Abstract
Chronic hepatitis B virus (HBV) infection is a major health issue, especially in Asia. A recent genome-wide association study (GWAS) has implicated genetic variants in the HLA-DP locus associated with chronic hepatitis B in Japanese and Thai populations. To confirm whether the polymorphisms at the HLA-DP genes are associated with persistent chronic hepatitis B virus infection in Han Chinese, we conducted an independent case-control study using 521 persistent chronic HBV carriers and 819 controls that included 571 persons with HBV natural clearance and 248 never HBV-infected (healthy) individuals. Eleven single nucleotide polymorphisms (SNPs) in a region including HLA-DPA and HLA-DPB and an adjacent SNP in strong linkage disequilibrium (LD) with a neighboring HLA-DR13 locus were genotyped using TaqMan SNP genotyping assay. Eleven variants at HLA-DP showed a strong association with persistent chronic HBV carrier status (p = 1.82×10−12 to 0.01). We also stratified the analysis by HBV clearance status to test the association between these polymorphisms and HBV natural clearance; similar results were obtained (p = 2.70×10−11 to 0.003). Included SNPs define highly structured haplotypes which were also strongly associated with HBV chronic infection (Block 1: odds ratio (OR) = 0.54, p = 8.73×10−7; block 2: OR = 1.98, p = 1.37×10−10). These results further confirm that genetic variants in the HLA-DP locus are strongly associated with persistent HBV infection in the Han Chinese population.
Keywords: Chronic hepatitis B, Haplotype association, GWAS, SNPs, Joint effects
Introduction
Hepatitis B virus (HBV) infection is a worldwide health problem that frequently leads to acute, fulminant, chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. More than 2 billion people have been infected with HBV worldwide, of whom 400 million are chronic carriers. HBV infection accounts for 600,000-1,200,000 deaths each year. The prevalence of hepatitis B virus varies greatly, but it is endemic in all countries. In China, Southeast Asia, the Western Pacific, and sub-Saharan Africa, where HBV infection is usually acquired perinatally or in early childhood, the prevalence is high and the carrier rate exceeds 8%(1, 2). In China about 120 million people are HBV chronic carriers, and 50-80% of cirrhosis patients are infected with HBV(3). Chronic infection with HBV has become a key cause of hepatocellular carcinoma. In North America and Europe, the prevalence of chronic HBV infection is low and primarily results from immigration from endemic areas, sexual transmission, injection drug use, or nosocomial infection(1, 2).
Persistent HBV infection is influenced by a complex combination of viral, environmental, and genetic components including HBV genomic variability, host age, sex, plus concurrent infection with hepatitis C virus, hepatitis D virus, and HIV (4-7). However, segregation analysis and twin studies strongly support the role of host genetic components in determining the chronicity of HBV infection (8, 9). Several studies revealed that variants in several host genes, including IFNG, TNF, MBP, VDR and ESR1, were associated with persistent HBV infection or HBV clearance (10-14). The human leukocyte antigen (HLA) class II loci also have been reported associated with HBV infection (15-18). Recently, a genome-wide association study (GWAS) demonstrated that genetic variants in the HLA-DP locus were strongly associated with chronic hepatitis B in Japanese and Thai populations (19). Han Chinese constitute about 92% of the population of China, 98% of Taiwan, 78% of Singapore, and about 20% of the world population (20). In our study we screened eleven single nucleotide polymorphisms (SNPs) within the HLA-DP genes and one SNP in strong linkage disequilibrium (LD) with a neighboring HLA-DR13 locus for association with persistent HBV chronic infection in Han Chinese from Hebei and Henan Provinces of northern China.
Materials and Methods
Participants
Cases and controls were recruited from Zhengding County in Hebei Province and Luohe city in Henan Province of northern China from May to September 2006. In 1983 Zhengding County established a database for epidemiological study of hepatitis B and assessment of hepatitis B vaccine. Specific details on HBV infection, liver function, disease outcome (including death related with HBV infection), hepatitis B vaccination, education, socioeconomic status etc. were collected each year in several areas across the county. Luohe city’s database was established in 2004, and the HBV markers were screened in several communities from 2004 to 2005. The individuals who were hepatitis B surface antigen (HBsAg) positive were tested again one year later in 2006; similar to the Zhengding database, other relevant information was also collected on persons in the Luohe city database. About 2/3 of cases were identified from the Zhengding database and 1/3 of cases were from records of Luohe database. Cases were persistent chronic HBV carriers who had been positive for both HBsAg and antibody to hepatitis B core antigen (anti-HBc), or positive for HBsAg only for at least 1 year. Among chronic HBV carriers, 97% were anti-HBc positive, 4% anti-HBs positive only, and about 11% had alanine aminotransferase levels (ATL) of more than 40 IU (Mean 105 IU, range 41-403 IU; see table 1). Controls were identified from the Zhengding database. Controls were at least 30 years of age with normal ATL and no history of hepatitis B vaccination (Note: HBV vaccine was not available 30 years ago) including HBV natural clearances and healthy individuals. Clinical criteria for HBV natural clearance were: negative for HBsAg, plus positive for both antibody to hepatitis B surface antigen (anti-HBs) and anti-HBc, or plus anti-HBs positive without history of hepatitis B vaccination. About 70% HBV natural clearances were anti-HBc positive in our cohort (table 1). Healthy controls were negative for HBsAg, anti-HBs and anti-HBc without hepatitis B vaccination.
Table 1.
Characteristics of participants in a study of persistent chronic HBV carriers, Han population from Northern China
| Cases |
Controls |
||
|---|---|---|---|
| Carriers | Clearances | Healthy individuals | |
| Age (years) | 41±14.1 (SD) | 50±11.2 (SD) | 47±10.2 (SD) |
| Male, % | 51.4 (268/521) | 42.6 (243/3572) | 37.1 (92/248) |
| HBsAg+, % | 100 | 0 | 0 |
| Anti-HBs+, % | 4.4 (23/521) | 100 | 0 |
| Anti-HBc+, % | 97.3 (507/521) | 70.1 (400/571) | 0 |
| ATL>40, % | 11.3 (59/521) | 0 | 0 |
| Total | 521 | 571 | 248 |
Age was the age of enrollment for all groups.
SD=standard deviation.
All participants self-identified as Han Chinese and self-reported 6 or more months of residency in Zhengding County of Hebei Province or Luohe city of Henan Province, China. Persons with blood relatives enrolled in the study were excluded. HBV markers including HBsAg, anti-HBs and anti-HBc were confirmed by solid radioimmunoassay at the time of study enrollment. Plasma ATLs were measured by Reitman-Frankel method using a commercial kit. Blood samples were obtained from 521 persistent chronic HBV carriers (268 males and 253 females) and 819 controls (335 males and 484 females). The mean age was 41 years ±14 for HBV chronic carriers and 49 years ±11 for controls. The controls included 571 persons with HBV natural clearance and 248 never HBV-infected (healthy) individuals (see table 1). Institutional review board approval was obtained from all participating institutions and informed consent was obtained from each study participant.
Genotyping
Genomic DNA was extracted from whole blood using Phenol/Chloroform with MaXtract high density tubes. Four SNPs on HLA-DPA1, 7 SNPs on HLA-DPB1 and one SNP in strong LD with HLA-DR13 were genotyped using a commercially available TaqMan SNP genotyping assay and GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA), in accordance with the manufacturer’s instructions. The sequence detection software was used for allelic discrimination. For quality control, 8 template-free controls, one Han Chinese family sample from another study cohort (21) and 5% of duplicate samples with a different extraction were included in each 384-well plate. The rates of successful genotyping calls were 96.3% to 99.4% for the 12 SNPs.
Statistical analysis
Hardy-Weinberg Equilibrium (HWE) assumptions were independently tested for each polymorphism. Chi-square tests were used for allele case-control comparisons, which test for additive allele effects on the disease penetrance. Odds ratios (OR) were calculated based on the 2×2 table of allele-by-trait counts. Differences between the theoretical binomially distributed genotypes and observed genotype frequencies were tested using a χ2 goodness of fit test. Odd ratios and 95% confidence intervals (CI) were computed by logistic regression and all results shown are adjusted for age and gender.
Haplotype frequencies were estimated with the expectation-maximization (EM) algorithm, as implemented in SAS PROC Haplotype. Genotypes with all missing alleles are dropped from calculations for haplotype frequencies. Omnibus tests of haplotype association and haplotype-specific ORs were calculated by haplotype replacement regression, assuming an additive model using the probability of carrying each pair of haplotypes provided by PROC Haplotype. The most common haplotype or joint haplotype were used as the reference respectively; all the haplotypes with frequency of less than 1% were combined to others. All statistics were calculated in the statistical package SAS and SAS Genetics version 9.1.3. The LD map, blocks and haplotypes were generated by Haploview software (22).
Results
Twelve SNPs in the HLA class II loci were genotyped in 521 persistent chronic HBV carriers and 819 controls (571 persons with HBV natural clearance and 248 healthy individuals). The genotype frequencies for 12 polymorphisms conformed to Hardy-Weinberg equilibrium (HWE) expectations for controls. Table 2 lists the SNP ID, risk allele, number of subjects, OR with 95% CI, and p value for cases versus controls; p value for cases versus HBV natural clearances, and gene information for 12 SNPs. ORs and p values were adjusted for age and sex. Allele frequencies of 11 SNPs located within HLA-DPA1 and HLA-DPB1 gene complexes were significantly different between cases and controls (p = 1.82 × 10−12 to 0.01). The 6 most significant variants were located at HLA-DPB1 (p = 1.82 × 10−12 to 1.65 × 10−6) (Table 2; Figure 1). The polymorphism of rs11752643 in strong LD with HLA-DR13 was not significantly associated with persistent chronic HBV infection (Table 2). To investigate the association of these polymorphisms with HBV clearance, we further analyzed the data using only the 571 HBV clearance individuals as controls. As shown in table 2, the p values were quite similar (p = 2.70 × 10−11 to 0.003) for 11 SNPs located at HLA-DP, while rs11752643 remained non-significant.
Table 2.
The association of SNP variant alleles within HLA-DP and HLA-DR13 regions with chronic hepatitis B, Han population from Northern China (See Figure 1 for map position and LD relationships)
| SNP-ID | Risk Allele |
No. Cases |
No. Controls |
No. Clearances |
OR (95%CI)a |
P valueb (Controls) |
P valuec (Clearance) |
Nearest Gene |
|---|---|---|---|---|---|---|---|---|
| rs2395309 | G | 496 | 805 | 564 | 0.71 (0.59-0.86) | 0.0005 | 0.0002 | HLA-DPA1 |
| rs3077 | G | 514 | 808 | 562 | 0.64 (0.53-0.78) | 4.92E-06 | 6.0E-06 | HLA-DPA1 |
| rs2301220 | T | 509 | 796 | 557 | 0.67 (0.56-0.81) | 3.19E-05 | 4.44E-05 | HLA-DPA1 |
| rs9277341 | C | 511 | 805 | 563 | 1.77 (1.39-2.25) | 3.99E-06 | 1.28E-05 | HLA-DPA1 |
| rs3135021 | G | 514 | 813 | 566 | 0.78 (0.64-0.94) | 0.01 | 0.003 | HLA-DPB1 |
| rs9277535 | G | 498 | 794 | 553 | 0.56 (0.47-0.68) | 4.24E-09 | 5.61E-09 | HLA-DPB1 |
| rs10484569 | A | 514 | 807 | 564 | 1.60 (1.33-1.93) | 1.65E-06 | 3.20E-06 | HLA-DPB1 |
| rs3128917 | G | 519 | 811 | 566 | 1.91 (1.59-2.30) | 5.71E-12 | 4.62E-11 | HLA-DPB1 |
| rs2281388 | A | 502 | 801 | 556 | 1.66 (1.38-2.01) | 3.37E-07 | 4.65E-07 | HLA-DPB1 |
| rs3117222 | T | 511 | 791 | 552 | 0.51 (0.42-0.61) | 1.82E-12 | 2.70E-11 | HLA-DPB1 |
| rs9380343 | T | 501 | 806 | 562 | 0.61 (0.50-0.73) | 5.23E-07 | 4.95E-07 | HLA-DPB1 |
| rs11752643 | 519 | 813 | 566 | 1.75 (0.71-4.30) | 0.24 | 0.29 | HLA-DR13 |
Odds ratios (OR) were calculated basis on the 2×2 table of allele counts from cases vs. controls. CI: confidence interval.
P value of Armitage’s trend test from cases vs. controls (controls included HBV natural clearance and healthy individuals).
P value of Armitage’s trend test from cases vs. clearances (clearances were HBV natural clearances).
The definition of HBV clearance and healthy individuals included in Materials and Methods for clinical criteria and table 1.
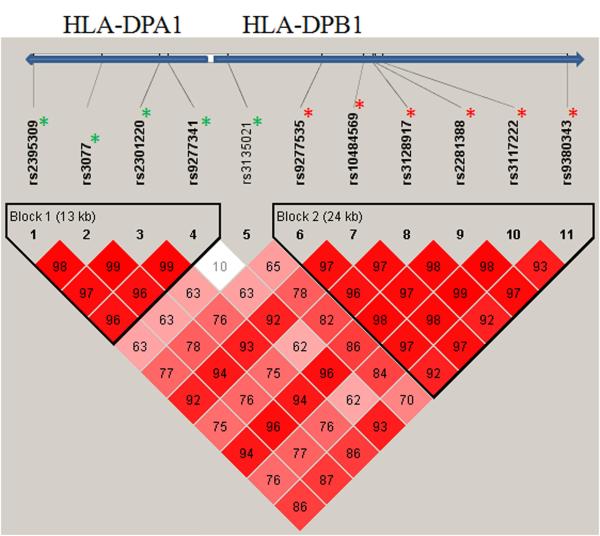
Figure 1.
LD map based on D’ was drawn using the genotype of the cases and controls
Red * indicated the minor alleles were risk alleles for those SNPs.
Green * indicated the minor alleles were protective alleles for those SNPs.
For 11 significant SNPs, we examined the association of genotype frequencies between cases and controls (both clearance and healthy combined), and also between cases and clearance controls only. Table 3 presents the genotype distribution in each group; OR with 95% CI and p values for carriers versus controls, and carriers versus clearances, respectively. As illustrated in Figure 1, the first five SNPs showed minor alleles (four in HLA-DPA1 and one adjacent within HLA-DPB1) associated with decreasing risk/protection of HBV chronic infection (Table 3; OR = 0.33 to 0.66, p = 6.7× 10−7 to 0.045 for homozygote, OR = 0.50 to 0.77, p = 4.6× 10−7 to 0.036 for heterozygote). The first 4 SNPs located in HLA-DPA1 formed haplotype block 1 (Fig. 1). The last six variants located on gene HLA-DPB1 had minor alleles significantly associated with increasing risk/susceptibility of HBV chronic infection (OR = 2.46 to 3.34, p = 5.7× 10−12 to 7.0 × 10−7 for homozygote, OR = 1.56 to 2.36, p = 6.0× 10−9 to 0.004 for heterozygote). These 6 SNPs with susceptibility minor alleles formed haplotype block 2 (Fig. 1). Similar significant associations were observed when we compared HBV carriers with HBV clearances (Table 3; column 8, 9).
Table 3.
Genotype distribution and association with chronic hepatitis B, Han population from Northern China
| SNP | Genotype | Carriers (%) |
Controls (%) |
Clearances (%) |
Carriers vs Controls | Carriers vs Clearances | ||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | |||||
| rs2395309 | GG | 226 (45.6) | 261 (32.4) | 175 (31.0) | 1.0 | 1.0 | ||
| AG | 209 (42.1) | 371 (46.1) | 262 (46.5) | 0.71 (0.55-0.92) | 0.0097 | 0.65 (0.49-0.87) | 0.0038 | |
| AA | 61 (12.3) | 173 (21.5) | 127 (22.5) | 0.47 (0.33-0.67) | 4E-05 | 0.42 (0.29-0.62) | 1.10E-05 | |
| rs3077 | GG | 247 (48.0) | 259 (32.0) | 180 (32.0) | 1.0 | 1.0 | ||
| AG | 211 (41.1) | 382 (47.3) | 262 (46.6) | 0.63 (0.48-0.81) | 0.0004 | 0.63 (0.47-0.83) | 0.0012 | |
| AA | 56 (10.9) | 167 (20.7) | 120 (21.4) | 0.39 (0.27-0.57) | 6.7E-07 | 0.37 (0.25-0.55) | 7.89E-07 | |
| rs2301220 | TT | 232 (45.6) | 241 (30.3) | 166 (29.8) | 1.0 | 1.0 | ||
| CT | 215 (42.2) | 384(48.2) | 270 (48.5) | 0.64 (0.49-0.83) | 0.0008 | 0.61 (0.46-0.81) | 0.0007 | |
| CC | 62 (12.2) | 171 (21.5) | 121 (21.7) | 0.42 (0.30-0.61) | 3E-06 | 0.40 (0.27-0.59) | 4.83E-06 | |
| rs9277341 | CC | 388 (75.9) | 472 (58.6) | 334 (59.3) | 1.0 | 1.0 | ||
| CT | 110 (21.5) | 284 (35.3) | 191 (33.9) | 0.50 (0.38-0.65) | 4.6E-07 | 0.51 (0.38-0.69) | 8.28E-06 | |
| TT | 13 (2.6) | 49 (6.1) | 38 (6.8) | 0.33 (0.18-0.64) | 0.0009 | 0.30 (0.16-0.60) | 0.0005 | |
| rs3135021 | GG | 260 (50.6) | 351 (43.2) | 239 (42.2) | 1.0 | 1.0 | ||
| AG | 206 (40.1) | 363 (44.6) | 249 (44.0) | 0.77 (0.60-0.98) | 0.036 | 0.77 (0.59-1.02) | 0.067 | |
| AA | 48 (9.3) | 99 (12.2) | 78 (13.8) | 0.66 (0.44-0.99) | 0.045 | 0.55 (0.36-0.85) | 0.007 | |
| rs9277535 | AA | 98 (19.7) | 266 (33.5) | 193 (34.9) | 1.0 | 1.0 | ||
| AG | 217 (43.6) | 364 (45.8) | 256 (46.3) | 1.56 (1.16-2.11) | 0.0035 | 1.69 (1.22-2.33) | 0.0014 | |
| GG | 183 (36.7) | 164 (20.7) | 104 (18.8) | 2.75 (1.98-3.82) | 1.9E-09 | 3.25 (2.26-4.68) | 2.06E-10 | |
| rs10484569 | GG | 175 (34.1) | 402 (49.8) | 281 (49.8) | 1.0 | 1.0 | ||
| AG | 237 (46.1) | 318 (39.4) | 229 (40.6) | 1.67 (1.29-2.17) | 9.3E-05 | 1.63 (1.23-2.15) | 0.0007 | |
| AA | 102 (19.8) | 87 (10.8) | 54 (9.6) | 2.46 (1.73-3.52) | 7E-07 | 2.80 (1.87-4.20) | 6.12E-07 | |
| rs3128917 | TT | 106 (20.4) | 320 (39.5) | 233 (41.2) | 1.0 | 1.0 | ||
| GT | 263 (50.7) | 358 (44.1) | 245 (43.3) | 2.18 (1.64-2.91) | 8.5E-08 | 2.40 (1.76-3.26) | 2.57E-08 | |
| GG | 150 (28.9) | 133 (16.4) | 88 (15.5) | 3.17 (2.26-4.45) | 2.4E-11 | 3.53 (2.43-5.12) | 3.24E-11 | |
| rs2281388 | GG | 175 (34.9) | 411 (51.3) | 288 (51.8) | 1.0 | 1.0 | ||
| AG | 238 (47.4) | 316 (39.5) | 222 (39.9) | 1.77 (1.37-2.29) | 1.5E-05 | 1.79 (1.35-2.37) | 4.93E-05 | |
| AA | 89 (17.7) | 74 (9.2) | 46 (8.3) | 2.67 (1.83-3.89) | 3.1E-07 | 3.09 (2.01-4.74) | 2.50E-07 | |
| rs3117222 | CC | 104 (20.3) | 319 (40.3) | 231 (41.8) | 1.0 | 1.0 | ||
| CT | 260 (50.9) | 344 (43.5) | 236 (42.8) | 2.36 (1.77-3.16) | 6E-09 | 2.56 (1.87-3.50) | 3.56E-09 | |
| TT | 147 (28.8) | 128 (16.2) | 85 (15.4) | 3.34 (2.37-4.71) | 5.7E-12 | 3.68 (2.52-5.36) | 1.30E-11 | |
| rs9380343 | CC | 165 (32.9) | 397 (49.3) | 280 (49.8) | 1.0 | 1.0 | ||
| CT | 239 (47.7) | 325 (40.3) | 231 (41.1) | 1.81 (1.39-2.35) | 8.3E-06 | 1.84 (1.39-2.44) | 2.36E-05 | |
| TT | 97 (19.4) | 84 (10.4) | 51 (9.1) | 2.65 (1.84-3.82) | 1.5E-07 | 3.12 (2.06-4.73) | 7.84E-08 | |
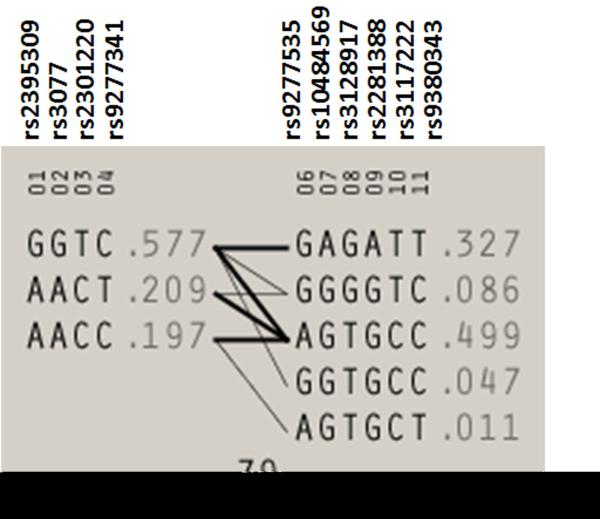
Next we examined haplotype association for block 1, block 2 and the two blocks combined. Table 4 lists the haplotype frequencies in cases and controls, OR with 95% CI and p values for block 1 and block 2. The haplotype AACT, which retains all rare protective alleles of block 1, was significantly associated with decreasing risk of chronic hepatitis B infection (OR = 0.54, = 8.7× 10−7). The haplotype GAGATT (which retains all rare susceptible alleles of block 2) and GGGGTC (which retains 3 rare susceptible alleles of block 2) were significantly associated with increased the risk of chronic hepatitis B infection (OR = 2.0, p= 1.4× 10−10 for GAGATT; OR=1.7 p= 0.002 for GGGGTC). Table 5 presents a combination of haplotype block 1 and block 2 considered together. The combined protective haplotypes of block 1 (AACT) and block 2 (AGTGCC) were very strongly associated with decreased risk of chronic hepatitis B (OR =0.36, p = 3.0× 10−11). The protective haplotype of block 2 (AGTGCC) combined with other haplotypes of block 1 were also significantly associated with decreased risk of chronic hepatitis B infection (OR =0.56 to −0.65, p = 0.002 to 0.0002).
Table 4.
Results of haplotype association analysis, Han population from Northern China (N=412 cases; N=680 controls)
| Block | Haplotype | Cases (Freq) | Controls (Freq) | OR (95%CI) | p value |
|---|---|---|---|---|---|
| Block 1 | |||||
| GGTC I | 538 (65.3) | 725 (53.3) | 1.0 | ||
| AACT | 122 (14.8) | 332 (24.4) | 0.54 (0.42-0.69) | 8.73E-07 | |
| AACC | 153 (18.6) | 276 (20.3) | 0.82 (0.64-1.04) | 0.10 | |
| Others | 11 (1.3) | 27 (2.0) | 0.61 (0.29-1.30) | 0.20 | |
| Block 2 | |||||
| AGTGCCI | 326 (39.6) | 757 (55.7) | 1.0 | ||
| GAGATT | 332 (40.3) | 382 (28.1) | 1.98 (1.61-2.44) | 1.37E-10 | |
| GGGGTC | 81 (9.8) | 106 (7.8) | 1.72 (1.23-2.40) | 0.002 | |
| GGTGCC | 45 (5.5) | 59 (4.3) | 1.54 (0.99-2.40) | 0.055 | |
| AGTGCT | 10 (1.2) | 14 (1.0) | 1.92 (0.80-4.61) | 0.14 | |
| Others | 30 (3.6) | 42 (3.1) | 1.80 (1.07-3.04) | 0.03 | |
The most common haplotype as the reference.
Bolded are risk alleles.
OR: odds ratio. CI: confidence interval.
SNPs order of block 1: rs2395309, rs3077, rs2301220 and rs9277341 within the HLA-DPA1 gene.
SNPs order of block 2: rs9277535, rs10484569, rs3128917, rs2281388, rs3117222 and rs9380343 within the HLA-DPB1 gene.
Table 5.
Joint effects of haplotype block 1 and block 2 with chronic hepatitis B
| Joint Haplotype | Cases (Freq) | Controls (Freq) | OR (95%CI) | p value |
|---|---|---|---|---|
| GGTC * GAGATTI | 311 (37.7) | 363 (26.7) | 1.0 | |
| AACC * AGTGCC | 136 (16.5) | 257 (18.9) | 0.65 (0.49-0.85) | 0.0020 |
| AACT * AGTGCC | 85 (10.3) | 292 (21.5) | 0.36 (0.27-0.49) | 3.03E-11 |
| GGTC * AGTGCC | 102 (12.4) | 196 (14.4) | 0.56 (0.41-0.75) | 0.0002 |
| GGTC * GGGGTC | 63 (7.7) | 80 (5.9) | 0.90 (0.61-1.32) | 0.58 |
| GGTC* GGTGCC | 43 (5.2) | 56 (4.1) | 0.81 (0.51-1.29) | 0.38 |
| AACT * GGGGTC | 17 (2.1) | 21 (1.5) | 0.92 (0.46-1.83) | 0.80 |
| Others | 67 (8.1) | 95 (7.0) | 0.89 (0.61-1.29) | 0.54 |
The most common joint haplotype as the reference.
OR: odds ratio. CI: confidence interval.
Bolded are risk alleles.
Discussion
In this study, twelve SNPs which were previously reported to be associated with chronic hepatitis B (18, 19) were interrogated in 521 persistent chronic HBV carriers and 819 controls in a Chinese Han population from northern China. Eleven SNPs located within HLA-DPA1 and HLA-DPB1 were strong significantly associated with persistent chronic HBV carrier status (Table 2). The genotype association analysis further confirmed this association (Table 3). The minor alleles of five SNP loci (four on HLA-DPA1 and one in the HLA-DPB1 region) were protective from risk of chronic hepatitis B. The minor alleles of six SNP loci in the HLA-DPB1 gene region were susceptible for chronic hepatitis B (Table 2 and 3). A closely adjacent SNP, rs11752643 that did not track HLA DPA1 or HLA-DPB2, but which did show strong LD with HLA-DR13, was not associated with chronic hepatitis B. Our results, from an independent Han Chinese population, replicated in SNP allele direction and statistical strength the reported Japanese/Thai GWAS association (19).
The haplotypes directly inform how alleles are organized along the chromosome and may provide additional power for mapping disease genes; haplotypes also provide insight on factors influencing the dependency among genetic markers. The haplotype-based methods can potentially capture cis-interactions between two or more causal variants. The haplotypes should be more informative than individual genotypes for revealing disease-causing mechanisms at a candidate gene (23). Based on these assumptions, we explored the haplotype and joint haplotype association of significant SNPs with chronic hepatitis B infection. The dominant (major) alleles were risk allele for rs2395309, rs3077, rs2301220, rs9277341 and rs3135021 in the Han Chinese population. The first 4 of these SNPs formed haplotype block 1; the haplotype GGTC combined by risk alleles was the most common haplotype (freq. =0.577) in our cohort (Table 4 and Fig. 2). The less common haplotype AACT (freq. =0.209) combined by protective alleles was significantly associated with decreasing risk of chronic hepatitis B infection (see table 2 and 4). The dominant alleles were protective allele for rs9277535, rs10484569, rs3128917, rs2281388, rs3117222 and rs9380343 located on HLA-DPB1 in our study population. These 6 SNPs formed haplotype block 2, the haplotype AGTGCC (freq. =0.499) combined by protective alleles was the most common haplotype (Table 4 and Fig. 2). The haplotype GAGATT (freq. =0.327) combined by risk alleles was significantly associated with increasing risk of HBV chronic infection (OR = 2.0, p = 1.4× 10−10, see table 4). The haplotype GGGGTC with 3 risk alleles was also associated with significant susceptibility to HBV chronic infection. We also tested the joint effects for haplotype block 1 and block 2. The susceptible haplotypes GGTC of block 1 and GAGATT of block 2 comprised the most common joint haplotype (Table 5). The joint haplotype of two protective haplotypes from block 1 and block 2 (AACT*AGTGCC) exerted a strong protective effect against HBV chronic infection (OR =0.4, p = 3.0× 10−11, table 5). The joint haplotypes including the protective haplotype of block 2 (AGTGCC) showed significant protective effects as well (Table 5). These findings supported single-SNP association results.
Figure 2.
Markers are shown across the top. Population frequencies are shown next to each haplotype and lines show the most common crossings from one block to the next, with thicker lines showing more common crossings than thinner lines. Shown beneath the crossing lines is multilocus D prime, which is a measure of the LD between two blocks.
Our analysis included 12 comparisons, but the 12 SNPs are not independent; 10 of them formed two haplotype blocks. A Bonferroni correction for multiple SNPs tested, which assumes the independence of all tests performed, is overly conservative. Nonetheless, after a correction for multiple SNP comparisons (current p value times 4), all 11 SNPs retain statistical significance (table 2 and 3).
Genes involved in the immune response including HLA loci are among the most numerous and diverse in the human genome. Classical HLA loci spanning 4-Mb on the short arm of chromosome 6p21 (24) include the class I and class II molecules identified for their role in presentation of antigen to CD8+ and CD4+ T cells, respectively. The HLA class II molecules are expressed as cell surface glycoproteins that bind and present short peptide epitopes to CD4+ T cells. Each HLA subtype has a particular binding motif that dictates a specific range of peptides that can physically bind in a groove on the surface of the HLA molecule (25). Human HLA class II molecules are classified in three isotypes: HLA-DR, -DQ and -DP. Compared to other class II molecules, very limited information is available concerning peptide interactions and the role of HLA-DP polymorphic positions both in peptide binding and T cell recognition. Functional analysis has shown that HLA-DP plays a key role in T cell allorecognition and peptide binding (26). There are no specific amino acids changes for 11 significant SNP variants, but these 11 SNPs located within or around the HLA-DPA1 and HLA-DPB1 locus, spanning a 52kb region of chromosome 6, were in very strong LD with HLA-DP alleles. The 11 SNPs are likely the proxy markers for adjacent, but yet to be identified, functional HLA-DP polymorphisms. Our finding suggests that variations in HLA-DP molecules would influence binding or presentation of viral peptides, perhaps regulating virus clearance and chronic hepatitis B pathogenesis. Further study should focus on how these variants impact gene expression and function.
In summary, our results further confirm that genetic variants in the HLA-DP locus are strongly associated with persistent HBV infection in the Han Chinese population of Northern China.
Acknowledgments
We thank all the participants in the cohorts. We thank Cheryl A. Winkler for invaluable discussion. We thank Michael Campsmith for review and editing the manuscript. We thank Man-Huei Chang and Quanhe Yang for sharing SAS genetics software.
Grant support: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- HLA
Human leukocyte antigen
- HBV
Hepatitis B virus
- GWAS
Genome-wide association study
- SNPs
Single nucleotide polymorphisms
- MAF
Minor allele frequency
- LD
Linkage disequilibrium
- OR
Odds ratios
- CI
Confidence interval
- HIV
Human immune deficiency virus
- HBsAg
Hepatitis B surface antigen
- HBeAg
Hepatitis e antigen
- anti-HBs
antibody to hepatitis B surface antigen
- anti-HBc
antibody to hepatitis B core antigen
References
- 1.Ocama P, Opio CK, Lee WM. Hepatitis B virus infection: current status. Am J Med. 2005;118:1413. doi: 10.1016/j.amjmed.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. Chronic viral hepatitis as a public health issue in the world. Best Pract Res Clin Gastroenterol. 2008;22:991–1008. doi: 10.1016/j.bpg.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Guo XC, Wu YQ. A review: progress of prevention and control on viral hepatitis in China. Biomed Environ Sci. 1999;12:227–232. [PubMed] [Google Scholar]
- 4.Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology. 2002;35:1274–1276. doi: 10.1053/jhep.2002.33161. [DOI] [PubMed] [Google Scholar]
- 5.Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641–644. doi: 10.3748/wjg.v9.i4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res. 2001;52:113–116. doi: 10.1016/s0166-3542(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 7.Frodsham AJ, Hill AV. Genetics of infectious diseases. Hum Mol Genet. 2004;13 Spec No 2:R187–194. doi: 10.1093/hmg/ddh225. [DOI] [PubMed] [Google Scholar]
- 8.Hann HW, Kim CY, London WT, Whitford P, Blumberg BS. Hepatitis B virus and primary hepatocellular carcinoma: family studies in Korea. Int J Cancer. 1982;30:47–51. doi: 10.1002/ijc.2910300109. [DOI] [PubMed] [Google Scholar]
- 9.Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, et al. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–741. [PubMed] [Google Scholar]
- 10.Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144–150. doi: 10.1111/j.1572-0241.2003.07179.x. [DOI] [PubMed] [Google Scholar]
- 11.Hohler T, Kruger A, Gerken G, Schneider PM, zum Buschenefelde KH Meyer, Rittner C. A tumor necrosis factor-alpha (TNF-alpha) promoter polymorphism is associated with chronic hepatitis B infection. Clin Exp Immunol. 1998;111:579–582. doi: 10.1046/j.1365-2249.1998.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas HC, Foster GR, Sumiya M, McIntosh D, Jack DL, Turner MW, Summerfield JA. Mutation of gene of mannose-binding protein associated with chronic hepatitis B viral infection. Lancet. 1996;348:1417–1419. doi: 10.1016/s0140-6736(96)05409-8. [DOI] [PubMed] [Google Scholar]
- 13.Bellamy R, Ruwende C, Corrah T, McAdam KP, Thursz M, Whittle HC, Hill AV. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis. 1999;179:721–724. doi: 10.1086/314614. [DOI] [PubMed] [Google Scholar]
- 14.Deng G, Zhou G, Zhai Y, Li S, Li X, Li Y, Zhang R, et al. Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology. 2004;40:318–326. doi: 10.1002/hep.20318. [DOI] [PubMed] [Google Scholar]
- 15.Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 16.Hohler T, Gerken G, Notghi A, Lubjuhn R, Taheri H, Protzer U, Lohr HF, et al. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol. 1997;26:503–507. doi: 10.1016/s0168-8278(97)80414-x. [DOI] [PubMed] [Google Scholar]
- 17.Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, Kim YS, et al. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000;31:1371–1373. doi: 10.1053/jhep.2000.7988. [DOI] [PubMed] [Google Scholar]
- 18.Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841–848. doi: 10.1111/j.1365-2893.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 19.Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 20.Chinese Han., editor. http://en.wikipedia.org/wiki/Han_Chinese.
- 21.Guo XC, Scott K, Liu Y, Dean M, David V, Nelson GW, Johnson RC, et al. Genetic factors leading to chronic Epstein-Barr virus infection and nasopharyngeal carcinoma in South East China: study design, methods and feasibility. Hum Genomics. 2006;2:365–375. doi: 10.1186/1479-7364-2-6-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Liu N, Zhang K, Zhao H. Haplotype-association analysis. Adv Genet. 2008;60:335–405. doi: 10.1016/S0065-2660(07)00414-2. [DOI] [PubMed] [Google Scholar]
- 24.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godkin A, Davenport M, Hill AV. Molecular analysis of HLA class II associations with hepatitis B virus clearance and vaccine nonresponsiveness. Hepatology. 2005;41:1383–1390. doi: 10.1002/hep.20716. [DOI] [PubMed] [Google Scholar]
- 26.Diaz G, Amicosante M, Jaraquemada D, Butler RH, Guillen MV, Sanchez M, Nombela C, et al. Functional analysis of HLA-DP polymorphism: a crucial role for DPbeta residues 9, 11, 35, 55, 56, 69 and 84-87 in T cell allorecognition and peptide binding. Int Immunol. 2003;15:565–576. doi: 10.1093/intimm/dxg057. [DOI] [PubMed] [Google Scholar]