene monooxygenase | 2903 | – | – | – |
| cymC | p-Cumic aldehyde dehydrogenase | 2904 | – | – | – |
| cymB | p-Cumic alcohol dehydrogenase | 2905 | – | – | – |
| cymR | Regulatory protein for cym and cmt operons | 2908 | – | – | – |
The gene annotations and comparisons were obtained using the MaGe annotation platform (Vallenet et al., 2006).
Although KT2440 has versatile metabolic pathways to degrade aromatic compounds, it is not able to grow on any aromatic hydrocarbon as a sole carbon source. In contrast, with a significantly wider range of growth substrates, P. putida F1 is best known for its capability of growth on several aromatic hydrocarbons, including benzene, toluene, ethylbenzene and p-cymene. Its toluene degradation (tod) pathway is featured by the toluene dioxygenase operon todABCDE (Zylstra et al., 1988; Gibson et al., 1990;) and the corresponding two-component regulatory system, todST (Lau et al., 1997). Besides the ability to use toluene, ethylbenzene or benzene as the sole carbon source, F1 is also able to grow on p-cymene (p-isopropyltoluene) and its acid derivative, p-cumate (Eaton, 1996, 1997). The mechanism uses two pathways: the cymAaAbBCDER pathway (Pput_2900-2905, 2908), responsible for the oxidation of p-cymene to p-cumate, and the adjacently located cmtAaAbAcAdBCDEFGHI pathway (Pput_2888-2899) to take p-cumate to isobutyrate, pyruvate and acetyl coenzyme A (Eaton, 1996, 1997). In F1, the cym/cmt and the tod pathways are located less than 3 kb apart on a putative genomic island (3260962–3302713), which is featured by a lack of synteny with other P. putida strains and surrounded by phage-related genes, an organization that is indicative that this region was acquired via transduction. A sepABC gene cluster is located upstream of this genomic island, encoding for solvent efflux or multidrug pumps (Phoenix et al., 2003). KT2440, W619 and GB-1 all have conserved homologs for this cluster, referred to as ttgABC (Duque et al., 2001). In addition, genomic sequence comparisons predicted the universal existence of other aromatic catabolic pathways in all four P. putida strains, including the protocatechuate (pca genes) and catechol (cat genes) branches of the β-ketoadipate pathway, and the phenylacetate pathway (pha genes) (see Table S5).
The genome of W619 contains two mph operons for the degradation of 3-hydroxyphenylpropionate (Ferrandez et al., 1997): one complete mhpRABCDFET operon encoding enzymes for the conversion from 3-HTT to acetyl coenzyme A (CoA) and another incomplete cluster consisting of mhpEFD. The mhpEFD genes encode three enzymes that are conserved with part of the cym/cmt pathway: a 4-hydroxy-2-oxovalerate aldolase, an acetaldehyde dehydrogenase and a 2-hydroxypenta-2,4-dienoate hydratase (Table 4), which are respectively able to catalyze the conversion of 2-keto-4-pentenoate to 4-hydroxy-2-ketovalerate, to pyruvate and acetaldehyde and finally to acetyl CoA. This indicates that W619 has a broader potential for the degradation of aromatic compounds as compared with KT2440 and GB-1, but is less versatile than F1. For instance, unlike F1, P. putida W619 was unable to metabolize toluene or TCE, a property that could be complemented via acquisition of the pTOM plasmid of Burkholderia cepacia BU61 (Taghavi et al., 2005; Weyens et al., 2009;). The W619 mhpRABCDFET operon is flanked by a truncated copy of IS1182 (PputW619_1976-1977) and an IS element from the IS30 family (IS1382; PputW619_1990), which are absent in KT2440, GB-1 and F1. The presence of these mobile genetic elements points toward the acquisition of this catabolic region via horizontal gene transfer.
Mn(II) oxidation by P. putida
In the environment, Mn cycles between the soluble reduced form Mn(II) and the insoluble oxidized forms Mn(III and IV) that can adsorb other trace metals from the environment and react as potent oxidizing agents. Thus, the Mn redox cycle has beneficial effects on the bioavailability and geochemical cycling of many essential or toxic elements (Tebo et al., 2005). Pseudomonas putida GB-1 is a Mn(II)-oxidizing model bacterium (Corstjens et al., 1992). Because Mn(III, IV) oxides are able to bind trace metals, this feature makes P. putida GB-1 a good candidate for bioremediation of heavy metal-contaminated sites. However, the mechanism for Mn(II) oxidation still remains to be elucidated. Several random transposon mutagenesis experiments led to the identification of genes important for Mn oxidation: the ccm operon coding for c-type cytochrome synthesis genes (Caspi et al., 1998; de Vrind et al., 1998;), the MCO cumA (Brouwers et al., 1999) and genes encoding a general secretory pathway (de Vrind et al., 2003). A recent paper showed that the MCO CumA is dispensable for Mn(II) oxidation. Instead, a two-component regulatory system (MnxS/R) was found to be essential for Mn(II) oxidation (Geszvain & Tebo, 2009). This is consistent with the fact that cumA is present in both Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains (Francis & Tebo, 2001). Moreover, through transposon mutagenesis of other Mn(II) oxidases, such as mnxG (PputGB1_2447) and PputGB1_2665, Mn(II) oxidation was only slowed, but not lost in the mutant strains (Yamaguchi, 2009). These findings suggest that P. putida GB-1 may have multiple Mn(II) oxidases, with the expression of alternate enzymes dominating under different environmental conditions. Comparative genomics further indicates that genes related to Mn(II) oxidation in GB-1 have homologs in P. putida KT2440, W619 and F1, except that W619 lacks the homolog for PputGB1_2665 (Table S6). The presence of these genes points to the potential for Mn(II) oxidation by KT2440, F1 and W619. Also, a link between Mn(II) oxidation and pyoverdine siderophore synthesis was reported for P. putida MnB1 (Parker et al., 2004). Thus, Mn(II) oxidation may influence the pyoverdine-mediated iron uptake by Mn(II)-oxidizing fluorescent P. putida strains.
Response to oxidative stress by P. putida
Pseudomonas putida strains thrive in environments that are characterized by oxidative stress including the rhizosphere, soils and sediments containing reactive metal species generated during the Mn-redox cycle, or reactive intermediates generated during the oxidative breakdown of aromatic hydrocarbons. As part of their adaptation, P. putida strains possess various enzymes including catalases, peroxidases and superoxide dismutases for defense against reactive oxygen species (ROS) (including superoxide, hydroperoxyl radical and hydrogen peroxide species), nitric oxide and phytoalexins (Hammond-Kosack & Jones, 1996; Zeidler et al., 2004;). Comparative genomic analysis shows that many of the enzymes involved in the oxidative stress response are common among the P. putida species (Table S7). For example, the W619 genome encodes three superoxide dismutases: SodA, a Mn superoxide dismutase (PputW619_4269), SodB, an Fe superoxide dismutase (PputW619_0981), and SodC, a Cu/Zn superoxide dismutase (PputW619_2485). The sodC gene is located on a putative genomic island (region 12) and it is absent from the other P. putida strains. Pseudomonas putida W619 is also predicted to contain five catalases, KatA (PputW619_4722), KatB (PputW619_2032), KatE (PputW619_5113), KatG (PputW619_2235) and one putative catalase (PputW619_2390); two alkyl hydroperoxide reductases, AhpF and AhpC (PputW619_3186-3187), four additional putative alkyl hydroperoxide reductases (one putative AhpC, PputW619_1113 and three having an AhpD domain, PputW619_3104, PputW619_3108, PputW619_3238); a chloroperoxidase (PputW619_1849); two thiol peroxidases (PputW619_2803 and PputW619_3977); and two putative glutathione peroxidases (PputW619_1244, PputW619_1483), a glutathione oxidoreductase (Gor, PputW619_3188) and a glutaredoxin (PputW619_3239). Among these enzymes, the KatB and AhpD domain proteins, whose genes are located on three genomic islands (regions 9, 19 and 20), are unique to P. putida W619. We also identified an organic hydroperoxide resistance protein (Ohr, PputW619_1469) located adjacent to its organic hydroperoxide resistance transcriptional regulator (OhrR, PputW619_1470). Moreover, P. putida W619 is likely able to detoxify free radical nitric oxide by the presence of a flavohemoprotein nitric oxide dioxygenase (PputW619_4378) with its anaerobic nitric oxide reductase transcription activator NorR (PputW619_4379) (Tucker et al., 2004).
The oxidative stress response system is controlled via complex regulatory networks (Storz & Imlay, 1999). One of the key regulators is the peroxide resistance protein PerR, which was identified as the major regulator of the inducible peroxide stress response in Bacillus subtilis (Mongkolsuk & Helmann, 2002). In P. putida W619, the PerR protein is encoded by PputW619_2615, a LysR family transcriptional regulator that shares 61.5% identity with PerR of E. coli K12. PerR regulates the expression of dps (DNA-binding stress protein, PputW610_4005), fur (the ferric uptake repressor, PputW619_0702), ahpCF and katA (Mongkolsuk & Helmann, 2002), all of which are present in P. Putida W619.
In some cases, the regulation of ROS-responsive genes is iron dependent and coupled to that of iron-binding proteins, such as bacterioferritin (Bfr). As in KT2440, the bacterioferritin α subunit (bfrA, PputW619_4721) in P. putida W619 is located next to catalase A (katA) (Dos Santos et al., 2004), and the bfrB and Bfd-associated ferredoxin gene (PputW619_1111-1112) are located adjacent to ahpC (PputW619_1113). Furthermore, various oxidation-resistant metabolic enzymes were putatively identified in W619, including acnA (encoding an aconitase, PputW619_1631), which is stable under oxidative stress, and fumC (PputW619_4271), located upstream of sodA (PputW619_4269), which encodes a hydroperoxide resistant isoform of fumarase. In P. aeruginosa, the fumC-sodA operon is shown to be negatively regulated in the presence of iron via the Fur protein (Polack et al., 1996).
Siderophore production by P. putida
The bioavailability of iron in the soil is limited by the very low solubility of the Fe3+ ion, and bacteria have developed diverse mechanisms for iron acquisition. One of the most important mechanisms is the production and release of siderophores to scavenge iron from the environment via the formation of soluble Fe3+ complexes, which can subsequently be taken up by active transport systems (Neilands, 1995; Ravel & Cornelis, 2003;). In addition to their own siderophores, bacteria may utilize a large number of heterologously produced siderophores that are taken up via various siderophore receptors (Dean et al., 1996). Many Pseudomonas strains show fluorescence under UV light, indicative of the production of the siderophore pyoverdine (PVD) (Meyer, 2000). In accordance with this observation, nonribosomal peptide synthetase (NRPS) genes for pyoverdine synthesis were found in all four P. putida strains (Table S8). A cluster of peptide NRPS genes was identified, together with the PVD ABC transporter gene pvdE and a TonB-dependent receptor gene fpvA. At a separate locus, the chromosphore NRPS gene psvA was found to be present in all four strains along with the σ factor pvdS and, with the exception of strain F1, the siderophore biosynthesis gene pvdZ (Moon et al., 2008). In F1, the pvdZ gene is located next to the pvdE gene. It should also be noted that the four P. putida strains are lacking the homologs of pseudomonine synthesis genes as identified in P. entomopila L48 (PSEEN_2500-2507), and therefore seem to be unable to produce this secondary siderophore (Matthijs et al., 2009).
For the transport of pyoverdine, P. putida W619 possesses multiple copies (20) of genes encoding TonB-dependent outer-membrane receptors, as reported previously by Cornelis & Bodilis (2009). Out of these 20 TonB-dependent receptors, 17 are putative ferric siderophore receptors, including a putative ferric enterobactin receptor (fepA) and a FecA protein for ferric citrate uptake (Table S8). Of the three remaining TonB-dependent receptors, one is a putative copper-regulated channel protein (OprC) (PputW619_4630) (Yoneyama & Nakae, 1996), while the specificity of the other two receptors remains unclear (PputW619_1083, PputW619_5001). As summarized in Cornelis & Bodilis (2009), the genomes of F1, KT2440 and P. entomophila L48 carry 29 to 31 putative TonB-dependent receptor genes, a number considerably higher than the 20 genes identified in W619. It has been shown experimentally that P. entomophila L48 is able to utilize a large variety of heterologous pyoverdines, but that in contrast, KT2440 can only use its own pyoverdine and the pyoverdine heterologously produced by P. syringae LMG 1247 (Matthijs et al., 2009). This observation is in accordance with the fact that L48 has the highest number of ‘orphan’ TonB-dependent receptor genes (11) that are not found on the genomes of the P. putida strains, while the P. putida KT2440, F1 and W619 genomes only contain 4, 3 and 5 ‘orphan’ genes, respectively. Thus, the capacity of W619 to utilize heterologous pyoverdine needs further investigation (Cornelis & Bodilis, 2009).
The TonB-dependent siderophore receptors genes in W619 are often located adjacent to genes coding a transmembrane anti-σ sensor (FecR family) and an ECF-σ70 transcriptional regulator of the FecI family, which might regulate the expression of receptors (Folschweiller et al., 2000; Braun & Endriss, 2007; Cornelis et al., 2009;). It should be noted that in the genomes of P. putida strains, up to 13 ECF-σ factors were found that showed similarity to the FecI-σ factor of E. coli (Martinez-Bueno et al., 2002). The specificity of the receptors for various ferric siderophores is not well understood, but the presence of multiple TonB-dependent siderophore receptors might confer P. putida strains with the ability to use a broad spectrum of these heterologously synthesized compounds (Paulsen et al., 2005). For example, the P. putida strains possess an iron uptake system that involves the genes coding for the outer-membrane ferric enterobactin receptor, PfeA/FepA, and its corresponding two-component regulator, PfeRS (Dean et al., 1996). This system may allow P. putida to compete with Enterobacteriaceae for iron by utilizing the ferric–enterobactin complex. In addition, a universal TonB-dependent heme receptor gene (phuR) was found in all four P. putida strains (PP_1006, PputW619_4218, Pput_1043, PputGB1_1005), while two other heme uptake-related genes, HasR and HxuC, typically found in the pathogens P. aeruginosa (Ochsner et al., 2000) and P. entomophila (Cornelis & Bodilis, 2009), were absent in P. putida. The redundant presence of systems that scavenge iron from the environment argues for the importance of iron acquisition in the ecology of this bacterium and its ability to colonize multiple niches.
Association of P. putida with plants
Both P. putida KT2440 and W619 were found to live in association with plants, either as a rhizospheric strain or as an endophyte, respectively. Comparative genomics, using P. putida W619 as the reference strain, was used to identify the functions that are important for the association of these bacteria and their host plant.
Motility
The P. putida W619 genome contains a large cluster of fifty-two genes involved in flagella biosynthesis (PputW619_3655-3737) (Table S9). This cluster is similarly organized in all four P. putida strains, except that W619 contains within the flagellar biosynthesis cluster a gene cluster involved in LPS biosynthesis and sporulation. Furthermore, the flagellar biosynthesis clusters in W619, KT2440 and F1 are interrupted by a region of atypical composition and lost synteny that contains d-Alanine and cystathionine ligases (genomic region 22; PputW619_3671-3677). Pseudomonas putida KT2440 is known to be a good swimmer; this in contrast to the solvent-tolerant P. putida DOT-T1E (Segura et al., 2004). It was demonstrated that the impaired swimming capability of DOT-T1E resulted from a mutation in the flhB gene, which resulted in higher solvent resistance (Segura et al., 2004). The P. putida W619 flhB gene is 91% identical to that of KT2440, suggesting a swimming capability similar to KT2440. In addition, it was demonstrated in KT2440 that mutations in the flagellar biosynthesis genes flgL (PputW619_3721), fliA (PputW619_3667), fleQ (PputW619_3698), fliL (PputW619_3691), fliN (PputW619_3683) and flgD (PputW619_3730) resulted in different degrees of impaired swimming motility, and swarming, and also affected the adhesion to seeds (Espinosa-Urgel et al., 2000; Yousef-Coronado et al., 2008;). The preliminary microarray result obtained with P. putida W619 showed that the transcription of genes encoding the flagellar proteins (PputW619_3724-3726) is induced when the cells are grown hydroponically in the presence of poplar roots, pointing toward a role of mobility in the plant–bacterial recognition/interaction process (data not shown).
Pili and curli fibers
Type I pili assembly occurs in the periplasm and involves the chaperone–usher pili assembly system (Thanassi et al., 1998). The four P. putida genomes all encoded a cluster of type I pili synthesis proteins, CsuABCDE, where CsuC and CsuD are predicted as the usher pathway chaperone and the usher protein, respectively (Table S10).
Type IV pili are typically 5-7 nm fibers that play a very important role in host colonization by a wide range of plant and animal pathogens (Mattick, 2002). The biogenesis and function of type IV pili are controlled by a large number of genes. On the P. putida genomes, three clusters of type IV pili biosynthesis genes were identified, pilMNPQ, pilACD and pilEXW/fimT, as well as four copies of the pilZ gene for pili assembly. The pili assembly might be linked to the cell cycle, because pilZ is transcriptionally coupled to holB, which encodes the δ subunit of DNA polymerase III (Alm et al., 1996). Also P. putida contains a complex pili biosynthesis regulatory system, pilGHIJLchpA, where pilL/chpA encode a large fusion protein that acts as a very sophisticated signal transduction protein, ChpA (Alm & Mattick, 1997). Moreover, type IV pili are involved in natural DNA uptake (transformation). There are two additional putative traX genes (PputW619_2699, PputW619_5167) in P. putida W619, related to the conjugal DNA transfer that are absent in KT2440, F1 and GB-1.
Curli fibers are involved in adhesion to surfaces, cell aggregation and biofilm formation and play an important role in host cell adhesion and invasion (Barnhart & Chapman, 2006). The P. putida strains possess two gene clusters for curli fiber biogenesis: csgEFG and csgAB. The CsgAB proteins provide the two curli structural subunits, while CsgEFG are accessory proteins required for curli assembly (Hammar et al., 1995).
Colonization of seeds by P. putida
In P. putida KT2440, colonization of seeds was affected by mutations in various genes (Yousef-Coronado et al., 2008). As expected, homologs of these genes were identified in P. putida W619: lapA (PputW619_5060), lapBCD (PputW619_5062-5064), hemN2 (PputW619_0364), coxE (PputW619_0130), galU (PputW619_3189) and a hypothetical protein (PputW619_0162) that was shown to be involved in the oxidative stress response. We hypothesize that these genes have the same functions in W619. For instance, the LapA protein involved in biofilm formation is exported to the outside of the cell via the type I LapBC secretion system (Hinsa et al., 2003). Furthermore, P. putida W619 contains an additional gene coding for an outer membrane component of the type I secretion system (PputW619_5061). The genome of P. putida W619 also encodes a putative adhesin (PputW619_3808) and a surface-adhesion calcium-binding outer membrane-like protein that was 24% identical to LapA. This large (3923 aa) protein is encoded next to its putative type I secretion system in a gene organization similar to the lapABC cluster. Other genes on the P. putida W619 genome putatively involved in adhesion include PputW619_1818 and PputW619_1489, both of which code for autotransporter proteins (secretion type V) with a pectin/lyase/pertactin domain. In contrast to the poplar endophyte Enterobacter sp. 638, no genes encoding a hemagglutinin protein were identified on the P. putida W619 genome. Among closely related Pseudomonas strains, we only found hemagglutinin-like adhesion genes in the insect pathogen P. entomophila L48 (PSEEN_0141, PSEEN_2177, PSEEN_3946), where they are predicted to be important virulence factors and involved in adhesion and type I or two-partner secretion systems (Vodovar et al., 2006).
Establishment of P. putida W619 in poplar
The ndvB gene (PputW619_2133) encodes a protein (2881 aa) involved in the production of β-(1,2)-glucan. The membrane-bound NdvB protein catalyzes three enzymatic activities: the initiation (protein glucosylation), elongation and cyclization in situ of β-(1,-2)-glucan, which is then released into the periplasm (Castro et al., 1996). It has been reported that cyclic β-(1,2)-glucan is involved in the attachment of Agrobacterium tumefaciens to plant cells (Douglas et al., 1985). In Rhizobium meliloti, mutations of the ndvB gene reduced the amounts of periplasmic β-(1,2)-glucan, which resulted in altered phenotypes related to phage and antibiotic sensitivity, motility, and growth in low-osmolarity media. Bacteroids produced by two of the downstream mutants were morphologically abnormal, indicating that ndvB is involved not only in invasion but also in bacteroid development (Ielpi et al., 1990). The ndvB gene is not present in P. putida GB-1, F1 or KT2440, but was identified in the other two poplar endophytes: Serratia proteamaculans 568 and Enterobacter sp. 638.
Plant growth-promoting properties of P. putida
Synthesis of plant growth-promoting hormones by P. putida
Indole-3-acetic acid
Many plant growth-promoting bacteria are capable of synthesizing phytohormones that affect the growth and development of their plant host. In vitro, P. putida W619 was the most efficient producer of IAA in comparison with other endophytic bacteria (Taghavi et al., 2009). Consistently, the P. putida W619 genome encodes two putative tryptophan-dependent IAA synthesis pathways. One is via tryptamine and indole-3-acetaldehyde and requires a tryptophan decarboxylase (PputW619_2223), an amine oxidase (PputW619_0482) and an indole-3-acetaldehyde dehydrogenase (encoded by many putative genes PputW619_0192/0213/0597/2257/2639/2872/2926/2546/3767). This pathway also exists in KT2440, F1 and GB-1. The alternate pathway is converting tryptophan to indole-3-acetamide via a tryptophan 2-monooxygenase (PputW619_1175/4820), which is subsequently converted into IAA by a putative deaminase (PputW619_2551). Pseudomonas putida KT2440, F1 and GB-1 lack homologs for PputW619_1175, but contain conserved genes for PputW619_4820. Furthermore, the step to convert indole-3-acetamide into IAA seems to be absent in KT2440; thus, this pathway might be incomplete and nonfunctional. Along with the high level of IAA production, P. putida W619 has three genes encoding putative auxin efflux carriers (PputW619_2484/2492/3829). Pseudomonas putida KT2440, F1 and GB-1 also have auxin efflux carriers, but only in two copies for F1 and KT2440.
ACC deaminase
A functional 1-aminocyclopropane-1-carboxylate deaminase (acd) (EC: 3.5.99.7), involved in counteracting the plant's ethylene stress response, is absent on the genomes of P. putida W619, GB-1, F1 and KT2440, as was confirmed by the failure of these strains to grow on 1-aminocyclopropane-1-carboxylate as the sole carbon or nitrogen source. Although putative ACC deaminases were identified, all lack the particular amino acids E296 and L323 (respectively, replaced by a T or S and a T) that constitute part of the signature sequence of the active site of the true ACC deaminases and are involved in substrate specificity (Glick et al., 2007).
Acetoin and 2,3-butanediol synthesis
Volatile compounds such as 3-hydroxy-2-butanone (acetoin) and 2,3-butanediol are emitted by rhizobacteria to enhance plant growth (Ryu et al., 2003). In contrast to the poplar endophyte Enterobacter sp. 638, whose genome was sequenced recently (Taghavi et al., 2010), the P. putida W619 genome does not encode the budABC operon involved in the conversion of pyruvate to acetoin and 2,3-butanediol. The P. putida W619 genome carries the gene poxB (PputW619_2979) encoding a pyruvate dehydrogenase (genomic region 18). While the principal function of this enzyme is to convert pyruvate into acetaldehyde, a small fraction of the pyruvate can be converted into acetoin as a byproduct of the hydroxyethyl-thiamin diphosphate reaction intermediate. Although the production of acetoin by P. putida W619 is probably very low, especially at a low pyruvate concentration, this plant growth hormone can be utilized and converted into 2,3-butanediol, another plant growth hormone, by the poplar tree.
On the genomes of P. putida F1 (Pput_0595-0591), GB-1 (PputGB1_0601-0597) and KT2440 (PP_0556-0552) genes (acoXABC adh) involved in the catabolic conversion of acetoin and 2,3-butanediol to central metabolites were identified. Interestingly, these genes were not present in P. putida W619. Therefore, P. putida W619 does not have an antagonist effect on the production of the plant growth-promoting phytohormones acetoin and 2,3-butanediol, which might be beneficial as these compounds stimulate root formation and indirectly the availability of carbon sources for KT2440 when residing in the plant rhizosphere.
Salicilic acid
The genome of P. putida W619 encodes a salicylate 1-monooxygenase (mahG, PputW619_2140) involved in the conversion of salicylate into catechol. Homologues of this gene were also found on the genomes of P. putida KT2440, F1 and GB-1. Salicylate is a phenolic phytohormone involved in plant growth and development, and plays a role in the resistance to pathogens by inducing the production of pathogenesis-related proteins (Van Huijsduijnen et al., 1986).
Defense against plant pathogens by P. putida
Many pathogenic fungi secrete mannitol, a powerful reactive oxygen scavenger, into the apoplast during plant infection (Jennings, 1984; Velez et al., 2007;). This process has been shown to be required for pathogenicity as it suppresses the reactive oxygen-mediated plant host defenses (Chaturvedi et al., 1996a,b; Velez et al., 2008;). The production of mannitol dehydrogenase (MTD, EC 1.1.1.255) was hypothesized to increase the effectiveness of the plant defense response via the conversion of pathogen-secreted mannitol into mannose (Jennings et al., 2002). Interestingly, the gene encoding for MTD was found in P. putida W619 (mltD, PputW619_2037) located on a putative genomic island (region 9), along with genes coding for a mannitol ABC transporter (mltKGFE, PputW619_2038-2041), a transcriptional activator (mltR, PputW619_2042), a fructokinase (mltZ, PputW619_2035) and a xylulokinase (mltY, PputW619_2036). It is therefore hypothesized that P. putida W619 can assist its plant host in its defense against pathogenic fungi, an important property for its commensal life style. The mltZYDKGFE cluster, which is very similar to the operon found in P. fluorescens DSM50106 (Brunker et al., 1998a,b;), is absent in P. putida KT2440, F1 and GB-1.
Virulence factors in P. putida
Like for the granted biosafety strain P. putida KT2440, strains W619, F1 and GB-1 also lack a number of key determinants required for virulence and virulence-associated traits (Nelson et al., 2002). There is no evidence in the four P. putida genome sequences for putative functions required for the biosynthesis of exotoxin A, phospholipase C or pectin lyase that are frequently present in plant and animal pathogens. Additionally, the type III secretion pathway present in the plant pathogens P. syringae pv. tomato DC3000 or pv. phaseolicola 1448A (Cornelis & Van Gijsegem, 2000; Feil et al., 2005; Vencato et al., 2006;) is missing in all four P. putida strains. Some individual homologs with putative functions were found, such as PputW619_0806 coding for a putative type III HopPmaJ effector. However, this is in sharp contrast to the presence of a 20-kb gene cluster encoding a putative type III secretion system in the rhizosphere bacterium P. fluorescens SBW25 (Preston et al., 2001). Components of the type VI secretion system (T6SS) were also identified in P. putida strains and compared with the three T6SS clusters described in P. aeruginosa (Filloux et al., 2008) (Table S11). The T6SS clusters are partially conserved among the P. putida strains, including the ATPase ClpV, and the secreted VgrG and Hcp proteins. In W619, one of the T6SS-associated gene clusters (PputW619_3242-43, 3245-46, 3255-56, 3260-61) is located in genomic region 20 (3574758–3603632). Although the majority of genomic region 20 appears to lack synteny with the other three P. putida strains, the T6SS-associated genes seem to be conserved among KT2440, F1 and GB-1. Furthermore, in region 20, GAF/GGDEF and EAL signaling domains (Galperin, 2004) were identified that are hypothesized to play a role in regulating bacteria–host interactions. It also needs to be examined how far the functions of the putative T6SS in P. putida differ from the systems described in other Pseudomonads. For instance, they do not seem to play a role in virulence, as they lack homologs of the serine–threonine protein kinase (STPK), a key virulence factor in P. aeruginosa (Bingle et al., 2008).
Concluding remarks
The present inventory of genes and operons and the comparative genome analysis of the genome sequences of P. putida KT2440, W619, F1 and GB-1 provided a powerful tool to gain new insights into the adaptation of this species to specific lifestyles and environmental niches. Although P. putida strains are generally known for their versatile metabolic properties, this comparison also clearly demonstrated that horizontal gene transfer played an important role in this adaption process, as many of the niche-specific functions were found to be encoded on clearly defined genomic islands or on regions with lost synteny between the closely related strains. Often the genomic islands are delineated by mobile elements, many of which seem to be strain specific. This further supports the notion that these genomic islands were acquired rather than being part of the P. putida core genome.
The presence of many incomplete pathways as well as the duplication of pathways on the genomes of all four P. putida strains is striking. Examples include pathways involved in heavy metal resistances such as the copper resistance systems in P. putida W619 or the duplication of czcCBA in P. putida F1, KT2440 and GB-1, the breakdown of aromatic compounds, such as the presence of a complete plus an incomplete mph operon for the degradation of 3-hydroxyphenylpropionate (3-HPP) in W619, or the presence of two alternative pathways for the synthesis of IAA in P. putida W619 compared with only one pathway in the other three strains. The presence of various complete and incomplete pathways indicates that these strains, before adapting to their current biotopes, have a rich history of colonizing various biotopes, each of which required specific adaptations that were facilitated via the acquisition and reshuffling of new pathways. These adaptations also required the dedicated management of incoming systems to avoid functional redundancy (i.e. under stress when resources are scarce), their optimization and the economical interplay between various pathways genes, supported by a large diversity of regulators typically found in P. putida. A similar duplication and interplay of functions was described recently for the adaptation of C. metallidurans CH34 to environments polluted by heavy metals (Janssen et al., 2010). Whole-genome expression studies combined with metabolite analysis and proteomics should help to better understand the mechanisms for the interplay and roles of functions essential for the niche adaptation of these strains, and when combined with genome-scale metabolic reconstruction models (Nogales et al., 2008; Puchalka et al., 2008;), will provide a nonempirical basis for the development of biotechnological applications that can be further tailored as a function of strain-specific metabolic pathways.
As expected from the unique features of the niches from which the strains were isolated, P. putida F1 was the best adapted to degrade a variety of aromatic compounds, while W619 was more competent with regard to heavy metal resistances and beneficial effects on plant. A comparison between P. putida W619 and KT2440 is of special interest, as it could provide valuable clues to identify key functions for an endophytic lifestyle in comparison with survival in the rhizosphere. Noticeable differences include the presence of the ndvB gene in W619 involved in the production of β-(1,2)-glucan, a putative adhesin, a surface-adhesion calcium-binding outer membrane-like protein that was 24% identical to LapA and two genes putatively involved in adhesion that code for autotransporter proteins (secretion type V) with a pectin/lyase/pertactin domain. In addition, P. putida W619 seems to be a better producer of the plant growth-promoting compound IAA. However, as this inventory mainly relies on the comparison of the two strains for already known genes and mechanisms in plant-colonizing bacteria, genuine novel genes or functions may be overlooked. A combination of transcriptomics, proteomics, metabolomics and mutagenesis to study the plant colonization process will be of invaluable help in this respect: it will allow assigning new functions to putative genes and pathways, help to detect new proteins and confirm the metabolic potential of the strains. A similar approach is also required to better understand the mechanisms underlying the Mn(II)-oxidization ability of the various P. putida strains, because comparative genomics indicated that genes related to Mn(II) oxidation in GB-1 have homologs in P. putida KT2440, W619 and F1.
Acknowledgments
This work was supported by the US Department of Energy, Office of Science, BER, project number KP1102010 under contract DE-AC02-98CH10886. X.W., S.M., S.T. and D.v.d.L were supported by Laboratory Directed Research and Development funds (LDRD09-005) at the Brookhaven National Laboratory under contract with the US Department of Energy. Thanks are due to Claudine Médigue and the team of the Genoscope annotation platform for their help and the use of the MAGE system for the annotation and comparison of P. putida W619. Sequencing of P. putida W619 was performed at the Joint Genome Institute (JGI) under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344 and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Thanks are due to Max Mergeay for providing precious feedback on heavy metal resistances and their organization in P. putida.
Authors' contribution
X.W. and S.M. contributed equally to the manuscript.
Statement
Re-use of this article is permitted in accordance with the Terms and Conditions set out at http://wileyonlinelibrary.com/onlineopen#OnlineOpen_Terms
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Putative genomic islands identified on the chromosomes of Pseudomonas putida W619, KT2440, F1 and GB-1.
Table S2. Putative orthologous relationships between the chromosomes of the four Pseudomonas putida strains.
Table S3. Comparative analysis of the functional group distribution among bacteria, Gammaproteobacteria, Pseudomonas, and the four P. putida strains (KT2440, F1, GB-1 and W619), based on Clusters of Orthologous Groups of proteins (COGs) classification.
Table S4. Incomplete or truncated remnants of IS elements identified among the genomes of Pseudomonas putida KT2440,W619, F1 and GB-1.
Table S5. Complete overview of putative genes involved in the degradation of aromatic compounds identified on the genomes of Pseudomonas putida W619, KT2440, F1 and GB-1.
Table S6. Putative genes involved in Mn(II) oxidation identified on the genomes of Pseudomonas putida W619, KT2440, F1 and GB-1.
Table S7. Putative genes involved in the oxidative stress responce in Pseudomonas putida W619, KT2440, F1 and GB-1.
Table S8. Putative genes involved in siderophore synthesis and iron uptake.
Table S9. Putative genes involved in the motility of Pseudomonas putida strains.
Table S10. Putative genes involved in the pili and curli fibers biosynthesis.
Table S11. Conserved genes of the type VI secretion system of Pseudomonas putida strains compared with the three T6SS clusters identified in Pseudomonas aeruginosa.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alm RA, Mattick JS. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Braun V, Endriss F. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals. 2007;20:219–231. doi: 10.1007/s10534-006-9072-5. [DOI] [PubMed] [Google Scholar]
- Brouwers GJ, de Vrind JP, Corstjens PL, Cornelis P, Baysse C, de Vrind-de Jong EW. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl Environ Microb. 1999;65:1762–1768. doi: 10.1128/aem.65.4.1762-1768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker P, Altenbuchner J, Mattes R. Structure and function of the genes involved in mannitol, arabitol and glucitol utilization from Pseudomonas fluorescens DSM50106. Gene. 1998a;206:117–126. doi: 10.1016/s0378-1119(97)00574-x. [DOI] [PubMed] [Google Scholar]
- Brunker P, Hils M, Altenbuchner J, Mattes R. The mannitol utilization genes of Pseudomonas fluorescens are regulated by an activator: cloning, nucleotide sequence and expression of the mtlR gene. Gene. 1998b;215:19–27. doi: 10.1016/s0378-1119(98)00274-1. [DOI] [PubMed] [Google Scholar]
- Canovas D, Cases I, de Lorenzo V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol. 2003;5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- Caspi R, Tebo BM, Haygood MG. C-type cytochromes and manganese oxidation in Pseudomonas putida MnB1. Appl Environ Microb. 1998;64:3549–3555. doi: 10.1128/aem.64.10.3549-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro OA, Zorreguieta A, Ielmini V, Vega G, Ielpi L. Cyclic beta-(1,2)-glucan synthesis in Rhizobiaceae: roles of the 319-kilodalton protein intermediate. J Bacteriol. 1996;178:6043–6048. doi: 10.1128/jb.178.20.6043-6048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JS, Cooksey DA. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. P Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JS, Cooksey DA. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl Environ Microb. 1993;59:1671–1674. doi: 10.1128/aem.59.5.1671-1674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V, Flynn T, Niehaus WG, Wong B. Stress tolerance and pathogenic potential of a mannitol mutant of Cryptococcus neoformans. Microbiology. 1996a;142:937–943. doi: 10.1099/00221287-142-4-937. [DOI] [PubMed] [Google Scholar]
- Chaturvedi V, Wong B, Newman SL. Oxidative killing of Cryptococcus neoformans by human neutrophils. Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. J Immunol. 1996b;156:3836–3840. [PubMed] [Google Scholar]
- Choi EN, Cho MC, Kim Y, Kim CK, Lee K. Expansion of growth substrate range in Pseudomonas putida F1 by mutations in both cymR and todS, which recruit a ring-fission hydrolase CmtE and induce the tod catabolic operon, respectively. Microbiology. 2003;149:795–805. doi: 10.1099/mic.0.26046-0. [DOI] [PubMed] [Google Scholar]
- Clarke PH. The metabolic versatility of pseudomonads. Antonie Van Leeuwenhoek. 1982;48:105–130. doi: 10.1007/BF00405197. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Cornelis P, Bodilis J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ Microbiol Rep. 2009;1:256–262. doi: 10.1111/j.1758-2229.2009.00041.x. [DOI] [PubMed] [Google Scholar]
- Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals. 2009;22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- Corstjens PL, de Vrind JP, Westbroek P, de Vrind-de Jong EW. Enzymatic iron oxidation by Leptothrix discophora: identification of an iron-oxidizing protein. Appl Environ Microb. 1992;58:450–454. doi: 10.1128/aem.58.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean CR, Neshat S, Poole K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J Bacteriol. 1996;178:5361–5369. doi: 10.1128/jb.178.18.5361-5369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrind J, De Groot A, Brouwers GJ, Tommassen J, de Vrind-de Jong E. Identification of a novel Gsp-related pathway required for secretion of the manganese-oxidizing factor of Pseudomonas putida strain GB-1. Mol Microbiol. 2003;47:993–1006. doi: 10.1046/j.1365-2958.2003.03339.x. [DOI] [PubMed] [Google Scholar]
- de Vrind JP, Brouwers GJ, Corstjens PL, den Dulk J, de Vrind-de Jong EW. The cytochrome c maturation operon is involved in manganese oxidation in Pseudomonas putida GB-1. Appl Environ Microb. 1998;64:3556–3562. doi: 10.1128/aem.64.10.3556-3562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Mergeay M. Czc/cnr efflux: a three-component chemiosmotic antiport pathway with a 12-transmembrane-helix protein. Mol Microbiol. 1994;14:185–187. doi: 10.1111/j.1365-2958.1994.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Dos Santos VA, Heim S, Moore ER, Stratz M, Timmis KN. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- Douglas CJ, Staneloni RJ, Rubin RA, Nester EW. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985;161:850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque E, Segura A, Mosqueda G, Ramos JL. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol Microbiol. 2001;39:1100–1106. doi: 10.1046/j.1365-2958.2001.02310.x. [DOI] [PubMed] [Google Scholar]
- Eaton RW. p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RW. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Urgel M, Salido A, Ramos JL. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;182:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil H, Feil WS, Chain P, et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. P Natl Acad Sci USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandez A, Garcia JL, Diaz E. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl) propionate catabolic pathway of Escherichia coli K-12. J Bacteriol. 1997;179:2573–2581. doi: 10.1128/jb.179.8.2573-2581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- Folschweiller N, Schalk IJ, Celia H, Kieffer B, Abdallah MA, Pattus F. The pyoverdin receptor FpvA, a TonB-dependent receptor involved in iron uptake by Pseudomonas aeruginosa (review) Mol Membr Biol. 2000;17:123–133. doi: 10.1080/09687680050197356. [DOI] [PubMed] [Google Scholar]
- Francis CA, Tebo BM. cumA multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl Environ Microb. 2001;67:4272–4278. doi: 10.1128/AEM.67.9.4272-4278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geszvain K, Tebo BM. Identification of a two-component regulatory pathway essential for Mn(II) oxidation in Pseudomonas putida GB-1. Appl Environ Microb. 2009;1:1–2. doi: 10.1128/AEM.02473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D, Zylstra GJ, Chauhan S. Biotransformations catalyzed by toluene dioxygenase from Pseudomonas putida F1. In: Silver S, Chakrabarty AM, Iglewski B, Kaplan S, editors. Pseudomonas: Biotransformations, Pathogenesis, and Evolving Biotechnology. Washington, DC: American Society for Microbiology Press; 1990. pp. 121–132. [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26:227–242. [Google Scholar]
- Grass G, Fan B, Rosen BP, Lemke K, Schlegel HG, Rensing C. NreB from Achromobacter xylosoxidans 31A Is a nickel-induced transporter conferring nickel resistance. J Bacteriol. 2001;183:2803–2807. doi: 10.1128/JB.183.9.2803-2807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse C, Grass G, Anton A, et al. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin PF, Stærfeldt H-H, Rotenberg E, Binnewies TT, Benham CJ, Ussery D. GeneWiz browser: an interactive tool for visualizing sequenced chromosomes. Stand Genomic Sci. 2009;1:204–215. doi: 10.4056/sigs.28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritha A, Sagar KP, Tiwari A, Kiranmayi P, Rodrigue A, Mohan PM, Singh SS. MrdH, a novel metal resistance determinant of Pseudomonas putida KT2440, is flanked by metal-inducible mobile genetic elements. J Bacteriol. 2009;191:5976–5987. doi: 10.1128/JB.00465-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Huijberts GNM, Eggink G. Production of poly(3-hydroxyalkanoates) by Pseudomonas putida KT2442 in continuous cultures. Appl Microbiol Biot. 1996;46:233–239. [Google Scholar]
- Huijberts GNM, Eggink G, Dewaard P, Huisman GW, Witholt B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl Environ Microb. 1992;58:536–544. doi: 10.1128/aem.58.2.536-544.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ielpi L, Dylan T, Ditta GS, Helinski DR, Stanfield SW. The ndvB locus of Rhizobium meliloti encodes a 319-kDa protein involved in the production of beta-(1,2)-glucan. J Biol Chem. 1990;265:2843–2851. [PubMed] [Google Scholar]
- Janssen PJ, Van Houdt R, Moors H, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One. 2010;5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings DB, Daub ME, Pharr DM, Williamson JD. Constitutive expression of a celery mannitol dehydrogenase in tobacco enhances resistance to the mannitol-secreting fungal pathogen Alternaria alternata. Plant J. 2002;32:41–49. doi: 10.1046/j.1365-313x.2001.01399.x. [DOI] [PubMed] [Google Scholar]
- Jennings DH. Polyol metabolism in fungi. Adv Microb Physiol. 1984;25:149–193. doi: 10.1016/s0065-2911(08)60292-1. [DOI] [PubMed] [Google Scholar]
- Jimenez JI, Minambres B, Garcia JL, Diaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol. 2002;4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- Lau PC, Wang Y, Patel A, et al. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. P Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol R. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Breier AM, Higgins NP. Microarray analysis of transposition targets in Escherichia coli: the impact of transcription. P Natl Acad Sci USA. 2004;101:9780–9785. doi: 10.1073/pnas.0400745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Bueno MA, Tobes R, Rey M, Ramos JL. Detection of multiple extracytoplasmic function (ECF) sigma factors in the genome of Pseudomonas putida KT2440 and their counterparts in Pseudomonas aeruginosa PA01. Environ Microbiol. 2002;4:842–855. doi: 10.1046/j.1462-2920.2002.00371.x. [DOI] [PubMed] [Google Scholar]
- Matthijs S, Laus G, Meyer JM, Abbaspour-Tehrani K, Schafer M, Budzikiewicz H, Cornelis P. Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals. 2009;22:951–964. doi: 10.1007/s10534-009-9247-y. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Mellano MA, Cooksey DA. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988;170:4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M, Monchy S, Vallaeys T, et al. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27:385–410. doi: 10.1016/S0168-6445(03)00045-7. [DOI] [PubMed] [Google Scholar]
- Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol. 2000;174:135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- Molina-Henares MA, García-Salamanca A, Molina-Henares J, de la Torre AJ, Herrera MC, Ramos JL, Duque E. Functional analysis of aromatic biosynthetic pathways in Pseudomonas putida KT2440. Microbial Biotech. 2009;2:91–100. doi: 10.1111/j.1751-7915.2008.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Henares MA, de la Torre J, Garcia-Salamanca A, Molina-Henares AJ, Herrera MC, Ramos JL, Duque E. Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome-wide mutant library. Environ Microbiol. 2010;12:1468–1485. doi: 10.1111/j.1462-2920.2010.02166.x. [DOI] [PubMed] [Google Scholar]
- Monchy S, Benotmane MA, Wattiez R, et al. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology. 2006;152:1765–1776. doi: 10.1099/mic.0.28593-0. [DOI] [PubMed] [Google Scholar]
- Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, Mergeay M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J Bacteriol. 2007;189:7417–7425. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- Moon CD, Zhang XX, Matthijs S, Schafer M, Budzikiewicz H, Rainey PB. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. 2008;8:7. doi: 10.1186/1471-2180-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T. Travels of a Pseudomonas, from Japan around the world. Environ Microbiol. 2002;4:782–786. doi: 10.1046/j.1462-2920.2002.00310.x. [DOI] [PubMed] [Google Scholar]
- Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Nogales J, Palsson BO, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt 1):185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- Okazaki M, Sugita T, Shimizu M, et al. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl Environ Microb. 1997;63:4793–4799. doi: 10.1128/aem.63.12.4793-4799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DL, Sposito G, Tebo BM. Manganese(III) binding to a pyoverdine siderophore produced by a manganese(II)-oxidizing bacterium. Geochim Cosmochim Ac. 2004;68:4809–4820. [Google Scholar]
- Paulsen IT, Press CM, Ravel J, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix P, Keane A, Patel A, Bergeron H, Ghoshal S, Lau PC. Characterization of a new solvent-responsive gene locus in Pseudomonas putida F1 and its functionalization as a versatile biosensor. Environ Microbiol. 2003;5:1309–1327. doi: 10.1111/j.1462-2920.2003.00426.x. [DOI] [PubMed] [Google Scholar]
- Polack B, Dacheux D, Delic-Attree I, Toussaint B, Vignais PM. The Pseudomonas aeruginosa fumC and sodA genes belong to an iron-responsive operon. Biochem Biophys Res Co. 1996;226:555–560. doi: 10.1006/bbrc.1996.1393. [DOI] [PubMed] [Google Scholar]
- Preston GM, Bertrand N, Rainey PB. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol. 2001;41:999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- Puchalka J, Oberhardt MA, Godinho M, et al. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol. 2008;4:e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta D, McCarty R, Bandarian V, Rensing C. The copper-inducible cin operon encodes an unusual methionine-rich azurin-like protein and a pre-Q0 reductase in Pseudomonas putida KT2440. J Bacteriol. 2007;189:5361–5371. doi: 10.1128/JB.00377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. P Natl Acad Sci USA. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosson RA, Nealson KH. Manganese binding and oxidation by spores of a marine Bacillus. J Bacteriol. 1982;151:1027–1034. doi: 10.1128/jb.151.2.1027-1034.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW. Bacterial volatiles promote growth in Arabidopsis. P Natl Acad Sci USA. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Segura A, Hurtado A, Duque E, Ramos JL. Transcriptional phase variation at the flhB gene of Pseudomonas putida DOT-T1E is involved in response to environmental changes and suggests the participation of the flagellar export system in solvent tolerance. J Bacteriol. 2004;186:1905–1909. doi: 10.1128/JB.186.6.1905-1909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain JC, Gibson DT. Oxidation of substituted phenols by Pseudomonas putida F1 and Pseudomonas sp strain Js6. Appl Environ Microb. 1988;54:1399–1404. doi: 10.1128/aem.54.6.1399-1404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain JC, Zylstra GJ, Blake CK, Gibson DT. Monohydroxylation of phenol and 2,5-dichlorophenol by toluene dioxygenase in Pseudomonas putida F1. Appl Environ Microb. 1989;55:2648–2652. doi: 10.1128/aem.55.10.2648-2652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, van der Lelie D. Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microb. 2005;71:8500–8505. doi: 10.1128/AEM.71.12.8500-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S, Garafola C, Monchy S, et al. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microb. 2009;75:748–757. doi: 10.1128/AEM.02239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S, van der Lelie D, Hoffman A, et al. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 2010;6:e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebo BM, Johnson HA, McCarthy JK, Templeton AS. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 2005;13:421–428. doi: 10.1016/j.tim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Saulino ET, Hultgren SJ. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr Opin Microbiol. 1998;1:223–231. doi: 10.1016/s1369-5274(98)80015-5. [DOI] [PubMed] [Google Scholar]
- Timmis KN. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol. 2002;4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- Tucker NP, D'Autreaux B, Studholme DJ, Spiro S, Dixon R. DNA binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse proteobacteria. J Bacteriol. 2004;186:6656–6660. doi: 10.1128/JB.186.19.6656-6660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussery DW, Kiil K, Lagesen K, Sicheritz-Ponten T, Bohlin J, Wassenaar TM. The Genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 2009;6:140–157. doi: 10.1159/000235768. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Huijsduijnen RAMH, Alblas SW, De Rijk RH, Bol JF. Induction by salicylic acid of pathogenesis-related proteins and resistance to alfalfa mosaic virus infection in various plant species. J Gen Virol. 1986;67:2135–2143. [Google Scholar]
- Velez H, Glassbrook NJ, Daub ME. Mannitol metabolism in the phytopathogenic fungus Alternaria alternata. Fungal Genet Biol. 2007;44:258–268. doi: 10.1016/j.fgb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Velez H, Glassbrook NJ, Daub ME. Mannitol biosynthesis is required for plant pathogenicity by Alternaria alternata. FEMS Microbiol Lett. 2008;285:122–129. doi: 10.1111/j.1574-6968.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- Vencato M, Tian F, Alfano JR, et al. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol Plant Microbe In. 2006;19:1193–1206. doi: 10.1094/MPMI-19-1193. [DOI] [PubMed] [Google Scholar]
- Villalobos M, Toner B, Bargar J, Sposito G. Characterization of the manganese oxide produced by Pseudomonas putida strain MnB1. Geochim Cosmochim Ac. 2003;67:2649–2662. [Google Scholar]
- Vodovar N, Vallenet D, Cruveiller S, et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- Wackett LP, Gibson DT. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microb. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel C, Schmidt A, Ruckert B, Lurz R, Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16:6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinel C, Nelson KE, Tummler B. Global features of the Pseudomonas putida KT2440 genome sequence. Environ Microbiol. 2002;4:809–818. doi: 10.1046/j.1462-2920.2002.00331.x. [DOI] [PubMed] [Google Scholar]
- Weyens N, Van Der Lelie D, Artois T, et al. Bioaugmentation with engineered endophytic bacteria improves contaminant fate in phytoremediation. Environ Sci Technol. 2009;43:9413–9418. doi: 10.1021/es901997z. [DOI] [PubMed] [Google Scholar]
- Williams PA, Jones RM, Shaw LE. A third transposable element, ISPpu12, from the toluene-xylene catabolic plasmid pWW0 of Pseudomonas putida mt-2. J Bacteriol. 2002;184:6572–6580. doi: 10.1128/JB.184.23.6572-6580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A. 2009. Genetic analysis reveals complex Mn(II) oxidation regulation in Pseudomonas putida GB-1. PhD Thesis, Oregon Health & Science University, Portland, OR.
- Yee TW, Smith DW. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. P Natl Acad Sci USA. 1990;87:1278–1282. doi: 10.1073/pnas.87.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama H, Nakae T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology. 1996;142(Pt 8):2137–2144. doi: 10.1099/13500872-142-8-2137. [DOI] [PubMed] [Google Scholar]
- Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol Lett. 2008;288:118–124. doi: 10.1111/j.1574-6968.2008.01339.x. [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zahringer U, Gerber I, et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. P Natl Acad Sci USA. 2004;101:15811–15816. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra GJ, McCombie WR, Gibson DT, Finette BA. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microb. 1988;54:1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


