Abstract
The immune correlates of protection for most of the currently used vaccines are based on long-lived humoral immunity. Vaccines based on humoral immunity alone are unlikely to protect against infections caused by intracellular pathogens and today's most pressing infectious diseases of public health importance are caused by intracellular infections that not only include Chlamydia trachomatis but also tuberculosis, malaria and HIV/AIDS. For these infections, vaccines that induce cellular immune responses are essential. Major impediments in developing such vaccines include difficulty in identifying relevant T-cell antigens and delivering them in ways that elicit protective cellular immunity. In turn this is compounded by the complexity and plasticity of T-cell developmental pathways that often correlate with specific aspects of protective immunity. Genomics and proteomics now provide tools to allow unbiased selection of candidate T-cell antigens. This review mainly focuses on an immunoproteomic approach used in our laboratory to identify Chlamydia T-cell antigens and how these T-cell antigens can be developed into a future human Chlamydia vaccine.
Key words: chlamydia, vaccine, immunoproteomics, MHC, T cell, dendritic cell, antigen, adjuvant
Why is a Chlamydia Vaccine Needed?
Public health programs have targeted Chlamydia trachomatis as a major problem mainly because of the ability of the organism to cause long-term disease sequelae such as blindness, infertility and ectopic pregnancy. The World Health Organization estimates that over 85 million cases of trachoma and 92 million cases of sexually transmitted C. trachomatis infection globally occur each year. C. trachomatis is the most common reportable communicable disease in Canada and the United States. The principle components of the control program for sexually transmitted C. trachomatis include case detection, antimicrobial treatment and contact tracing. In jurisdictions where programs have been systematically introduced, C. trachomatis case rates have substantially fallen. This has been the experience in British Columbia (BC), Canada where case rates were reduced by more than 50% between 1991 and 1997 following the introduction of a control program.1 However, BC like other parts of Canada and several Scandinavian countries has subsequently seen a substantial rise in rates2 concomitant with a substantial rise in reinfection rates.1 We have hypothesized that control programs based on case identification and treatment interfere with the effects of immunity on population susceptibility to infection and termed this the “arrested immunity hypothesis”. In effect, the control program appears to increase the incidence of infection by increasing susceptibility to reinfection. Interestingly parallel results have also been reported for trachoma control programs based on antimicrobial drug therapy.3 These data suggest that the current treatment-based approach to C. trachomatis transmission is likely to provide incomplete control and that an effective vaccine may be the best strategy.
Chlamydia vaccine research was first initiated shortly after the initial isolation of C. trachomatis from cases of trachoma in the early 1960s.4 Trials used different strains (serovars) of C. trachomatis as whole inactivated bacterial cells termed elementary bodies administered intramuscularly with oil-in-water adjuvants to children at risk for trachoma. Vaccines were also tested in primate models of experimental ocular infection.5 In aggregate, the studies generated results that have endured as the fundamental paradigm of C. trachomatis vaccinology. Critical observations concluded that partial protection occurs in both humans and primates but is short lived; protection is strain (serovar) specific; and infection following vaccination in primates is associated with worse inflammation.6,7 The organism was speculated to be composed of antigens that elicited protective immunity and others that engendered immunopathology. Thus, these trials yielded important lessons for the modern era of Chlamydia vaccinology, namely that an effective C. trachomatis vaccine would need to be molecularly defined and engender long-lived protective immune responses. Since then Chlamydia research has focused on understanding the details of how cellular immune responses are generated and maintained in vivo during Chlamydia infection both in humans and in mice.
IFNγ-Mediated Th1 Immune Response is Essential for Chlamydia Clearance
Although Chlamydia is clearly able to persist in hosts after the initial development of an adaptive immune response, there is strong evidence for immunity. Bailey et al.8 have collected some of the best clinical data during studies of trachoma where they observed that host resistance dramatically increases with age and that clearance of infection correlated with enhanced cell-mediated immune responses.9 We have studied resistance to C. trachomatis infection among a unique cohort of highly exposed sex workers in Nairobi, Kenya and collected immunoepidemiologic evidence for immunity. Women whose peripheral blood lymphocytes had IFNγ responses to Chlamydia antigen were highly resistant to infection whereas women who made interleukin-10 (IL-10) responses had a nearly 6-fold increased risk of acquiring infection.10 Women with low CD4 T cell counts due to HIV had lower IFNγ responses and a greater risk of developing chlamydial pelvic inflammatory disease.11 Collectively the clinical data show that the immune correlates for protection against C. trachomatis infection are cell-mediated IFNγ responses; IL-10 responses correlate with susceptibility.
The above clinical data correlating susceptibility and resistance with cell-mediated immune responses mirror data obtained in the mouse model.12 Mice and humans are clearly different but mouse biology has proven surprisingly informative of human biology especially with respect to the molecular basis for disease. This has been true for murine C. muridarum infection informing the immunobiology of human C. trachomatis infection. C. muridarum is a mouse adapted strain of Chlamydia whose genome is highly syntenic to that of C. trachomatis and which differs in composition in only six genes.13 In the C. muridarum murine model, clearance and resistance to infection were correlated with CD4 T cell immune responses14 and IL-10 antagonized protective immunity.15 B cells and antibody also played a role in protection against reinfection although that role strongly depended on CD4 T cell-mediated adaptive changes that occurred in the local genital tract during primary infection.16
Overall both the clinical and experimental data demonstrate that cell-mediated IFNγ immune responses are correlated with clearance of Chlamydia infection and resistance to reinfection. Genomics has uncovered the total repertoire within which protective Chlamydia antigens are to be found; and the new tools of proteomics offer heretofore-unavailable sensitivity in detecting immunologically relevant antigens. Furthermore, there has been substantial progress in rationalizing the whole process for the development of a vaccine based upon cellular immunity. In particular, the essential roles for dendritic cells (DCs) and MHC molecules in cell-mediated immunity have been elucidated in molecular detail.
Dendritic Cell Based Immunoproteomic Discovery of Chlamydia T-Cell Antigens
DCs are at the centre of the initiation of immune responses by naïve T cells and appear to be particularly important to the development of Chlamydia immunity. DCs capture antigen in the periphery and migrate to regional lymph nodes where they present processed antigen on MHC molecules to naïve T cells. DCs undergo a maturation process during migration to lymph nodes and express new surface molecules that act as co-stimulants to naïve T cells causing activation and polarization of the cytokine secretion pattern. Therefore, in addition to containing appropriate molecular antigens, a successful Chlamydia vaccine will need to activate professional antigen-presenting cells and polarize T-cell responses to the protective type cytokine secretion pattern and generate long-lived T-cell memory. Initial work from our laboratory demonstrated that expression of GM-CSF—a cytokine known to mobilize DCs—in the mouse airway compartment significantly enhanced systemic Th1 cellular immune responses following immunization with inactivated C. muridarum.17 This suggested that DCs recruited by GM-CSF contributed to the development of a Th1 immune response. Further in vitro observations demonstrated that DCs exposed to live versus to UV-irradiated C. muridarum develop distinct phenotypes, such that DCs exposed to live Chlamydia become mature and effectively present antigen to Chlamydia-specific CD4 T cells, while DCs exposed to UV-irradiated Chlamydia were not immunologically mature.18 Since DCs are essential to induce Chlamydia immunity via presentation of chlamydial antigens to naïve T cells, we hypothesized that Chlamydia peptide antigens presented by the surface MHC molecules of Chlamydia infected DCs could be explored as potential source for the identification of T-cell antigens.
An approach called immunoproteomics, in which peptides presented by MHC molecules are identified by tandem mass spectrometry (MS/MS),19,20 allows genomic information to guide the delineation of the complete T-cell immunoproteome of the organism. These methods have been applied to several immunological problems21 but instrument sensitivity has prevented its applicability to pathogens. Recent advancements in MS/MS technology now provide sensitivity limits near one femtomole (fmol) and are able to measure peptide masses to within one part-per-million accuracy.22 This brings the detection technology into a range compatible with the levels of microbial peptides that can reasonably be purified from MHC molecules presented on the surface of antigen presenting cells such as DCs.
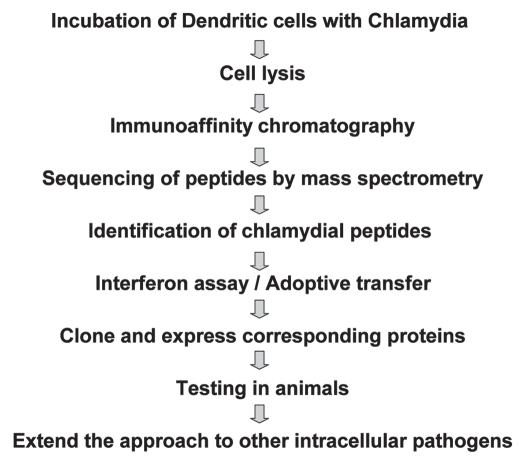
The immunoproteomic approach that we used to identify Chlamydia T-cell antigens involved multiple steps (Fig. 1). First, bone marrow cells isolated from the femurs and tibias of mice are grown in the presence of GM-CSF and IL-4. These bone marrow derived DCs (BM-DCs) are pulsed with Chlamydia. Pulsed BM-DCs are lysed and MHC molecules isolated using allele-specific anti-MHC monoclonal antibody affinity columns. Purified MHC molecules are then washed and peptides eluted with acetic acid and separated from high molecular weight material by ultrafiltration through 5-kDa cut-off membranes. The purified MHC-bound peptides are initially analyzed qualitatively using an LTQ-OrbitrapXL (Thermo Electron) online coupled to a nanoflow HPLC using a nanospray ionization source. The mass spectrometer is set to fragment the five most intense multiply-charged ions per cycle. Fragment spectra are extracted using DTASuperCharge (http://msquant.sourceforge.net) and searched using the Mascot algorithm against a database comprised of the protein sequences from C. muridarum. DC adoptive transfer method is employed to deliver peptides that are recognized by Chlamydia specific CD4 T cells to identify those that are protective. The selected peptides are further characterized by cloning the parent proteins of these MHC-binding peptides as vaccine candidates and tested in a genital tract mouse model.
Figure 1.
Schematic depiction of the sequence of steps involved in the immunoproteomic approach for Chlamydia T cell vaccine development.
We used the immunoproteomic approach to identify 13 Chlamydia peptides derived from 8 novel epitopes presented by MHC class II molecules from BM-DCs infected with Chlamydia (Table 1).23 These MHC class II-bound peptides were recognized in vitro by Chlamydia specific CD4 T cells harvested from immune mice recovered from Chlamydia infection and adoptive transfer of DCs pulsed ex vivo with the peptides partially protected mice against intranasal and genital tract Chlamydia infection. We further investigated these peptides by cloning recombinant proteins corresponding to the MHC binding peptides. Recombinant Chlamydia proteins were also found to be recognized in vitro by immune T cells suggesting that these proteins can be processed to generate the immunologically relevant peptides. These proteins were also able to protect mice against Chlamydia infection in vivo as shown by DC adoptive transfer experiments.24 Based on these results, three of the 8 source proteins (RplF, PmpG and PmpE/F) were deemed suitable for further evaluation. However, only PmpE/F and PmpG were selected for further animal studies as RplF has significant homology to the human homolog and thus may not be suitable as a human vaccine.
Table 1.
Chlamydia T cell antigens identified by immunoproteomic approach
| Peptide sequence | Source protein | Abbreviation |
| KGNEVFVSPAAHIIDRPG | Ribosomal protein L6 | RplF |
| SPGQTNYAAAKAGIIGFS | 3-oxoacyl-(acyl carrier protein) reductase | FabG |
| KLDGVSSPAVQESISE | Anti-anti-sigma factor | Aasf |
| ASPIYVDPAAAGGQPPA | Polymorphic membrane protein G | PmpG-1 |
| DLNVTGPKIQTDVD | Hypothetical protein TC0420 | TC0420 |
| IGQEITEPLANTVIA | ATP-dependent Clp protease, proteolytic subunit | ClpP-1 |
| AFHLFASPAANYIHTG | Polymorphic membrane protein F | Pmpe/F-2 |
| MTTVHAATATQSVVD | Glyceraldehyde 3-phosphate dehydrogenase | Gap |
The immunoproteomic approach described above directly identifies T cell epitopes presented by antigen presenting cells resulting in a vast improvement in the positive validation rate. Another advantage in using immunoproteomics is that the peptides identified are the result of physiological processing and presentation pathways and are based on both the affinity for the MHC molecules as well as the frequency of their presentation.
Pathogens contain a large number of possible antigens for immune responses but only a few immunodominant antigens are typically recognized following infection or immunization. The reasons for immunodominance are complex and can be intrinsic to the targets (i.e., differences in affinity for immune receptors) or extrinsic (i.e., competition that suppresses the response to one target in favor of another).25 We hypothesized that the antigens we identified through the immunoproteomic approach are immunodominant and can be recognized in varying MHC backgrounds. We tested whether the peptides (originally identified in C57BL/6) and their source proteins are also recognized by mouse strains other than C57BL/6. As expected, proteins and peptides were readily recognized by T cells from C57BL/6 mice. Proteins, but not peptides, were also recognized by Chlamydia-specific T cells from Balb/c and C3H/HeN mice (unpublished data). These data suggest that there are T cell epitopes within the identified proteins that are presented by MHC haplotypes other than I-Ab and that these Chlamydia proteins are immunologically recognized in genetically different strains of mice, arguing that they are immunodominant.
Delivery Systems and Immunomodulators
Since a Chlamydia vaccine cannot be practically delivered via adoptive transfer of DCs, it is essential to identify a delivery system together with an immunomodulator that elicits protective cell-mediated immunity exhibiting a Th1 bias. The three major adjuvant systems that we have evaluated include DDA/TDB (Dimethyldioctadecylammonium Bromide/D-(+)-trehalose 6,6′-dibehenate), AbISCO (an effective immune stimulating complex) and CpG ODN (CpG oligonucleotide). Among the three adjuvants tested, CpG ODN formulation was not able to engender protection against Chlamydia infection at any level in vaccinated mice and the AbISCO formulation conferred only moderate protection. Strikingly, studies using DDA/TDB substantially improved the protective efficacy of chlamydial protein antigens in murine models of genital tract C. muridarum infection.26
DDA/TDB is a recently discovered adjuvant that consists of cationic liposomes as delivery system and a synthetic mycobacterial cord factor as immunomodulator. One of the important observations that we made during the evaluation of DDA/TDB in our adjuvant preparations is that it induced Th17 in addition to strong Th1 immune response.26 It is known that TDB selectively activates the FcRγ-Syk-Card9 pathway in antigen presenting cells to induce a unique innate immune activation program that directs protective Th1 and Th17 immunity after subunit Mycobacterium tuberculosis vaccination27 demonstrating that TDB adjuvant with this mode of action is distinct from TLR-triggering adjuvants such as CpG engaging MyD88.28 TDB was recently shown to use the monocyte-inducible C-type lectin (Mincle) as its cell receptor on the surface of APCs.29 A reason why the Th17 Chlamydia antigen-specific response was not found in mice vaccinated with CpG formulation might be that CpG may inhibit Th17 differentiation through IL-12 and IFNγ.30 Based on these observations, we speculate that an adjuvant that triggers a Th17 response in addition to Th1 may be the optimum choice for Chlamydia vaccine. Interestingly, a recent study from our laboratory also showed that differences in Th1/Th17 responses could explain differences in susceptibility and resistance to C. muridarum infection in Balb/c and C57BL/6 mice.31
We evaluated protection against Chlamydia infection mice immunized with both individual proteins as well as in combination formulated with different adjuvants. The protein combination consist of PmpG, PmpE/F and MOMP (MOMP is the major outer membrane protein and a reference chlamydial antigen used in many vaccine immunological studies) gave promising results approaching the levels of protection observed in positive controls using live Chlamydia infection.26 The superior protection obtained in the protein combination group compared with individual protein group suggests that a successful Chlamydia vaccine will need to be composed of multiple proteins in order to provide broad coverage in an outbred population, to cross protect against multiple variants of C. trachomatis as well as to maximize immunogenicity.
Since all the protection results described above were observed in C57BL/6 mouse, the strain in which the antigens were originally discovered by immunoproteomics, we also tested the protection efficacy of multiple Chlamydia protein antigens and DDA/TDB in Balb/c mice that has a different MHC genetic background. Remarkably the vaccine combination engendered significant protection in Balb/c mice suggesting the T-cell antigens identified through the immunoproteomic approach are truly immunodominant.
Future Work
Even though vaccine formulated with three Chlamydia T-cell antigens and DDA/TDB adjuvant generated significant protection against infection, it remains important to evaluate protection against tissue pathology. Importantly, mice primed by a prior intranasal infection were completely protected against the development of oviductal pathology following intravaginal challenge with Chlamydia. We therefore have initiated experiments with different adjuvant formulations together with multiple antigens in mice to examine for pathology as determined by visually apparent hydrosalpinx in oviducts. It remains important to discover the mechanism behind the different outcomes for oviductal pathology following infection-induced versus vaccine-induced immunity. Clearly an ongoing challenge for Chlamydia vaccine research remains the discovery of strategies that maximize the protective effects of immune T cells while simultaneously preventing such cells from causing immune-mediated tissue damage. Finally it also appears likely that the immunoproteomic approach we are exploring in Chlamydia vaccine research may be useful for other problematic intracellular pathogens such as tuberculosis, malaria and HIV for which vaccine solutions are desperately sought.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (grant no. R01AI076483).
Footnotes
Previously published online: www.landesbioscience.com/journals/vaccines/article/12299
References
- 1.Brunham RC, Pourbohloul B, Mak S, White R, Rekart ML. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J Infect Dis. 2005;192:1836–1844. doi: 10.1086/497341. [DOI] [PubMed] [Google Scholar]
- 2.Gotz H, Lindback J, Ripa T, Arneborn M, Ramsted K, Ekdahl K. Is the increase in notifications of Chlamydia trachomatis infections in Sweden the result of changes in prevalence, sampling frequency or diagnostic methods? Scand J Infect Dis. 2002;34:28–34. doi: 10.1080/00365540110077001. [DOI] [PubMed] [Google Scholar]
- 3.Atik B, Thanh TT, Luong VQ, Lagree S, Dean D. Impact of annual targeted treatment on infectious trachoma and susceptibility to reinfection. JAMA. 2006;296:1488–1497. doi: 10.1001/jama.296.12.1488. [DOI] [PubMed] [Google Scholar]
- 4.Grayston JT, Wang SP. The potential for vaccine against infection of the genital tract with Chlamydia trachomatis. Sex Transm Dis. 1978;5:73–77. doi: 10.1097/00007435-197804000-00011. [DOI] [PubMed] [Google Scholar]
- 5.Wang SP, Grayston JT, Alexander ER. Trachoma vaccine studies in monkeys. Am J Ophthalmol. 1967;63:1615–1630. doi: 10.1016/0002-9394(67)94155-4. [DOI] [PubMed] [Google Scholar]
- 6.Grayston JT, Gale JL, Yeh LJ, Yang CY. Pathogenesis and immunology of trachoma. Trans Assoc Am Physicians. 1972;85:203–211. [PubMed] [Google Scholar]
- 7.Schachter J, Caldwell HD. Chlamydiae. Annu Rev Microbiol. 1980;34:285–309. doi: 10.1146/annurev.mi.34.100180.001441. [DOI] [PubMed] [Google Scholar]
- 8.Bailey R, Duong T, Carpenter R, Whittle H, Mabey D. The duration of human ocular Chlamydia trachomatis infection is age dependent. Epidemiol Infect. 1999;123:479–486. doi: 10.1017/s0950268899003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey RL, Holland MJ, Whittle HC, Mabey DC. Subjects recovering from human ocular chlamydial infection have enhanced lymphoproliferative responses to chlamydial antigens compared with those of persistently diseased controls. Infect Immun. 1995;63:389–392. doi: 10.1128/iai.63.2.389-392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen CR, Koochesfahani KM, Meier AS, Shen C, Karunakaran K, Ondondo B, et al. Immunoepidemiologic profile of Chlamydia trachomatis infection: importance of heat-shock protein 60 and interferon-gamma. J Infect Dis. 2005;192:591–599. doi: 10.1086/432070. [DOI] [PubMed] [Google Scholar]
- 11.Kimani J, Maclean IW, Bwayo JJ, MacDonald K, Oyugi J, Maitha GM, et al. Risk factors for Chlamydia trachomatis pelvic inflammatory disease among sex workers in Nairobi, Kenya. J Infect Dis. 1996;173:1437–1444. doi: 10.1093/infdis/173.6.1437. [DOI] [PubMed] [Google Scholar]
- 12.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 13.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infect Immun. 2002;70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Gartner J, Zhu L, Wang S, Brunham RC. IL-10 gene knockout mice show enhanced Th1-like protective immunity and absent granuloma formation following Chlamydia trachomatis lung infection. J Immunol. 1999;162:1010–1017. [PubMed] [Google Scholar]
- 16.Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol. 2005;175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Xing Z, Brunham RC. GM-CSF transgenebased adjuvant allows the establishment of protective mucosal immunity following vaccination with inactivated Chlamydia trachomatis. J Immunol. 2002;169:6324–6331. doi: 10.4049/jimmunol.169.11.6324. [DOI] [PubMed] [Google Scholar]
- 18.Rey-Ladino J, Koochesfahani KM, Zaharik ML, Shen C, Brunham RC. A live and inactivated Chlamydia trachomatis mouse pneumonitis strain induces the maturation of dendritic cells that are phenotypically and immunologically distinct. Infect Immun. 2005;73:1568–1577. doi: 10.1128/IAI.73.3.1568-1577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 20.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, et al. Characterization of peptides bound to the class I MHC molecule HLAA2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 21.de Jong A. Contribution of mass spectrometry to contemporary immunology. Mass Spectrom Rev. 1998;17:311–335. doi: 10.1002/(SICI)1098-2787(1998)17:5<311::AID-MAS1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, et al. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol. 2008;180:2459–2465. doi: 10.4049/jimmunol.180.4.2459. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol. 2009;182:1602–1608. doi: 10.4049/jimmunol.182.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sette A, Peters B. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Curr Opin Immunol. 2007;19:106–110. doi: 10.1016/j.coi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, et al. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFNgamma)/tumor necrosis factor alpha and IFNgamma/interleukin-17 double-positive CD4+ T cells. Infect Immun. 2010;78:2272–2282. doi: 10.1128/IAI.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werninghaus K, Babiak A, Gross O, Holscher C, Dietrich H, Agger EM, et al. Adjuvanticity of a synthetic cord factor analogue for subunit Mycobacterium tuberculosis vaccination requires FcRgamma-Syk-Card9-dependent innate immune activation. J Exp Med. 2009;206:89–97. doi: 10.1084/jem.20081445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwissa M, Amara RR, Robinson HL, Moss B, Alkan S, Jabbar A, et al. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J Exp Med. 2007;204:2733–2746. doi: 10.1084/jem.20071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV. CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th1) immunity. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Shen C, Yu H, Karunakaran KP, Brunham RC. Differences in innate immune responses correlate with differences in murine susceptibility to Chlamydia muridarum pulmonary infection. Immunology. 2010;129:556–566. doi: 10.1111/j.1365-2567.2009.03157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]