Abstract
Transcription of the herpes simplex virus 1 (HSV–1) immediate early (IE) genes is determined by multiprotein enhancer complexes. The core enhancer assembly requires the interactions of the POU-homeodomain protein Oct–1, the viral transactivator αTIF and the cellular factor C1 (HCF). In this context, the C1 factor interacts with each protein to assemble the stable enhancer complex. In addition, the IE enhancer cores contain adjacent binding sites for other cellular transcription factors such as Sp1 and GA-binding protein (GABP). In this study, a direct interaction of the C1 factor with GABP is demonstrated, defining the C1 factor as the critical coordinator of the enhancer complex assembly. In addition, mutations that reduce the GABP transactivation potential also impair the C1–GABP interaction, indicating that the C1 factor functions as a novel coactivator of GABP-mediated transcription. The interaction and coordinated assembly of the enhancer proteins by the C1 factor may be critical for the regulation of the HSV lytic–latent cycle.
Keywords: coactivator/herpes simplex virus/protein interactions/transcription
Introduction
After an initial infection with herpes simplex virus (HSV), the virus undergoes lytic replication and subsequently establishes a latent state in the neurons of the host's sensory ganglia. Various stimuli such as stress, hormonal alterations, UV exposure or tissue damage can result in the reactivation of lytic replication and recurrent disease. While the mechanisms governing the establishment of the latent state and the reactivation process remain unknown, the lytic replication cycle has been well characterized and has been a model for the biochemical analysis of gene expression studies (reviewed in Roizman and Sears, 1996).
Upon infection, the viral immediate early (IE) genes are expressed under the control of reiterated inducible enhancer elements (Mackem and Roizman, 1982; Kristie and Roizman, 1984). These elements nucleate the assembly of multiprotein complexes containing both viral (αTIF) and cellular proteins [Oct–1, GA-binding protein (GABP), Sp1 and C1] (Jones and Tjian, 1985; McKnight et al., 1987; Gerster and Roeder, 1988; O'Hare and Goding, 1988; Preston et al., 1988; Triezenberg et al., 1988; Kristie et al., 1989, 1995; Kristie and Sharp, 1990, 1993; Wilson et al., 1993a,b). Oct–1 binds the common octamer sequence in the enhancer elements while the viral transactivator, αTIF, determines the HSV–1 site specificity by interaction with both Oct–1 and HSV-specific DNA sequences directly adjacent to the octamer core (Stern et al., 1989; Kristie and Sharp, 1990; Stern and Herr, 1991; Pomerantz et al., 1992). This interaction is of relatively low affinity, and the formation of the stable enhancer complex further requires the cellular C1 factor (HCF) (Gerster and Roeder, 1988; Kristie et al., 1989; Katan et al., 1990; Kristie and Sharp, 1990; Xiao and Capone, 1990). This factor does not bind the enhancer sequences but rather interacts via direct protein–protein interactions with αTIF (Hayes and O'Hare, 1993; Kristie and Sharp, 1993; Wilson et al., 1993b) and Oct–1 (data not published). A unique protein, the C1 factor is actually a family of related polypeptides resulting from site-specific proteolytic processing at reiterated amino acid sequences in the central domain of the precursor (Wilson et al., 1993a, 1995; Kristie et al., 1995). In addition to its role in the regulation of HSV–1 IE gene expression, this factor has also been implicated in the control of cell cycle (Goto et al., 1997), although the biochemical mechanisms remain unclear.
Additional cellular transcription factors (Sp1 and GABP) have also been shown to contribute significantly to the activation of the viral IE gene expression, and DNA recognition sequences for each of these can be found adjacent to the IE enhancer cores (Jones and Tjian, 1985; Triezenberg et al., 1988; LaMarco and McKnight, 1989). For GABP, point mutations in the GA-binding site result in decreased αTIF-dependent expression, suggesting that the protein plays a role in the enhancer-mediated transcription of the IE genes (Triezenberg et al., 1988). A member of the ETS domain transcription factor family, GABP (NRF2/E4TF1) is a heterodimeric factor consisting of a DNA-binding subunit (α) and a transcription activation subunit (β) (Thompson et al., 1991; Virbasius et al., 1993; Watanabe et al., 1993; Gugneja et al., 1995; Batchelor et al., 1998). This factor has been the focus of many studies that have defined functional domains of the protein as well as implicated the factor in the regulation of a large number of viral and cellular promoters (Triezenberg et al., 1988; Virbasius et al., 1993; Watanabe et al., 1993; Rosmarin et al., 1995; Flory et al., 1996; Ouyang et al., 1996; Hoffmeyer et al., 1998; Vassias et al., 1998). In addition, the activation of the transactivation potential of GABP has been shown to be regulated by stress-activated and mitogen-activated protein kinase pathways (Flory et al., 1996; Ouyang et al., 1996; Hoffmeyer et al., 1998). However, while synergistic activation by GABP in conjunction with other transcription factors has been demonstrated for numerous promoters (Rosmarin et al., 1995, 1998; Boccia et al., 1996; Vassias et al., 1998; Nuchprayoon et al., 1999), no direct interaction of this factor with other proteins has been shown.
In the studies presented here, a direct interaction of GABP with the C1 factor is demonstrated, defining the C1 factor as the critical coordinator of the HSV IE enhancer assembly. In addition, residues that are critical for the transactivation potential of GABP are also critical for the GABP–C1 interaction, implying that transactivation by GABP is mediated by the novel C1 coactivator. While this interaction plays a role in the regulation of the IE genes during HSV–1 lytic replication, it may also play a critical role in the expression of the IE genes during reactivation of the virus from the latent state.
Results
Interaction of GABP with the C1 factor in a yeast two-hybrid system
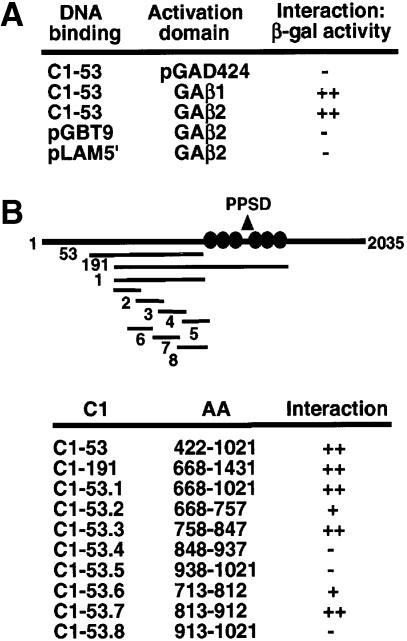
While numerous studies have focused upon the roles of Oct–1 and αTIF in the regulation of HSV–1 IE genes, little is known about the function of the C1 factor due to its relatively recent isolation. To determine the roles that this protein may play in various cellular processes, the protein was divided into segments, fused to the GAL4 DNA-binding domain, and used to screen for cellular interacting proteins in the yeast two-hybrid system. In a screen using the C1-53 (amino acids 422–1021) region of the C1 protein, three positive clones were isolated. As shown in Figure 1A, one cDNA was identified as coding for amino acids 60–347 of the transcription factor GABPβ2. The interaction was specific for C1-53 as no interaction was detected between GABPβ2 and either the GAL4 DNA-binding domain alone or the control GAL4–lamin fusion protein (Figure 1A). Furthermore, several studies have previously shown that GABP binds to sequences adjacent to the enhancer core in the HSV–1 IE regulatory domain and that these elements are critical for the enhanced αTIF-dependent transcription of these genes (Triezenberg et al., 1988; Wu et al., 1994; Douville et al., 1995). The C1–GABP interaction and the assembly of GABP into the core enhancer complex could therefore provide a biochemical mechanism for the observed GABP–αTIF synergistic enhancement of IE gene expression.
Fig. 1. The C1 factor interacts with GABPβ in the yeast two-hybrid system. (A) The indicated GAL4 DNA-binding and activation domain fusion pairs were assayed for β-galactosidase expression in the yeast strain HF7c. pGBT9 and pLAM5′ represent the vector and lamin fusion protein controls, respectively. (B) Schematic representation of the C1 factor indicating the fragments of the C1 factor that were fused to the GAL4 DNA-binding domain. The circles represent the sites of processing in the C1 proteolytic processing domain (PPSD). The indicated fusions were transformed into HF7c containing the GAL4 activation domain–GABPβ2 fusion and assayed for β-galactosidase expression. (–), (+) and (++) indicate the relative levels of β-galactosidase expression.
As previously noted, GABP is a heterodimeric member of the ETS transcription factor family with an α DNA-binding subunit and a β transactivation subunit. In addition, several forms of GABPβ (GAβ1 and GAβ2) have been identified that have distinct C–termini (Thompson et al., 1991; de la Brousse et al., 1994; Gugneja et al., 1995; Batchelor et al., 1998). GAβ1 contains a leucine zipper and can homodimerize to form heterotetramers with GAα, while GAβ2 does not and forms only heterodimers. Therefore, GAβ1 was tested to determine if the C–terminal domain of GAβ2 was significant for the C1–GAβ interaction. As shown in Figure 1A, both GAβ1 and GAβ2 were fully capable of interacting with C1-53, suggesting that this divergent domain did not contain determinants that are critical for this interaction.
The C1 factor domain that is important for the C1–GAβ interaction was mapped using clones encoding segments of the C1 protein as shown in Figure 1B. The overlapping clone C1-191 (C1 amino acids 668–1431) also exhibited a strong interaction with GAβ. Therefore, a series of clones encoding segments of the C1-53/C1-191 sequences was tested. C1-53.3 and C1-53.7 interacted with GAβ as strongly as the original C1-53 protein. C1-53.2 and C1-53.6 exhibited a significantly weaker interaction, while C1-53.4 and C1-53.5 did not interact. These results defined the primary determinants for C1–GAβ interaction as amino acids 813–847, with sequences directly N–terminal (amino acids 758–812) contributing weakly to the interaction.
Interaction of GABP with C1 in vitro: requirement for the GAβ transactivation domain
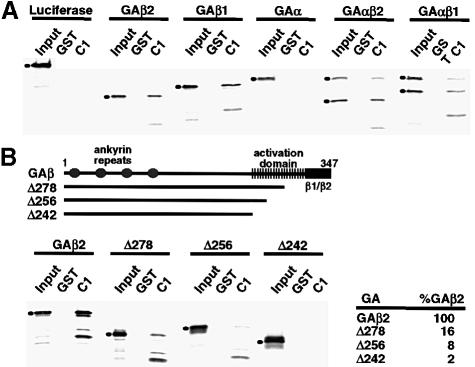
The interaction of GABP and C1 was confirmed by GST pull-down assay using GST–53.1 (amino acids 668–1021) and in vitro synthesized GAβ1 or GAβ2. Equivalent molar amounts of GST or GST–53.1 were bound to glutathione beads and incubated with 3–5 fmol of the appropriate in vitro translated protein. After extensive washing, the proteins were eluted, resolved by SDS–PAGE and quantitated. As shown in Figure 2A, both GAβ2 and GAβ1 bound the GST–53.1 with the same relative affinity (5.7 or 6.6% of the input protein, respectively). In contrast, no significant interaction was seen between GST–53.1 and GAα (0.1% of input). However, when GAα was co-translated with either GAβ1 or GAβ2, the heteromeric GABP was bound efficiently by the C1 protein, suggesting that the interaction is determined primarily by the GAβ subunit.
Fig. 2. The C1 factor interacts with the GABPβ transactivation domain. GST pull-down reactions were done as described in Materials and methods using the in vitro transcription–translation products indicated at the top and either GST or GST–53.1 (C1). The filled circles indicate the full-length GABP and luciferase control proteins. An aliquot (10%) of the input protein extract and bound proteins were resolved in 12% SDS–PAGE gels, transferred to nitrocellulose and quantitated as described. (A) The indicated labeled protein (3 fmol) was used in each reaction. (B) The deletions of GAβ are shown below a schematic representation of the wild-type protein. The regions containing the ankyrin repeats, transcription activation domain and sequences that distinguish GAβ1 and GAβ2 are illustrated. The amount of each deletion protein that was precipitated with GST–53.1 is expressed as a percentage of the full-length protein. In each reaction, 5 fmol of the indicated labeled protein was used.
The general architecture of GAβ is shown in Figure 2B. The protein contains a series of ankyrin repeats at the N–terminus for interaction with GAα (Thompson et al., 1991; Batchelor et al., 1998), a region that is divergent between β1 and β2 at the C–terminus (Thompson et al., 1991; Watanabe et al., 1993; Gugneja et al., 1995) and a well characterized 69 amino acid transactivation domain (amino acids 246–315) (Gugneja et al., 1995, 1996). As shown in Figures 1A and 2A, the divergent region is not a primary determinant for C1–GAβ interaction. Therefore, to define the interaction domain further, a series of deletions in GAβ were produced by in vitro synthesis and tested for their ability to interact with GST–53.1 (Figure 2B, top). As shown (Figure 2B, bottom), GAΔ278 deletes 37 amino acids of the transactivation domain and exhibits only 16% of the C1–GAβ binding activity as compared with the full-length protein. Deletion of additional residues within the transactivation domain (GAΔ256) or the entire domain (GAΔ242) further compromises the ability of the protein to bind the C1 factor (8 and 2% of the full-length protein, respectively), indicating that the transactivation domain contains the primary determinants for the GA–C1 interaction.
Residues required for GAβ–C1 interaction correlate directly with those required for GABP transactivation
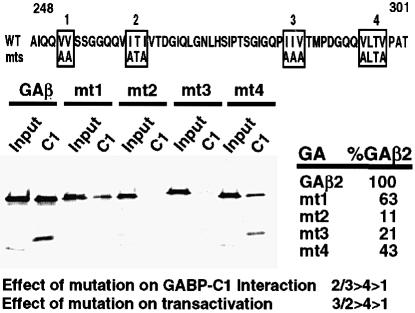
The transactivation domain of GAβ (Figure 3, top) contains hydrophobic clusters and multiple glutamine residues (Gugneja et al., 1995). While alteration of the glutamines had no effect upon the transactivation activity, mutations in four hydrophobic clusters significantly reduce the transactivation potential (Gugneja et al., 1996). To determine if these residues were also significant for GA–C1 interaction, these mutant proteins were assayed for their ability to bind GST–53.1 (Figure 3, bottom). GAβ proteins containing mutations in box 1 have little effect (63% of wild-type activity) upon either GA–C1 interaction or GABP transactivation. In contrast, mutations in boxes 2, 3 and 4 significantly affect both C1–GA binding and GAβ transactivation. In addition, the severity of the mutation for transactivation directly correlates with the impairment of C1–GA interaction and further suggests that the hydrophobic residues in these boxes are the major determinants for both activities.
Fig. 3. Residues required for GAβ transactivation define the interaction with the C1 factor. The amino acid sequence of the GAβ transactivation domain is shown. The hydrophobic residues that are critical for GAβ transactivation are boxed (1–4), with the sequence of the point mutations indicated below (mts) (Gugneja et al., 1996). Wild-type or point mutant proteins (4 fmol) were used in pull-down assays as described with 3 μg of GST–53.1. The percentage of precipitated protein is shown relative to the wild-type GAβ2 protein. The significance of each set of residues for transactivation and for C1 interaction is summarized.
Specific repression of GABP-mediated transcription by the C1–GABP interaction domain
The significance of the C1–GABP interaction for GA-mediated transcription in vivo was assessed by co-transfection of HeLa cells with constructs expressing a Gal4–GA activation domain fusion protein, Gal4–luciferase reporter and C1-53.1a (C1–GABP interaction domain). As illustrated in Figure 4A, expression of the C1–GABP interaction domain significantly inhibited the Gal4–GA-mediated expression of the reporter gene in a dose-dependent manner. In contrast, expression of C1-53.1a had no effect upon the control Gal4–VP16-mediated transcription of the reporter, indicating that the repression was GABP activation domain specific. As C1.53.1a contains only the C1–GABP interaction domain and not the transactivation functions, repression of Gal4–GA-dependent transcription suggests that C1.53.1a functions as a competitive inhibitor for wild-type endogenous C1 factor.
Fig. 4. The domain of the C1 factor required for C1–GABP interaction can inhibit GABP-dependent transcriptional activation. (A) HeLa cells were co-transfected with plasmids encoding Gal4–GA activation domain fusion protein, Gal4–luciferase reporter and increasing amounts of C1-53.1a. The results of five independent experiments are plotted relative to the expression of the reporter in the absence of C1-53.1a. The control experiment contained Gal4–VP16 in place of Gal4–GA activation domain. (B) Gel mobility assays were done as described in Materials and methods. Reactions contained nuclear extracts of HeLa cells transfected with control vector (lanes 1–3), Gal4 DNA-binding domain (Gal4, lanes 4–6) or Gal4–GA (lanes 7–9). Purified GST–53.1 (0, 100 or 200 ng) was included in the reactions as indicated at the top of the gel.
In similar experiments, co-transfection of a construct expressing the full-length C1 factor resulted in a consistent but low level stimulation (1.7- to 2.3-fold) of the Gal4–GA-mediated transcription, possibly due to the presence of high levels of endogenous C1 factor in tissue culture cells (data not shown).
The specific interaction of these proteins is illustrated by gel shift analysis of reactions containing extracts of mammalian cells transfected with control vector, the Gal4 DNA-binding domain or the Gal4–GA fusion (Figure 4B). Inclusion of increasing amounts of purified Cl.53.l in these reactions results in the formation of a novel complex only in reactions containing the Gal4–GA protein (Figure 4B, lanes 8 and 9).
Interaction of C1 with GABP derived from nuclear extracts of mammalian cells and bacterial expression strains
As shown in Figure 5A, the interaction of native GABP and C1 can be detected using mammalian cell nuclear extracts. GST and GST53.1 were incubated with nuclear extracts from HEK293 cells and the bound proteins were analyzed by Western blotting using anti-GAα and anti-GAβ antisera. Interestingly, all forms of GAβ were precipitated efficiently from the extracts. In addition, GAα was also precipitated, probably due to its association with GAβ in the heterodimer. No specific protein was detected using a control anti-Oct–1 serum.
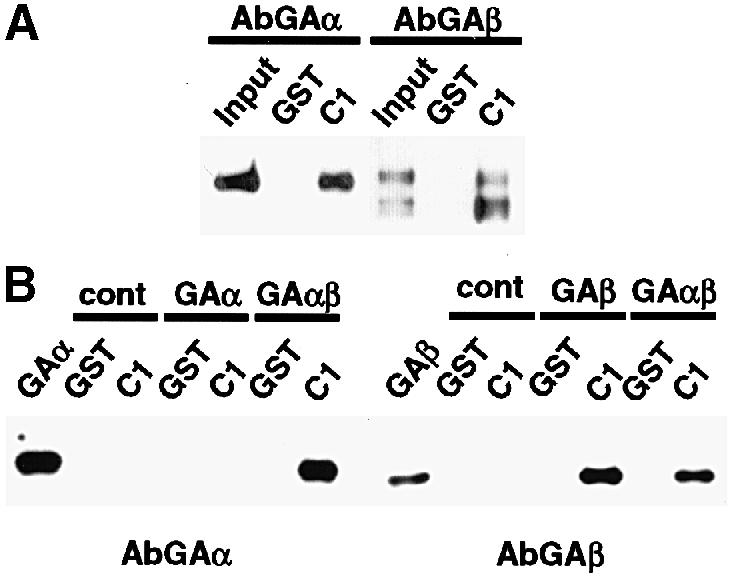
Fig. 5. The C1 factor interacts with GABP derived from either mammalian cell or bacterial expression extracts. GST pull-downs were done using GST or C1 (GST–53.1). The source of GABP was nuclear extracts of HEK293 cells (A) or bacterial strains expressing the appropriate subunits (B). The reactions were resolved on 8–16% SDS–PAGE gels, transferred to Immobilon and probed with affinity-purified antibodies to either GAα (AbGAα) or GAβ (AbGAβ). Control reactions (cont) contained no added source of GABP.
In each of the experiments presented here, the source of the GABP subunit proteins has been reticulocyte lysates or nuclear extracts from mammalian cells. Therefore, it remains possible that modification of the GABP subunits may be required for the interaction or that additional cellular proteins may play a role. It has been demonstrated that GABP transactivation activity is regulated by MAPK and SAPK pathways (Flory et al., 1996; Ouyang et al., 1996; Hoffmeyer et al., 1998). In contrast to some other transcription factors, this regulation is not at the level of subcellular localization or DNA-binding activity, suggesting that regulation of GABP protein–protein interactions may be the target. To determine if modification or additional cellular proteins are essential for C1–GABP interaction, GABP subunits were expressed and purified from extracts of bacterial expression strains. Purified GAβ, GAα or GAα/β were tested in GST pull-down assays using either GST or GST–53.1 (Figure 5B). In these assays, GAβ bound efficiently to GST–53.1. Similarly to the results presented above, GAα did not exhibit significant binding to C1 (1% of input protein) unless present in heterodimeric form with GAβ (12% of input). The data demonstrate that while GAα may contribute to the C1 interaction, the primary interaction is determined by GAβ. Furthermore, this interaction does not require either modification of GABP subunits or additional cellular proteins.
Discussion
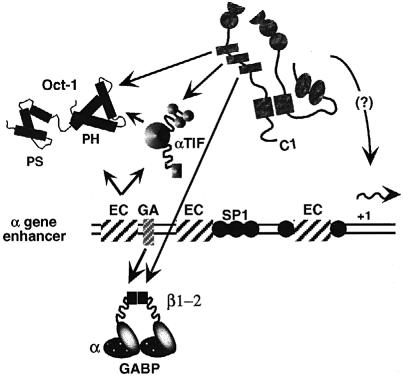
Regulation of the HSV–1 IE genes is determined by reiterated inducible enhancer elements that nucleate core multiprotein assemblies containing both viral (αTIF) and cellular proteins (Oct–1 and C1 factor). In addition, the regulatory domain contains recognition sites for other cellular factors such as GABP and Sp1 adjacent to the minimal enhancer core (Figure 6). The analysis of these assemblies has been a model for the regulation of RNA polymerase II-directed eukaryotic gene expression. The assembly of the enhancer core requires the binding of the POU-homeodomain protein, Oct–1, which recognizes the octamer motif in the enhancer by virtue of cooperative binding of the POU-specific and POU-homeo subdomains (Sturm et al., 1988; Kristie and Sharp, 1990; Verrijzer et al., 1990; Klemm et al., 1994). The viral transactivator, αTIF, binds HSV-specific DNA sequences directly adjacent to the octamer motif and interacts directly with helix 2 of the Oct–1 POU-homeobox (Stern and Herr, 1991; Pomerantz et al., 1992). However, the affinity of the ternary complex is low, requiring the interaction of the cellular C1 factor for stable assembly. This unusual factor is a family of related polypeptides that are derived from a 220 kDa precursor by site-specific proteolysis at a series of 20 amino acid sites within the central domain of the protein. The factor interacts with αTIF with high affinity and with Oct–1 with low affinity (T.M.Kristie, unpublished observations), resulting in the stabilization of the quaternary or minimal enhancer complex.
Fig. 6. Schematic representation of an HSV IE gene enhancer domain and the protein interactions involved in the complex assembly. The location of the reiterated enhancer cores (EC) and binding sites for GABP and Sp1 are shown. The subdomains of the Oct–1 POU-homeodomain (PS, POU-specific and PH, POU-homeo) are illustrated.
Several studies have contributed to the analysis of the domain structure of the C1 factor. In addition to the proteolytic processing domain, the protein contains an N–terminal ‘kelch’ domain that interacts with αTIF (La Boissiere et al., 1997; Wilson et al., 1997; Hughes et al., 1999), L-ZIP (Luman) (Freiman and Herr, 1997; Lu et al., 1997, 1998) and several cellular transcription factors (J.L.Vogel and T.M.Kristie, unpublished observations). This domain (amino acids 1–380) is sufficient for the assembly of the minimal enhancer complex in vitro; however, additional sequences may play a significant role in the assembly and activation of the full enhancer complex. Interestingly, a temperature-sensitive mutation in the kelch domain (amino acid 134) has also implicated C1 in the control of cell cycle progression (Goto et al., 1997), although rescue of the cycle block requires sequences C–terminal to the kelch region (Wilson et al., 1997), again suggesting that additional domains may be critical. The C–terminus of the protein contains a nuclear localization signal that also provides a mechanism for the nuclear import of associated proteins such as αTIF (La Boissiere et al., 1999) as well as a putative transcription activation domain (J.L.Vogel and T.M.Kristie, unpublished observations).
As noted, the IE regulatory domain also contains sites for additional transcription factors such as GABP and Sp1. Several studies have indicated the significance of these sites for the enhancer-dependent stimulation of HSV–1 IE transcription by either site-directed mutagenesis or in vitro transcription assays (Jones and Tjian, 1985; Triezenberg et al., 1988; Wu et al., 1994). In the studies presented here, it has been demonstrated that the C1 factor interacts directly with GABP. In addition, studies in progress indicate that C1 also interacts directly with Sp1 (J.L.Vogel and T.M.Kristie, unpublished observation), suggesting that the enhancer core assembly makes protein contacts with additional transcription factors to form a more complex regulatory unit. Interestingly, the GA-binding sites do not contribute significantly to the basal level expression of the IE genes in the absence of an induced TAATGARAT site (Wu et al., 1994), suggesting that GABP activation is dependent upon the formation of the C1 enhancer complex (i.e. the presence of the C1 factor). In addition, the juxtaposition of TAATGARAT and GA sites is conserved in the five IE gene promoters, and alteration of this spacing results in loss of the GA-dependent stimulation of the IE genes (Bailey and Thompson, 1992). These studies support the hypothesis that the GABP transcriptional activity is dependent upon the assembly of the C1 factor enhancer complex and the proximity of the C1 factor. The interaction of C1 and GABP demonstrated here thereby further defines the role of the C1 protein as the critical coordinator of the enhancer-regulatory protein complex.
GABP, a member of the ETS family of transcription factors, is a heterodimeric protein consisting of a DNA-binding subunit (α) and a transactivation subunit (β). It has been well characterized and demonstrated to be an important component of the regulation of numerous viral and cellular promoters. Interestingly, GABP has been shown to stimulate transcription synergistically with other factors such as Sp1 (Rosmarin et al., 1998; Vassias et al., 1998; Nuchprayoon et al., 1999), C/EBPα (Boccia et al., 1996) and PU.1 (Rosmarin et al., 1995), although no direct interaction between these proteins has been demonstrated. In contrast, attempts to demonstrate interactions between GABP and Sp1 have been unsuccessful (Rosmarin et al., 1998; J.L.Vogel and T.M.Kristie, unpublished observation). The interaction of GABP and Sp1 with the C1 factor suggests that the synergism may be a result of interactions with other factors or coactivators such as C1.
Several studies have defined the GABP transactivation domain in the GAβ subunit, and mutational analysis has indicated residues that are critical for the transactivation potential of the protein (Gugneja et al., 1995, 1996). Significantly, these same residues are required for direct interaction of GAβ and the C1 factor, and the severity of the mutation for GABP transactivation correlates directly with its effect upon GABP–C1 interaction. Furthermore, the domain of the C1 factor that is required for C1–GABP interaction can significantly inhibit GA-mediated transcription in vivo. The data support the model that transactivation by GABP is, at least in part, mediated by the C1 factor, and defines this protein as a novel coactivator.
Interestingly, the C1 factor interacts with all of the defined proteins in the enhancer complex. Proteins such as αTIF interact with C1 via residues distinct from their independent transcription activation domains (Hayes and O'Hare, 1993; Wu et al., 1994; Lai and Herr, 1997) and require the C1 protein for stable assembly with other enhancer components. In contrast, the transactivation domain of GABP itself represents the site of C1 interaction and, in this case, the function of C1 may be to mediate directly the interaction of GABP and the basal transcription apparatus. It remains to be determined if other GABP-dependent promoters also require the C1 factor for transcriptional stimulation. Similarly, it is important to note that transcription factors such as Luman (LZIP) have also been shown to interact with the C1 factor (Freiman and Herr, 1997; Lu et al., 1997, 1998). These interactions may represent additional cellular regulatory circuits in which the C1 factor plays an important role in mediating or facilitating transcriptional activation.
While the significance of the enhancer complex assembly in regulation of the HSV–1 IE genes during the lytic replication cycle is clear, the role of these proteins during the reactivation of HSV–1 from the latent state remains to be addressed. It is likely that activation of the IE genes would be a critical stage in the initiation of reactivation. However, the viral transactivator, αTIF, is not present at this time in sensory neurons (Roizman and Sears, 1996). This suggests that cellular factors play the primary role in activation of the IE genes during the initial stages of the reactivation process. The coordinated assembly of other transcription factors such as Oct–1, Sp1 and GABP by the C1 factor may represent an important mechanism for activation of these genes in an αTIF-independent manner. Significantly, in sensory neurons, nuclear localization of the C1 factor is dependent upon signals that initiate the reactivation of HSV–1 (Kristie et al., 1999). In addition, these signals also induce SAPK and MAPK pathways, which have been demonstrated to regulate the transcriptional activity of GABP. These observations have led to the hypothesis that modification of GABP may alter the C1–GABP interaction in these cells, resulting in activation of HSV–1 IE genes during reactivation of the virus from the latent state (Kristie et al., 1999). Regardless of the role of the C1 factor in the reactivation process, the coordination of the HSV–1 IE enhancer proteins and the direct mediation of the transcriptional activity of GABP by the C1 factor clearly define this protein as a critical coactivator.
Materials and methods
Two-hybrid analyses
A DNA fragment encoding amino acids 422–1021 of the C1 factor was inserted into pGBT9 (pGAL53). The yeast strain HF7c was transformed with pGAL53 and the resulting strain (scGAL53) was transformed with a human HeLa MATCHMAKER cDNA library (Clontech). Positive clones were selected for by growth on His– plates and screened for expression of β-galactosidase by filter lifts according to the manufacturer's instructions. DNAs from positive colonies were isolated and their nucleotide sequences were determined. GABPβ1 and β2 subunits (amino acids 10–383 and 10–347, respectively) were cloned into pGAD424, transformed into scGAL53, scGBT9 and scLAM5′. The resulting strains were tested for growth on His– plates and, where appropriate, for β-galactosidase activity as described. Fragments of pGAL53 were constructed by PCR with the appropriate oligonucleotide primers according to standard procedures.
Purification of GABP polypeptides and production of anti-GABP antisera
Bacterial vectors for the expression of GABP subunits [pET3DNRF-2α (GABPα), pET3DNRF-2β2 (GABPβ1) and pET3DNRF-2γ2 (GABPβ2)] were a gift of R.Scarpulla (Gugneja et al., 1995). Expressed proteins were purified essentially as described (Thompson et al., 1991; Gugneja et al., 1995). Polyclonal antisera were produced as previously described (Kristie et al., 1995) using purified GAα/GAβ subunits. Antisera were purified by protein affinity chromatography as described (Kristie et al., 1995) using GST fusion proteins GAα, amino acids 1–320 and GAβ, amino acids 121–330.
Protein interactions
A DNA fragment encoding amino acids 668–1021 of the C1 factor was cloned into pGEX2T (pGEX53.1). GST and GST–53.1 proteins were purified as described (Kristie et al., 1995). Luciferase, GAα, GAβ1, GAβ2 and GAβ2 deletions were labeled in vitro with [35S]methionine (Promega TNT) according to the manufacturer's recommendations. Aliquots of the labeled proteins were resolved in 12% SDS–PAGE gels, transferred to nitrocellulose and quantitated by scintillation counting of the full-length products. A 3 μg aliquot of purified GST or GST–53.1 was adsorbed to 10 μl of glutathione beads in 200 μl of G buffer (20 mM HEPES pH 7.9, 100 mM NaCl, 0.1% NP–40, 10 mM β-mercaptoethanol, 10% glycerol) for 30 min at 4°C and washed with 0.5 ml of buffer G. Then 3–5 fmol of the appropriate labeled protein were added to the GST or GST–53.1 beads in a total of 200 μl of buffer G containing 200 μg of bacterial protein extract. After incubation at 4°C for 1 h, the beads were washed three times with 0.5 ml of buffer, and the proteins were eluted in SDS–PAGE loading buffer. Aliquots of each input and elution protein sample were resolved by SDS–PAGE, transferred to nitrocellulose and quantitated by phosphoimaging (Storm, Molecular Dynamics) prior to autoradiography. For pull-downs from nuclear extracts, 1.5 μg of GST or GST–53.1 were adsorbed to 10 μl of glutathione beads and mixed with 200 μg of HEK293 cell nuclear extract protein in 200 μl of buffer G. Bound proteins were eluted with SDS–PAGE loading buffer, resolved in an 8–16% SDS–polyacrylamide gel and transferred to Immobilon. Western blots were probed with Ab2271 (AbGAα) or Ab2275 (AbGAβ) and developed using ECL-Plus (Amersham). Pull-downs using bacterially expressed and purified GABP proteins were as described above using 2 μg of GST or GST–53.1 and 100 ng of purified GAα, GAβ or GAα/GAβ complex.
GABP deletions and site-directed mutagenesis
GAΔ278, Δ256 and Δ242 were generated by restriction enzyme truncation with PflM1, EcoRI and PstI, respectively. Site-directed mutagenesis of NRF2γ was done using the Gene Editor System (Promega Biotech) and the following mutagenic primers: Mt1, GCCATTCAGCAAGCAGCTAGTTCAGGGGGTC; Mt2, TCCATCTGTAAACTGCTGTGGCGACTT– GCTGACC; Mt3, GGAATTGGTCAGCCCGCCGCTGCGACCATG– CCAGATGGAC; and Mt4, GATGGACAACAAGCATTAACAGCA– CCAGCAACAGAC.
Cell transfections and reporter assays
The wild-type GAβ activation domain encoding amino acids 246–315 was inserted into pM (Clontech) to produce the GAL4 DNA-binding–GAβ activation domain fusion pMGA/Ad. A fragment encoding the C1–GABP interaction domain (amino acids 668–933 of the C1 factor) was inserted into pCMV/myc/nuc (Invitrogen) to create pCMV53.1a. Transfections of 1 × 105 HeLa cells were performed according to the manufacturer's instructions (Superfect, Qiagen). Each transfection contained 800 ng of pG5luc (Promega) reporter plasmid, 20 ng of pRL-TK (Promega) as an internal control for transfection efficiency, 25 ng of pM, pMGA/AD or pM3-VP16 (Clontech) and 100–400 ng of pCMV/myc/nuc or pCMV53.1a. Dual luciferase assays (Promega) were carried out 46–50 h post-transfection according to the manufacturer's directions, and the resulting data points were normalized to the internal control.
Gel electrophoretic mobility shift assays
Gel electrophoretic mobility shift assays were done essentially as described (Kristie and Sharp, 1993) with reactions containing buffer A (20 mM HEPES pH 7.9, 6.25 mM MgCl2, 0.1% NP–40, 0.2 mM EDTA, 4% ficoll), 300 μg/ml bovine serum albumin, 100 μg/ml poly(dI–dC), 7.5 fmol Gal4 UAS DNA probe and 6.5 μg of nuclear extract protein derived from HeLa cells transfected with the appropriate plasmid constructs.
Acknowledgments
Acknowledgements
We thank R.Scarpulla for the gift of GABP expression plasmids, B.Moss, P.Sharp, A.Sears and members of the Laboratory of Viral Diseases for helpful discussions, R.Dashner and M.DeenLevsink for technical assistance, K.Potter for the partial purification of GABP subunits from bacterial extracts, and B.Moss and A.McBride for critical reading of this manuscript. These studies were supported by the Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health (T.M.K.).
References
- Bailey A.C. and Thompson, R. (1992) A sequence-specific DNA-binding protein recognising a GA-rich element cooperates with Oct–1 at the herpes simplex virus type 1 IE3 promoter. Intervirology, 34, 74–85. [DOI] [PubMed] [Google Scholar]
- Batchelor A.H., Piper, D.E., de la Brousse, F.C., McKnight, S.L. and Wolberger, C. (1998) The structure of GABPα/β: an ETS domain–ankyrin repeat heterodimer bound to DNA. Science, 279, 1037–1041. [DOI] [PubMed] [Google Scholar]
- Boccia L.M., Lillicrap, D., Newcombe, K. and Mueller, C.R. (1996) Binding of the Ets factor GA-binding protein to an upstream site in the factor IX promoter is a critical event in transactivation. Mol. Cell. Biol., 16, 1929–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Brousse F.C., Birkenmeier, E.H., King, D.S., Rowe, L.B. and McKnight, S.L. (1994) Molecular and genetic characterization of GABP β. Genes Dev., 8, 1853–1865. [DOI] [PubMed] [Google Scholar]
- Douville P., Hagmann, M., Georgiev, O. and Schaffner, W. (1995) Positive and negative regulation at the herpes simplex virus ICP4 and ICP0 TAATGARAT motifs. Virology, 207, 107–116. [DOI] [PubMed] [Google Scholar]
- Flory E., Hoffmeyer, A., Smola, U., Rapp, U.R. and Bruder, J.T. (1996) Raf–1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J. Virol., 70, 2260–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R.N. and Herr, W. (1997) Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev., 11, 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T. and Roeder, R.G. (1988) A herpesvirus trans-activating protein interacts with transcription factor OTF–1 and other cellular proteins. Proc. Natl Acad. Sci. USA, 85, 6347–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Motomura, S., Wilson, A.C., Freiman, R.N., Nakabeppu, Y., Fukushima, K., Fujishima, M., Herr, W. and Nishimoto, T. (1997) A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev., 11, 726–737. [DOI] [PubMed] [Google Scholar]
- Gugneja S., Virbasius, J.V. and Scarpulla, R.C. (1995) Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol. Cell. Biol., 15, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugneja S., Virbasius, C.M. and Scarpulla, R.C. (1996) Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol. Cell. Biol., 16, 5708–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S. and O'Hare, P. (1993) Mapping of a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J. Virol., 67, 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer A., Avots, A., Flory, E., Weber, C.K., Serfling, E. and Rapp, U.R. (1998) The GABP-responsive element of the interleukin-2 enhancer is regulated by JNK/SAPK-activating pathways in T lymphocytes. J. Biol. Chem., 273, 10112–10119. [DOI] [PubMed] [Google Scholar]
- Hughes T.A., La Boissiere, S. and O'Hare, P. (1999) Analysis of functional domains of the host cell factor involved in VP16 complex formation. J. Biol. Chem., 274, 16437–16443. [DOI] [PubMed] [Google Scholar]
- Jones K.A. and Tjian, R. (1985) Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro.Nature, 317, 179–182. [DOI] [PubMed] [Google Scholar]
- Katan M., Haigh, A., Verrijzer, C.P., van der Vliet, P.C. and O'Hare, P. (1990) Characterization of a cellular factor which interacts functionally with Oct–1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res., 18, 6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J.D., Rould, M.A., Aurora, R., Herr, W. and Pabo, C.O. (1994) Crystal structure of the Oct–1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell, 77, 21–32. [DOI] [PubMed] [Google Scholar]
- Kristie T.M. and Roizman, B. (1984) Separation of sequences defining basal expression from those conferring α gene recognition within the regulatory domains of herpes simplex virus 1 α genes. Proc. Natl Acad. Sci. USA, 81, 4065–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie T.M. and Sharp, P.A. (1990) Interactions of the Oct–1 POU subdomains with specific DNA sequences and with the HSV α-trans-activator protein. Genes Dev., 4, 2383–2396. [DOI] [PubMed] [Google Scholar]
- Kristie T.M. and Sharp, P.A. (1993) Purification of the cellular C1 factor required for the stable recognition of the Oct–1 homeodomain by the herpes simplex virus α-trans-induction factor (VP16). J. Biol. Chem., 268, 6525–6534. [PubMed] [Google Scholar]
- Kristie T.M., LeBowitz, J.H. and Sharp, P.A. (1989) The octamer-binding proteins form multi-protein–DNA complexes with the HSV αTIF regulatory protein. EMBO J., 8, 4229–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie T.M., Pomerantz, J.L., Twomey, T.C., Parent, S.A. and Sharp, P.A. (1995) The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem., 270, 4387–4394. [DOI] [PubMed] [Google Scholar]
- Kristie T.M., Vogel, J.L. and Sears, A.E. (1999) Nuclear localization of the C1 factor in sensory neurons correlates with initiation of reactivation of HSV from latency. Proc. Natl Acad. Sci. USA, 96, 1229–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Boissiere S., Walker, S. and O'Hare, P. (1997) Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol. Cell. Biol., 17, 7108–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Boissiere S., Hughes, T. and O'Hare, P. (1999) HCF-dependent nuclear import of VP16. EMBO J., 18, 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.S. and Herr, W. (1997) Interdigitated residues within a small region of VP16 interact with Oct–1, HCF and DNA. Mol. Cell. Biol., 17, 3937–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarco K.L. and McKnight, S.L. (1989) Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev., 3, 1372–1383. [DOI] [PubMed] [Google Scholar]
- Lu R., Yang, P., O'Hare, P. and Misra, V. (1997) Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol., 17, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Yang, P., Padmakumar, S. and Misra, V. (1998) The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol., 72, 6291–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S. and Roizman, B. (1982) Structural features of the herpes simplex virus α gene 4, 0 and 27 promoter-regulatory sequences which confer α regulation on chimeric thymidine kinase genes. J. Virol., 44, 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight J.L., Kristie, T.M. and Roizman, B. (1987) Binding of the virion protein mediating α gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl Acad. Sci. USA, 84, 7061–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuchprayoon I., Shang, J., Simkevich, C.P., Luo, M., Rosmarin, A.G. and Friedman, A.D. (1999) An enhancer located between the neutrophil elastase and proteinase 3 promoters is activated by Sp1 and an ets factor. J. Biol. Chem., 274, 1085–1091. [DOI] [PubMed] [Google Scholar]
- O'Hare P. and Goding, C.R. (1988) Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell, 52, 435–445. [DOI] [PubMed] [Google Scholar]
- Ouyang L., Jacob, K.K. and Stanley, F.M. (1996) GABP mediates insulin-increased prolactin gene transcription. J. Biol. Chem., 271, 10425–10428. [DOI] [PubMed] [Google Scholar]
- Pomerantz J.L., Kristie, T.M. and Sharp, P.A. (1992) Recognition of the surface of a homeo domain protein. Genes Dev., 6, 2047–2057. [DOI] [PubMed] [Google Scholar]
- Preston C.M., Frame, M.C. and Campbell, M.E. (1988) A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell, 52, 425–434. [DOI] [PubMed] [Google Scholar]
- Roizman B. and Sears,A.E. (1996) Herpes simplex viruses and their replication. In Fields,B., Knipe,D.M. and Howley,P.M. (eds), Fundamental Virology. Lippincott-Raven, Philadelphia, PA, pp. 1043–1107. [Google Scholar]
- Rosmarin A.G., Caprio, D.G., Kirsch, D.G., Handa, H. and Simkevich, C.P. (1995) GABP and PU.1 compete for binding, yet cooperate to increase CD18 (β2 leukocyte integrin) transcription. J. Biol. Chem., 270, 23627–23633. [DOI] [PubMed] [Google Scholar]
- Rosmarin A.G., Luo, M., Caprio, D.G., Shang, J. and Simkevich, C.P. (1998) Sp1 cooperates with the ets transcription factor, GABP, to activate the CD18 (β2 leukocyte integrin) promoter. J. Biol. Chem., 273, 13097–13103. [DOI] [PubMed] [Google Scholar]
- Stern S. and Herr, W. (1991) The herpes simplex virus trans-activator VP16 recognizes the Oct–1 homeo domain: evidence for a homeo domain recognition subdomain. Genes Dev., 5, 2555–2566. [DOI] [PubMed] [Google Scholar]
- Stern S., Tanaka, M. and Herr, W. (1989) The Oct–1 homeodomain directs formation of a multiprotein–DNA complex with the HSV transactivator VP16. Nature, 341, 624–630. [DOI] [PubMed] [Google Scholar]
- Sturm R.A., Das, G. and Herr, W. (1988) The ubiquitous octamer-binding protein Oct–1 contains a POU domain with a homeo box subdomain. Genes Dev., 2, 1582–1599. [DOI] [PubMed] [Google Scholar]
- Thompson C.C., Brown, T.A. and McKnight, S.L. (1991) Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science, 253, 762–768. [DOI] [PubMed] [Google Scholar]
- Triezenberg S.J., LaMarco, K.L. and McKnight, S.L. (1988) Evidence of DNA:protein interactions that mediate HSV–1 immediate early gene activation by VP16. Genes Dev., 2, 730–742. [DOI] [PubMed] [Google Scholar]
- Vassias I., Hazan, U., Michel, Y., Sawa, C., Handa, H., Gouya, L. and Morinet, F. (1998) Regulation of human B19 parvovirus promoter expression by hGABP (E4TF1) transcription factor. J. Biol. Chem., 273, 8287–8293. [DOI] [PubMed] [Google Scholar]
- Verrijzer C.P., Kal, A.J. and van der Vliet, P.C. (1990) The oct–1 homeo domain contacts only part of the octamer sequence and full oct–1 DNA-binding activity requires the POU-specific domain. Genes Dev., 4, 1964–1974. [DOI] [PubMed] [Google Scholar]
- Virbasius J.V., Virbasius, C.A. and Scarpulla, R.C. (1993) Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev., 7, 380–392. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Sawada, J., Yano, K., Yamaguchi, K., Goto, M. and Handa, H. (1993) cDNA cloning of transcription factor E4TF1 subunits with Ets and notch motifs. Mol. Cell. Biol., 13, 1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.C., LaMarco, K., Peterson, M.G. and Herr, W. (1993a) The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell, 74, 115–125. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Cleary, M.A., Lai, J.S., LaMarco, K., Peterson, M.G. and Herr, W. (1993b) Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harb. Symp. Quant. Biol., 58, 167–178. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Peterson, M.G. and Herr, W. (1995) The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Freiman, R.N., Goto, H., Nishimoto, T. and Herr, W. (1997) VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol., 17, 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.J., Monokian, G., Mark, D.F. and Wobbe, C.R. (1994) Transcriptional activation by herpes simplex virus type 1 VP16 in vitro and its inhibition by oligopeptides. Mol. Cell. Biol., 14, 3484–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P. and Capone, J.P. (1990) A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein–DNA complex assembly with Oct–1. Mol. Cell. Biol., 10, 4974–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]