Abstract
Using fusions of green fluorescent protein to subunits of RNA polymerase (RNAP) and ribosomes, we have investigated the subcellular localization of the transcriptional and translational machinery in the bacterium Bacillus subtilis. Unexpectedly, we found that RNAP resides principally within the nucleoid. Conversely, ribosomes localized almost exclusively outside the nucleoid, concentrating particularly towards sites of cell division. This zonal localization was not dependent on cell division and is probably due, at least in part, to exclusion from the nucleoid. Dual labelling of RNAP and ribosomes was used to confirm the spatial separation of the two processes. We conclude that, even in the absence of a nuclear membrane, transcription and translation occur predominantly in separate functional domains. At higher growth rates, concentrations of RNAP developed, probably representing the sites of rRNA synthesis. These may represent a further spatial specialization, possibly equivalent to the eukaryotic nucleolus.
Keywords: Bacillus subtilis/compartmentalization/green fluorescent protein/ribosomes/RNA polymerase
Introduction
In recent years, it has become clear that the bacterial cytoplasm is not a homogeneous, undifferentiated medium in which proteins are free to diffuse. Rather, it appears that many proteins, such as those involved in cell division, DNA replication and chromosome segregation, have specific subcellular addresses (Jacobs and Shapiro, 1999; Losick and Shapiro, 1999). Classical experiments performed in the 1970s by Miller and co-workers indicated that transcription and translation are tightly linked, based on their ability to isolate DNA–RNA polymerase (RNAP)–RNA–ribosome complexes (Miller et al., 1970). Additional electron microscopy studies, using ultrathin sections of cryofixed cells, led others to suggest that the bacterial nucleoid forms a dense central mass from which DNA loops extend out into the cytoplasm (Hobot et al., 1985; Robinow and Kellenberger, 1994). Subsequently, it was proposed that transcription occurs on these loops, permitting coupled transcription and translation with the transcription machinery and ribosomes in close proximity (Ryter and Change, 1975; Woldringh and Nanninga, 1985; Woldringh et al., 1995; Figure 1A). On the basis of these observations, RNAP might be expected to lie at the periphery of the nucleoid in a zone available for interaction with ribosomes.
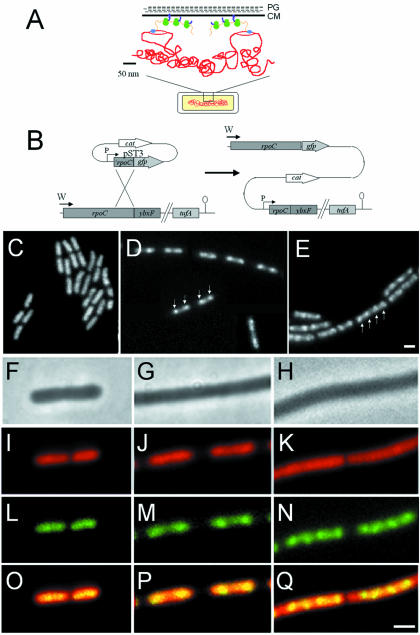
Fig. 1. (A) Schematic representation of transcription and translational organization in bacteria (derived from Woldringh et al., 1995). PG, peptidoglycan; CM, cytoplasmic membrane. Gram-positive organisms do not contain an outer membrane. The boxed region of the cell (bottom) is shown in expanded form above. Transcription by RNAP (blue) from DNA (red) that has looped out from the dense central mass of the nucleoid is closely linked to translation by ribosomes (green) via mRNA (orange). In this figure, the translated proteins (purple) are being inserted simultaneously into the cell membrane, but cytoplasmic proteins have also been proposed to be transcribed and translated in a similar peripheral subcellular location (Woldringh et al., 1995). (B) Construction of the fusion strain 1048 by insertion of pST3 into the chromosome. Integration of pST3 fuses the full-length rpoC gene, under the control of its natural promoter (W), to gfp. The plasmid provides a xylose-inducible promoter (P) to drive transcription of downstream genes, beginning with a truncated copy of rpoC. tufA is the last gene of the operon and encodes elongation factor Tu. (C–E) Images of RNAP–GFP in cells grown at 37°C in S medium [(C) doubling time 70 min], CH medium [(D) doubling time 45 min] and 2TY medium [(E) doubling time 20 min]. (D) is a composite image due to the low density of cells in the sample. (F–Q) Further magnified images of typical cells. (F), (I), (L) and (O) Cells grown in S medium. (G), (J), (M) and (P) Cells grown in CH medium. (H), (K), (N) and (Q) Cells grown in 2TY medium. (F–H) Phase contrast images; (I–K) DAPI images false coloured red; (L–N) RNAP–GFP images false coloured green; (O–Q) overlays of DAPI and GFP images. Scale bar, 2 μm.
As with all bacteria, Bacillus subtilis RNAP is a large multi-subunit enzyme comprising a catalytic core composed of 2α, β and β′ subunits, together with a variable regulatory factor (σ) that determines promoter specificity (Losick and Chamberlin, 1976). RNAP undergoes a transcription cycle involving promoter binding and clearance, elongation, termination and non-specific strand exchange until a promoter is re-bound, and so it is associated continuously with DNA. In addition, it is thought that RNAP is present at subsaturating levels during exponential growth (∼2000 copies per chromosome), so all of it will be involved in this transcription cycle (Jensen and Pedersen, 1990; Bremer and Dennis, 1996).
Ribosomes also cycle during translation. Mature 70S ribosomes translate mRNA and, on completion, dissociate into their component 30S and 50S subunits. The 30S subunits then bind initiation factors and a new mRNA, prior to re-association with a 50S subunit and initiation of a new round of translation (Gualerzi and Pon, 1990; Janosi et al., 1996; Karimi et al., 1999). Multiple ribosomes are also able to assemble on a single mRNA to form polysomes (Miller et al., 1970; Figure 1A). Since translation of a message takes much longer than recycling and assembly of a 70S–mRNA ribosome, ∼80% of subunits are present as mature ribosomes over a range of growth rates in Escherichia coli (Forchammer and Lindhal, 1971; Bremer and Dennins, 1996; Asai et al., 1999). Due to the high conservation of the translation apparatus, the proportion of 70S ribosomes to subunits is likely to be similar in B.subtilis and E.coli.
In this report, we have examined the distribution of green fluorescent protein (GFP) fusions to RNAP and ribosomes, together with deconvolution microscopy, to test the hypothesis shown in Figure 1A. Surprisingly, we found that RNAP was concentrated within the nucleoid rather than in the nucleoid periphery. Furthermore, at higher growth rates, RNAP became concentrated into foci that may be similar in function to eukaryotic nucleoli, in carrying out rRNA synthesis. Conversely, ribosomes were deficient in the part of the cytoplasm occupied by the nucleoid and were distributed around the cell periphery, being particularly concentrated at the cell poles. Thus, transcription and translation appear to occur predominantly in spatially separated domains within the bacterial cell.
Results
RNA polymerase localization, and its growth rate-dependent recruitment into foci
To determine the localization of RNAP in living cells, we constructed fusions of GFP to the α and β′ subunits of the core enzyme. Similar results were obtained with both fusions, so detailed results are only presented for the rpoC–gfp fusion (β′ subunit; strain 1048; Table I). In this strain, the only functional copy of the essential rpoC gene was the one fused to gfp (Figure 1B). The growth rate of strain 1048 in different media was indistinguishable from that of 168 (wild-type). Development was unaffected as the cells appeared to sporulate normally (spo+), and Western blotting with anti-GFP antibodies revealed only full-length fusion protein (not shown). We conclude that the fusion protein is fully functional.
Table I. Strains and plasmids used in this study.
| Strain/plasmid | Genotype | Source/construction |
|---|---|---|
| 168 | trpC2 | C.Anagnostopoulos |
| 1032 | trpC2 amyE:: (spc Pxyl-spo0J–gfp) | Lewis and Marston (1999) |
| 1510 | trpC2 amyE chr::pSG1517 (spo0J–gfpS65T cat) | Glaser et al. (1997) |
| 1517 | trpC2 amyE chr::pSG1223 (spo0J′–gfpS65T cat) | Glaser et al. (1997) |
| 1758 | trpC2 chr::pSG1045 (diIVA′–gfpS65T diIVA+ cat) | Edwards and Errington (1997) |
| 1960 | trpC2 amyE chr::pSG1726 (soj–gfp cat) | Marston and Errington (1999b) |
| 1989 | trpC2 chr::pSG1732 (ΩminC lacZ ermC Pspac-minD) Ω(amyE::spc Pxyl–gfp–minC)1731 | Marston and Errington (1999a) |
| BB11 | chr::pJSIZΔpble (Pspac-ftsZ ble) | Beall and Lutkenhaus (1991) |
| 1048 | trpC2 chr::pST3 (rpoC–gfpa cat Pxyl-′rpoC) | 168 transformed with pST3 |
| 1049 | trpC2 amyE:: (spc Pxyl-rpsB–gfp) | 168 transformed with pST7 |
| 1054 | trpC2 Pspac-ftsZ amyE::(spc Pxyl-rpsB–gfp) | BB11 transformed with pST7 |
| 1055 | trpC2 chr::pSG1188 (rpsB–gfp cat Pxyl-rpsB) | 168 transformed with pSG1188 |
| 1056 | trpC2 amyE:: (spc Pxyl-rpsB–gfp) chr::pST5 (rpoC–gfpuv cat ′rpoC) | 1049 transformed with pST5 |
| 1057 | trpC2 amyE:: (spc Pxyl-spo0J–gfp) chr::pST5 (rpoC–gfpuv cat ′rpoC) | 1032 transformed with pST5 |
| pSG1151 | bla cat gfp | Lewis and Marston (1999) |
| pSG1154 | bla amyE3′ spc Pxy–′gfp amyE5′ | Lewis and Marston (1999) |
| pSG1156 | bla cat gfpuv | Lewis and Marston (1999) |
| pSG1164 | bla cat Pxyl–gfp | Lewis and Marston (1999) |
| pSG1188 | bla cat Pxyl-rpsB–gfp | this work |
| pST3 | bla cat Pxyl-′rpoC–gfp | this work |
| pST5 | bla cat ′rpoC–gfpuv | this work |
| pST7 | bla amyE3′ spc Pxyl-rpsB–gfp amyE5′ | this work |
aFor all but strains 1510, 1517 and 1758 (and gfpuv in dual labelled strains), the GFP used was the F64L S65T variant described by Cormack et al. (1996).
In S medium [doubling time (Td) 75 min] the RNAP–GFP signal was generally undifferentiated with a morphology similar to 4′,6-diamidino-2-phenylindole (DAPI)-stained nucleoids (Figure 1C). However, at moderate (CH medium; Td 45 min; Figure 1D) or high (2TY medium; Td 20 min; Figure 1E) growth rates, RNAP became concentrated into sub-regions we have named transcription foci (TFs; arrows, Figure 1D and E) whose intensity and frequency became progressively greater, indicating that they are generated in a growth rate-dependent manner. Magnified images of cells co-stained with DAPI to visualize their nucleoids are shown in Figure 1F–Q. Co–localization of DNA and RNAP in S medium cells showed that the signals were coincident, with no RNAP signal visible beyond the boundaries of the nucleoid (Figure 1O). In CH medium (Figure 1P), TFs localized toward the outer ends of the nucleoid, whereas multiple TFs appeared evenly distributed along the nucleoid in 2TY medium (Figure 1Q). In all media, whilst the RNAP and DAPI signals were coincident, the polymerase appeared to be concentrated particularly within the boundaries of the DNA stain (Figure 1O–Q), supporting the biochemical data indicating that it is always bound to DNA (Bremer and Dennis, 1996).
Although the data presented above suggested that RNAP was concentrated in the core of the nucleoid, due to the relatively low resolution of the light microscope, it was still possible that the signal observed arose from RNAP distributed predominantly around the nucleoid periphery. Also, the concentration of RNAP into foci at higher growth rates could simply reflect non-specific binding to DNA in the origin-proximal region of the chromosome as this is present in multiple copies during exponential growth. As a control in these experiments, we used strain 1960 (Table I) containing an Soj–GFP fusion in an spo0J mutant background. Normally, Soj behaves in a dynamic manner, jumping from one nucleoid to another in an unusual irregular manner, but in the absence of Spo0J it binds uniformly to all nucleoids (Marston and Errington, 1999b).
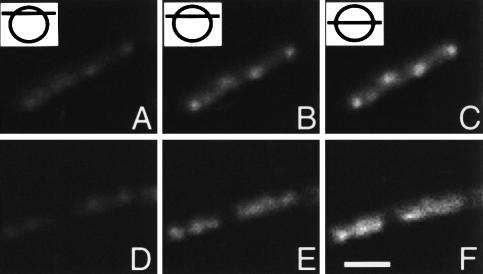
Image stacks of cells from strains 1048 and 1960 grown in CH were obtained and processed to remove out of focus light (deconvolved). Three optical sections from the image stacks are shown taken at high, intermediate and median levels in the cell as indicated (RNAP, Figure 2A–C; Soj, Figure 2D–F). The RNAP signal increased in intensity from the top through to the middle of the cell, and there was no indication that it was concentrated towards the outer edges of the nucleoid (Figure 2A–C). TFs were also clearly visible in the image slices as regions of intense signal, growing in magnitude from the top to the middle of the cell. The distribution of Soj–GFP throughout the nucleoid was not similar to RNAP, and it appeared to be distributed heterogeneously throughout the nucleoid (Figure 2D–F). Even if patterns of staining were observed that bore a passing resemblence to TFs (e.g. see right hand cell, Figure 2D), they represented local heterogeneities as they were not present throughout the cell (Figure 2E and F). Thus, contrary to expectation, RNAP seems to lie mostly within the bulk of the nucleoid, rather than being concentrated in a peripheral zone. Moreover, its concentration into foci seems to be a feature of this particular protein, and is not due simply to the increased dosage of origin regions at higher growth rates.
Fig. 2. High resolution localization of RNAP and Soj. Strains 1048 (rpoC–gfp) and 1960 (soj–gfp) were grown in CH medium at 37°C, and image slices obtained and processed to remove out of focus light as detailed in Materials and methods. Three slices taken from the image stacks are shown and cartoons with lines indicating the relative positions of the sections selected from the image stacks shown in the top left hand corners of (A–C). (A–C) Strain 1048. (D–F) Strain 1960. Scale bar, 2 μm.
RNA polymerase foci represent sites of rRNA synthesis
Since the majority of RNA synthesized at high growth rates is rRNA (Jensen and Pedersen, 1990; Bremer and Dennis, 1996), it seemed likely that TFs represented sites of rRNA synthesis. Since seven of the 10 rRNA (rrn) gene clusters are located within a 200 kbp region of DNA to one side of oriC (Kunst et al., 1997), TFs should be similar in number and subcellular position to oriC regions, which can be visualized conveniently using an Spo0J–GFP fusion (Glaser et al., 1997; Lewis and Errington, 1997; Lin et al., 1997). Spo0J binds to at least eight sites that span a region of ∼15% of the chromosome, centred approximately around oriC (Lin and Grossman, 1998). At a slow growth rate (S medium; Table II), when the rate of rRNA synthesis should be low, only a small proportion of the cells contained visible TFs, but those that did almost always contained two, corresponding to the two oriC regions that are present through most of the cell cycle under these conditions (Sharpe et al., 1998). However, there was a strong correlation between the average numbers of TF and Spo0J foci in both CH and 2TY media (Table II). Nearly all of the cells contained visible TFs, and the average number per nucleoid was similar to that calculated for Spo0J. As a further confirmation that TFs and Spo0J occupied similar subregions of the nucleoid, strain 1057 (Table I) was constructed containing rpoC–gfpuv and spo0J–gfp fusions. These fusions can be visualized separately due to their separate excitation spectra (Lewis and Marston, 1999). Analysis of cells grown in CH at 37°C indicated that nearly all TFs and Spo0J foci co-localized: 79% were perfectly coincident, 17% were partially coincident and 3% were separate (not shown). These results strongly suggested that the rrn gene clusters on each chromosome associate into discrete zones into which large amounts of RNAP are recruited.
Table II. Number of RNAP foci per nucleoid in different growth media.
| Medium | Nucleoids counted | Nucleoids with TFs (%) | TF/nucleoida | Spo0J foci/nucleoid |
|---|---|---|---|---|
| S | 686 | 6.7 | 2.0 | 2.2b |
| CH | 782 | 88.5 | 2.0 | 2.2c |
| 2TY | 560 | 93.0 | 4.2 | 4.1d |
Strain 1048 was grown in three different media and samples were examined during exponential growth. Nucleoids were visualized by staining the DNA with DAPI, and RNAP was visualized directly by GFP fluorescence. The numbers of copies of oriC per nucleoid in equivalent cultures was estimated by measuring Spo0J–GFP foci.
aFor those nucleoids with visible foci.
bTaken from Sharpe and Errington (1998).
cGlaser et al. (1997) reported an average of 4.4 Spo0J foci per cell in CH medium, and for nearly all of the cell cycle such cells contain two nucleoids (Sharpe et al., 1998). Hence, an average of 2.2 foci per nucleoid.
dStrain 1510 (spo0J–gfp) (Glaser et al., 1997) was grown in 2TY at 37°C and processed as detailed previously (Glaser et al., 1997). The number of foci/nucleoid was calculated from 366 nucleoids.
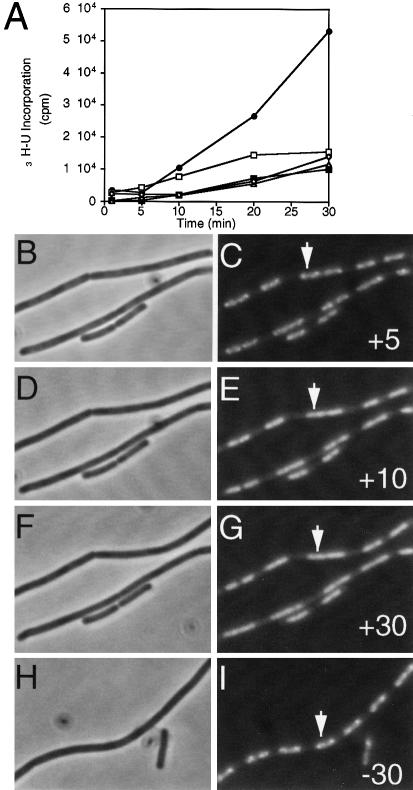
If the RNAP–GFP foci did indeed represent specialized regions of rRNA synthesis, they might be dissipated by induction of the stringent response. This response occurs on amino acid starvation and results in a strong down-regulation of stable RNA (i.e. rRNA and tRNA) synthesis, whereas transcription of many genes encoding biosynthetic enzymes is up-regulated (Cashel et al., 1996; Wendrich and Marahiel, 1997). In CH medium, the minimum concentration of the amino acid analogue arginine hydroxamate (R-HX) needed to induce the stringent response was determined to be 500 μg/ml, and the effect of this concentration of inhibitor became apparent within 10 min (Figure 3A, black squares). Phase contrast and fluorescence images of cells treated with R–HX at this concentration were then acquired over a period of 30 min (Figure 3B–G). After 5 min, TFs were still clearly visible (Figure 3C, arrow). However, after 10 min, they had disappeared and a more homogeneous staining pattern became apparent (Figure 3E), and this was still evident after 30 min (Figure 3G). Control cells not treated with R-HX still contained TFs 30 min after mounting and similar imaging (Figure 3I, arrow). Therefore, the loss of TFs was not due to oxygen starvation or to UV damage while acquiring the images.
Fig. 3. TFs disappear on induction of the stringent response. (A) The rate of incorporation of [3H]uridine into stable RNA in strain 1048 in various concentrations of R-HX: •, none (positive control); □, 250 μg/ml; ▪, 500 μg/ml; ▵ 1000 μg/ml; ⋄, 2000 μg/ml. (B–I) Cells with or without added R-HX. (B–G) A time course of a field of cells acquired between 5 and 30 min after the addition of 500 μg/ml R-HX. (H and I) Images of control cells not treated with R-HX, 30 min after placing on an agarose slide. Times and the presence (+) or absence (–) of R-HX are indicated in the bottom right hand corners of the GFP images. (B), (D), (F) and (H) Phase contrast images. (C), (E), (G) and (I) GFP images. The arrow in (C), (E) and (G) indicates a cell displaying a typical bifocal RNAP distribution prior to induction of the stringent response; in (I), a control cell with a bifocal RNAP distribution is shown.
Ribosomes localize predominantly towards the cell poles
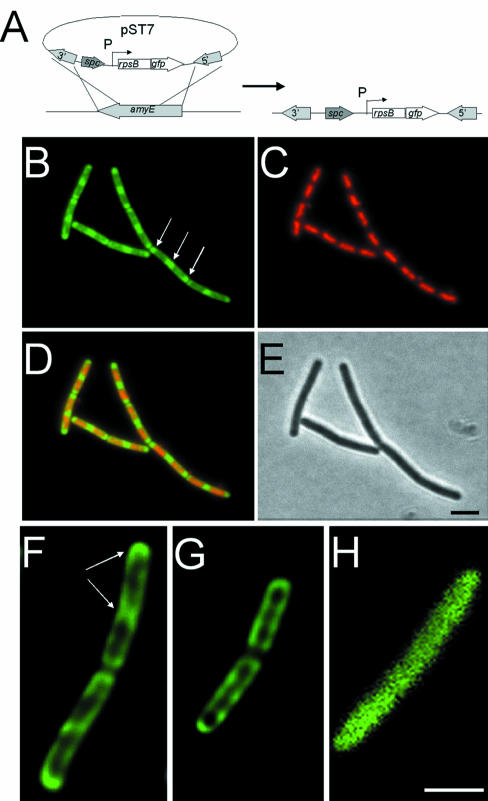
To determine the localization of ribosomes, GFP was fused to the C–terminus of the S2 protein (encoded by rpsB) of the 30S subunit. The fusion was integrated into the chromosome by double crossover at the amyE locus and placed under the control of a xylose-inducible promoter, giving strain 1049 (Figure 4A). Western blots using anti-GFP antibodies showed a band of expected molecular weight for an S2–GFP fusion with no evidence of degradation, indicating that the fusion protein was stable (not shown). Strain 1055 (Table I) in which the rpsB–gfp fusion was integrated into the chromosome by single crossover at rpsB was also constructed. This strain was spo+, and gave localization patterns identical to strain 1049, although it grew more slowly (approximately half the rate) than strain 168 (wild-type) in the absence of xylose (not shown). Therefore, results are presented for strain 1049 that grew with kinetics identical to 168 in CH medium at 37°C.
Fig. 4. Ribosome distribution in exponentially growing and stationary phase cells. (A) Construction of the fusion strain 1049 by insertion of pST7 into the chromosome. Integration of pST7 into the amyE gene allows induction of rps–gfp expression from the xylose-inducible promoter (P). (B–D) Distribution of ribosomes in strain 1049 grown in CH medium. (B) RpsB–GFP image false coloured green. Polar ribosomal concentrations are indicated with arrows. (C) DAPI image false coloured red. (D) Image overlays. (E) Phase contrast image. (F–H) Image slices taken from the middle of exponentially growing (F) and stationary phase (G) strain 1049, and exponentially growing strain 1758 (H). The image in (H) has been magnified 2.5 times as it was obtained without the optovar used to acquire the images in (F) and (G), due to the lower signal intensity in this sample. Scale bar, 2 μm.
Strain 1049 was grown in CH medium at 37°C containing xylose (Figure 4B–E) and, since ∼80% of the ribosomes are present as 70S units (Forchammer and Lindahl, 1971; Bremer and Dennis, 1996; Asai et al., 1999), nearly all of the signal should represent actively translating ribosomes. Figure 4B shows the distribution of the GFP signal in mid-exponential phase cells. Strikingly, fluorescence was localized predominantly towards the cell poles and mid-cell future division sites (Figure 4B, arrows), although a weaker signal was also distributed throughout the whole cell. Counterstaining the DNA with DAPI (Figure 4C) and overlaying the signals (Figure 4D) showed that there was little overlap between the DNA and ribosomal signals. Thus, the bulk of the ribosomal signal appeared to be outside the nucleoid. As the cultures reached stationary phase, the polar localization of ribosomes was less apparent, and the signal was both weaker and more uniformly distributed throughout the cell, perhaps reflecting the reduction in ribosome content and translational activity in these cells (see below).
Deconvolution methods were used to examine the distribution of ribosomes in more detail. Median optical sections from pairs of exponentially growing (Figure 4F) and stationary phase cells (Figure 4G) are shown. These images highlight the concentration of the GFP signal towards the poles of the cell and mid-cell sites in the exponentially-growing cells (Figure 4F, arrows) and show that the ribosomes are also distributed around the inner periphery of the cell (Figure 4F and G). However, the most striking feature of these images was the near absence of signal in the central core of the cells. Thus, the ribosomes are probably distributed primarily in the cytoplasm outside the nucleoid, around the periphery of the cell.
To ensure that this pattern of distribution was specific to ribosomes, strains 1517, 1758 and 1989 (Table I) were grown and image stacks obtained and deconvolved. Strains 1517 and 1758 contain non-functional truncations of spo0J and divIVA fused to gfp (Edwards and Errington, 1997; Glaser et al., 1997). Strain 1989 contains a fully functional gfp–minC fusion that is unable to localize correctly due to the absence of the division site selection protein MinD (Marston and Errington, 1999a). Results with all three strains were indistinguishable. Figure 4H shows an image of a median section of a cell of strain 1758 (DivIVA truncation) grown in CH at 37°C. The signal was distributed throughout the cell, and there was no obvious exclusion of the fusion from the region of cytoplasm occupied by the nucleoid, unlike the GFP fusion to the ribosomal protein.
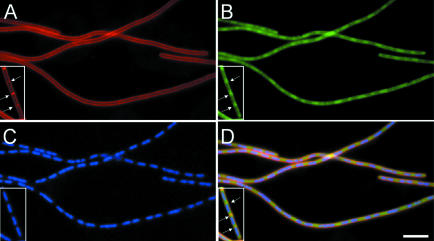
The enrichment of the ribosomal signal at the cell poles and potential division sites during exponential growth suggested that they might be targeted specifically to this portion of the cell. Alternatively, the ribosomal distribution could be influenced somehow by the centrally located nucleoid. To investigate these possibilities, the ribosomal distribution was examined in a strain (1054) in which expression of the essential cell division gene ftsZ (Lutkenhaus and Addinall, 1997) could be repressed. To identify residual division septa, the cells were also stained with the vital membrane stain FM4-64 (Molecular Probes; Lemon and Grossman, 1998; Pogliano et al., 1999). When strain 1054 was grown in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG), the cell size distribution was normal and septa were clearly identifiable by FM4-64 (Figure 5A, inset, arrows). Examination of the distribution of ribosomes in these cells showed that, as expected, they were concentrated adjacent to these septa (Figure 5B and D, inset, arrows).
Fig. 5. Ribosome distribution is due to nucleoid exclusion effects. Images of cells from strain 1054 grown in CH medium in the presence (inserts) or absence of IPTG are shown. (A) FM4-64 membrane stain showing cytoplasmic membranes. (B) RpsB–GFP distribution. (C) DAPI stain showing chromosomes. (D) Overlay of all three images. Cell division sites are marked by arrows in the insert. No septa were visible in the filaments after depletion of the division protein FtsZ. Scale bar, 5 μm.
Three hours after the removal of IPTG, depletion of FtsZ had resulted in nearly all of the cells becoming highly elongated filaments, with few, if any, visible division septa (Figure 5A). The nucleoids were positioned normally in the filaments because chromosome segregation is maintained in the absence of cell division (Figure 5C). Despite the block in division, the distribution of the ribosomal GFP signal was strikingly similar to that of normally dividing cells, with the signal concentrated in the inter-nucleoid spaces along the filament (Figure 5B and D). An additional experiment was also performed in which strain 1049 was grown in the presence of the DNA gyrase inhibitor, nalidixic acid. One hour after the addition of antibiotic, cells were slightly filamentous with respect to a control culture and nucleoids were highly condensed and often bisected by division septa. Examination of the ribosome distribution in such cells revealed that they occupied the whole of the cell except for the small region occupied by the condensed nucleoid (not shown). Thus, the ribosomes are not targeted to cell poles, and instead it seems more likely that they are somehow excluded from the region of the cell occupied by the nucleoid.
Spatial separation of RNA polymerase and ribosomes in dual labelled cells
The striking differences between the RNAP and ribosomal distribution were surprising. Two well known mechanisms should give rise to considerable overlap between these signals. First, the coupling of transcription and translational (Miller et al., 1970), and secondly, the assembly of ribosomes at sites of high level transcription of rrn genes (Miller et al., 1970; French and Miller, 1989). To compare the localization of the transcriptional and translational apparatuses more directly, a strain (1056) was constructed containing fusions (rpoC–gfpuv and rpsB–gfp) that could be visualized separately by using different excitation wavelengths (Lewis and Marston, 1999).
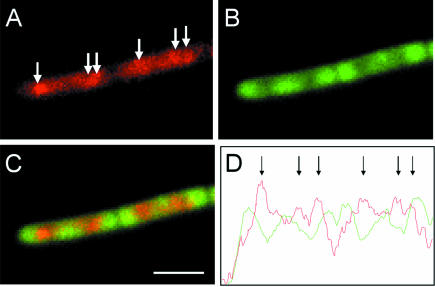
Images of a typical pair of cells of strain 1056 are shown in Figure 6. RpoC–GFPuv (Figure 6A) showed a similar distribution to that of RpoC–GFP in strain 1048 (Figure 1D), indicating that use of the UV spectral variant of GFP had no effect on the localization pattern of the protein. As expected for cells grown in CH medium, TFs were visible above the general polymerase signal (Figure 6A, arrows). The distribution of the RpsB–GFP fusion protein was also unaffected by the presence of the RpoC–GFPuv fusion (Figure 6B), and gave results indistinguishable from those obtained with strain 1049 containing the single fusion (Figure 4C). Image overlays (Figure 6C) showed that there was only slight overlap between the signals representing RNAP and ribosomes. A more quantitative indication of this spatial separation is shown in Figure 6D, in the form of fluorescence intensity plots from the centre line of the cell images. The peaks in fluorescence intensity corresponding to RNAP foci were clearly visible (arrows in Figure 6D) and, apart from the rightmost one, they were interdigitated completely between the peaks in the ribosome signal. Analysis of multiple fields of cells indicated that only 11% of the RpoC–GFP and S2–GFPuv fusion signals were (perfectly or partially) coincident. We conclude that the transcriptional and translational machineries mainly occupy separate subcellular compartments.
Fig. 6. Dual labelling of RNAP and ribosomes indicates that they occupy separate subcellular domains. (A–C) Images of a pair of cells from strain 1056 grown in CH medium. (A) RpoC–GFPuv distribution, false coloured red. The arrows indicate the positions of TFs. (B) RpsB–GFP distribution. (C) Image overlays. (D) Fluorescence intensity profile along a line through the image overlay, with Rpo–GFPuv (red) and RpsB–GFP (green) signals. Peaks in the RpoC–GFPuv signal are indicated with arrows. Scale bar, 2.5 μm.
Discussion
In this study, we have shown that, contrary to expectation, transcription and translation occur predominantly in separate subcellular domains. The finding that most RNAP localizes within the nucleoid was surprising, as it had generally been thought that RNAP would be associated with DNA loops on the outer perimeter of the nucleoid, allowing close transcriptional and translational coupling (Woldringh and Nanninga, 1985; Woldringh et al., 1995). However, our results suggest that most of the RNAP lies within the nucleoid and that the peripheral region is relatively deficient in transcriptional activity. Ribosomes, on the other hand, were found to localize around the periphery of the cell, being concentrated particularly at the cell poles and future division sites at higher growth rates. These results do not preclude the coupling of transcription and translation as this could occur at the interface between the two domains. However, it appears that the level of coupling may be less than was predicted on the basis of electron microscopic studies (Miller et al., 1970), and that most nascent mRNA is produced at sites that are relatively distant from the bulk of the ribosomes.
At higher growth rates, a further level of organization in the RNAP distribution became apparent, with the concentration of most of the fluorescence into subregions we have named transcription foci. Four strands of evidence support the hypothesis that these concentrations of RNAP correspond to sites of intensive transcription of the rRNA (rrn) gene clusters in the oriC region of the chromosome. First, electron microscopic and biochemical data indicate that rrn genes have unusually large amounts of RNAP bound during rapid growth (French and Miller, 1989; Jensen and Pedersen, 1990; Gotta et al., 1991; Bremer and Dennis, 1996). Secondly, the number and intensity of TFs increased in proportion to the growth rate, and, therefore, to the copy number of oriC (Figure 1). Accordingly, the frequency and distribution of TFs were almost identical to those of the chromosome segregation protein, Spo0J, which is known to bind multiple sites around oriC (Lewis and Errington, 1997; Lin and Grossman, 1998; Table II). Thirdly, comparison of the distribution of RNAP with the DNA-binding protein Soj showed that this concentation into foci reflected a specific differential loading of RNAP to a subregion of the chromosome (Figure 2). This is consistent with the distribution of HBsu, the histone-like protein in B.subtilis, which was shown to bind to the nucleoid with no reported heterogeneity or formation of structures similar to the TFs we have observed (Kohler and Marahiel, 1997). Fourthly, on induction of the stringent response, which results in down-regulation of rrn transcription (Cashel et al., 1996; Wendrich and Marahiel, 1997), the TFs disappeared in parallel with the inhibition of stable RNA synthesis (Figure 3).
In addition to the cluster of rrn operons in the oriC region, three further operons are distributed over a region corresponding to almost half a chromosome (Kunst et al., 1997). Currently, we do not know whether these operons are also recruited into TFs observed at higher growth rates. Assembly of such a widely spaced collection of operons into a single region within the cell would suggest that rRNA synthesis is organized into ‘transcription factories’ similar in function to eukaryotic nucleoli, which are discrete sites within the nucleus where RNA pol I transcribes 45S rrn genes and ribosomes assemble (Watson et al., 1987). We are currently investigating the rrn operon composition of the RNAP foci. It is tempting to speculate that concentration of RNAP into such sites could be important in optimizing the efficiency of transcription of these highly active genes. In particular, re-initiation of transcription by a second RNAP molecule immediately after promoter clearance by the preceding molecule would be favoured by having a high local concentration of RNAP. Then, the maximum attainable density of RNAP molecules on each gene could be achieved.
The zonal distribution of ribosomes during exponential growth was also striking, and image processing showed that this pattern was not an artefact caused by the curvature of cells at their poles. Analysis of the distribution of control fusion proteins also showed that they were free to diffuse throughout the cell, unlike that of the ribosomal protein fusion (Figure 4). Although we could not distinguish between ribosomal subunits and actively transcribing ribosomes, the majority of the signal detected was likely to be due to 70S units as these comprise ∼80% of the population at a variety of growth rates (Forchammer and Lindahl, 1971; Bremer and Dennis, 1996; Asai et al., 1999). Experiments in which cell division was inhibited showed that this zonal localization was not dependent on the formation of a division septum. Ribosomes (and presumably polysomes) could localize to the cell periphery simply because they are too large to penetrate the dense, highly folded DNA mass of the nucleoid. However, it is also possible that more active mechanisms control the topology of the ‘translational domain’ in the cell. It will now be interesting to determine whether the peripheral localization of ribosomes is associated with an enrichment of other components of the translational machinery, such as the translation factors and proteins required for precursor synthesis, which might be predicted to be enriched in a specialized subcellular zone involved in translation.
Finally, it is interesting to speculate on the implications of our findings, particularly with RNAP, for the organization of the nucleoid. The clustering of widely spaced rRNA genes (see above) to a single site per chromosome may help to arrange the DNA into a structure that facilitates chromosome segregation, particularly at high growth rates. By analogy with the transcription factory model for eukaryotes (Cook, 1999), bacterial RNAP may be relatively fixed within such ‘transcription domains’ through which loops of actively transcribed DNA are fed. The DNA in the outer regions of the nucleoid could represent either untranscribed genes or correspond to loops of DNA being fed in and out of active RNAP sites in the nucleoid core. If so, the path of gene expression may begin with transcription in the central core of the cell, followed by migration or extension of the mRNA transcript to the ribosomes producing nascent protein in the peripheral part of the cell. (The small amounts of RNAP found towards the periphery of the nucleoid could be involved in coupled transcription and translation.) Such a pathway would be much more similar to the organization of transcription and translation in eukaryotic cells than hitherto has been assumed for prokaryotes. Clearly, we have to reappraise our impression of the bacterial cytoplasm as an undifferentiated medium, and begin investigating the mechanisms that underlie its intricate spatial organization.
Materials and methods
Culture conditions
Table I lists the strains and plasmids used in this study. Strains were grown at 30 or 37°C in S, CH (Sharpe et al., 1998) or 2TY (Sambrook et al., 1989) media as detailed in the text. Where necessary, strains were supplemented with 1% (w/v) xylose or 0.5 mM IPTG.
Construction of GFP fusions
The complete rpoC gene was amplified from B.subtilis 168 chromosomal DNA by PCR (GeneAmp XL long-range PCR, Perkin Elmer) using the following primers: forward, 5′–AGGGAGGTAGGTACCTTGCTAGATGTG–3′; reverse, 5′–GAGTTAAATGAATTCTTCAACCGGGAC–3′. The 3.6 kb product was gel purified, digested with XhoI and EcoRI, the 0.6 kb fragment repurified and inserted into XhoI–EcoRI-cut pSG1164. The resulting plasmid, pST3, was transformed into competent B.subtilis 168 using standard techniques (Anagnastopoulos and Spizizen, 1961; Jenkinson, 1983) to give strain 1048. Since rpoC is situated within a large operon, expression of essential downstream genes could be driven on addition of xylose from the inducible Pxyl promoter on pSG1164. Normal growth required the presence of 1% (w/v) xylose. To construct pST5, the 0.6 kb XhoI–EcoRI fragment of rpoC was purified from pST3 and inserted into XhoI–EcoRI-cut pSG1156 producing an ′rpoC–gfpuv fusion (Table I).
To construct a fusion to the S2 protein of the 30S ribosomal subunit, the entire rpsB gene was amplified by PCR using the primers 5′–TTAGGAGGAGGTACCATGTCAGTCATT–3′ and 5′–CCTTTGAATCTCGAGCGCAGTTGTTGT–3′. After purification, the PCR product was cut with Acc651 and XhoI and inserted into Acc651- and XhoI-cut pSG1154 or pSG1164 to give pST7 (Table I).
Stringent response assays
Stable RNA synthesis was determined following the procedure of A.Levine and S.Autret (personal communication). Strain 1048 was grown at 37°C in CH medium to A6000.3. Aliquots of 2 ml of the culture were then removed, and 10 μCi of [3H]uridine (Amersham) and cold uridine (0.1 mM final concentration) were added to each tube. R-HX was added to final concentrations of 0, 250, 500, 1000 and 2000 μg/ml to respective tubes. Aliquots of 250 μl were removed and added to 1 ml of ice-cold 10% (w/v) trichloroacetic acid (TCA) 1, 5, 10, 20 and 30min after addition of 3H. Samples were left on ice for at least 30 min, prior to filtering onto glass fibre discs, washing with excess ice-cold 10% (w/v) TCA and drying with ethanol. 3H incorporation was determined by scintillation counting.
FtsZ depletion studies
Strain 1054 (Table I) was grown overnight at 30°C in CH supplemented with 1% (w/v) xylose and 0.5 mM IPTG. The culture was diluted back in the same medium to an A600of 0.05 and grown at 37°C to an A600of 0.3. The culture was then pelleted by centrifugation, resuspended, and diluted in CH and divided into two portions. Half of the culture was supplemented with 1% (w/v) xylose and 0.5 mM IPTG, whereas the other half was supplemented only with 1% (w/v) xylose. FM4-64 (Molecular Probes) was added to each culture to a concentration of 10 ng/ml 30 min before sampling.
Microscopy
Images were acquired essentially as detailed previously (Glaser et al., 1997; Lewis and Marston, 1999). For image stack acquisition, the following modifications were made. A 2.40 cm2Gene Frame (Advanced Biotechnologies, Epsom, UK) was stuck onto the slide. The frame was then filled with molten 1.2% (w/v) agarose and a Gene Frame polythene top liner was placed on top until the agarose had solidified. After removal of the liner, the cell suspension was placed on the agarose, and a coverslip was placed on top. Images were acquired using a Zeiss Axiovert 135TV microscope fitted with a 2.5× optovar (for RpsB–GFP images), a Lambda-10/2 filter wheel (Sutter Instruments; Novato, CA) and either a triple bandpass filter (Chroma set 61000v2 with a 365 nm DAPI exciter) or an Endow GFP filter (Chroma set 41018) with a Princeton Instruments MicroMax 1300Y/HS CCD camera. Incremental slices of 0.02 or 0.05 μm were obtained using a PIFOC PI P-721.10 microscope focus drive controlled via MetaMorph v. 3.6 software (Universal Imaging Corporation, West Chester, PA). Image stacks were deconvolved using AutoDeblur v. 5.1 software (AutoQuant Imaging Inc., Watervliet, NY).
Acknowledgments
Acknowledgements
P.J.L. would like to dedicate this paper to Toby Lewis, born November 22, 1999. We also thank M.Buttner, P.Cook, M.El Karoui and S.Autret for their helpful comments and discussions during the preparation of this manuscript. This work was supported by grants from the Biotechnology and Biological Sciences Research Council.
References
- Anagnostopoulos C. and Spizizen, J. (1961) Requirements for transformation in Bacillus subtilis.J. Bacteriol., 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Condon, C., Voulgaris, J., Zaporojets, D., Shen, B., Al-Omar, M., Squires, C. and Squires, C.L. (1999) Construction and initial characterisation of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol., 181, 3803–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B. and Lutkenhaus, J. (1991) FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev., 5, 447–455. [DOI] [PubMed] [Google Scholar]
- Bremer H. and Dennis,P.D. (1996) Modulation of chemical composition and other paramerters of the cell by growth rate. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Megasanik,B., Schaecter,M. and Umbarger.H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edn. American Society for Microbiology Press, Washington, DC, pp. 1553–1569. [Google Scholar]
- Cashel M., Gentry,D.R., Hernandez,V.J. and Vinella,D. (1996) The stringent response. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Megasanik,B., Schaecter,M. and Umbarger.H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edn. American Society for Microbiology Press, Washington, DC, pp. 1458–1496. [Google Scholar]
- Cook P.R. (1999) The organisation of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia, R.H. and Falkow, S. (1996) FACS-optimised mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Edwards D.H. and Errington, J. (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol., 24, 905–915. [DOI] [PubMed] [Google Scholar]
- French S.L. and Miller, O.L.,Jr (1989) Transcription mapping of the Escherichia coli chromosome by electron microscopy. J. Bacteriol., 171, 4207–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer J. and Lindahl, L. (1971) Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J. Mol. Biol., 55, 563–568. [DOI] [PubMed] [Google Scholar]
- Glaser P., Sharpe, M.E., Raether, B., Perego, M., Ohlsen, K. and Errington, J. (1997) Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev., 11, 1160–1168. [DOI] [PubMed] [Google Scholar]
- Gotta S.L., Miller, O.L.,Jr and French, S.L. (1991) rRNA transcription rate in Escherichia coli.J. Bacteriol., 173, 6647–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon, C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Hobot J.A., Villiger, W., Escaig, J., Maeder, M., Ryter, A. and Kellenberger, E. (1985) Shape and fine structure of nucleoids observed on sections of ultrarapidly frozen and cryosubstituted bacteria. J. Bacteriol., 162, 960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C. and Shapiro, L. (1999) Bacterial cell division: a moveable feast. Proc. Natl Acad. Sci. USA, 96, 5891–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L., Hara, H., Zhang, S. and Kaji, A. (1996) Ribosome recycling by ribosome recycling factor (RRF): an important but overlooked step of protein synthesis. Adv. Biophys., 32, 121–201. [DOI] [PubMed] [Google Scholar]
- Jenkinson H. (1983) Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis.J. Gen. Microbiol., 129, 1945–1958. [DOI] [PubMed] [Google Scholar]
- Jensen K.F. and Pedersen, S. (1990) Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol. Rev., 54, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov, M.Y., Buckingham, R.H. and Ehrenberg, M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kohler P. and Marahiel, M.A. (1997) Association of the histone-like protein Hbsu with the nucleoid of Bacillus subtilis.J. Bacteriol., 179, 2060–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., et al. (1997)The complete genome sequence of the Gram positive Bacillus subtilis bacterium. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- Lemon K.P. and Grossman, A.D. (1998) Localisation of bacterial DNA polymerase: evidence for a factory model of replication. Science, 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Errington, J. (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localisation with the Spo0J partitioning protein. Mol. Microbiol., 25, 945–954. [DOI] [PubMed] [Google Scholar]
- Lewis P.J. and Marston, A.L. (1999) GFP vectors for controlled expression and dual labeling of protein fusions in Bacillus subtilis.Gene, 227, 101–109. [DOI] [PubMed] [Google Scholar]
- Lin D.C.H. and Grossman, A.D. (1998) Identification and characterisation of a bacterial chromosome partitioning site. Cell, 92, 675–685. [DOI] [PubMed] [Google Scholar]
- Lin D.C.H., Levin, P.A. and Grossman, A.D. (1997) Bipolar localisation of a chromosome partition protein in Bacillus subtilis.Proc. Natl Acad. Sci. USA, 94, 4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R. and Chamberlin,M.J. (1976) RNA Polymerase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Losick R. and Shapiro, L. (1999) Changing views on the nature of the bacterial cell: from biochemistry to cytology. J Bacteriol., 181, 4143–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. and Addinall, S.G. (1997) Bacterial cell division and the Z ring. Annu. Rev. Biochem., 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Errington, J. (1999a) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localisation and activation of MinC. Mol. Microbiol., 33, 84–96. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Errington, J. (1999b) Dynamic movement of the ParA-like Soj protein of B.subtilis and its dual role in nucleoid organisation and developmental regulation. Mol. Cell, 4, 673–682. [DOI] [PubMed] [Google Scholar]
- Miller O.L. Jr, Hamkalo, B.A. and Thomas, C.A.,Jr (1970) Visualisation of bacterial genes in action. Science, 169, 392–395. [DOI] [PubMed] [Google Scholar]
- Pogliano J., Osborne, N., Sharp, M.D., Abanes-De Mello, A., Perez, A., Sun, Y.-L. and Pogliano, K. (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol., 31, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C. and Kellenberger, E. (1994) The bacterial nucleoid revisited. Microbiol. Rev., 58, 211–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter A. and Chang, A. (1975) Localisation of transcribing genes in the bacterial cell by means of high resolution autoradiography. J. Mol. Biol., 98, 797–810. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sharpe M.E. and Errington, J. (1998) A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for co-ordination of chromosome segregation and cell division. Mol. Microbiol., 28, 981–990. [DOI] [PubMed] [Google Scholar]
- Sharpe M.E., Hauser,P.M., Sharpe,R.G. and Errington,J. (1998) Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J. Bacteriol., 180, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.D., Hopkins,N.H., Roberts,J.W., Steitz,J.A. and Weiner,A.M. (1987) Molecular Biology of the Gene. 4th edn. Benjamin/Cummings Publishing Co. Inc, Menlo Park, CA. [Google Scholar]
- Wendrich T.M. and Marahiel, M.A. (1997) Cloning and characterization of a relA/spoT homologue from Bacillus subtilis.Mol. Microbiol., 26, 65–79. [DOI] [PubMed] [Google Scholar]
- Woldringh C.L. and Nanninga,N. (1985) Structure of nucleoid and cytoplasm in the intact cell. In Nanninga,N. (ed.), Molecular Cytology of E.coli. Academic Press, London, UK, pp. 161–197. [Google Scholar]
- Woldringh C.L., Jensen, P.R. and Westerhoff, H.V. (1995) Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces?FEMS Lett., 131, 235–242. [DOI] [PubMed] [Google Scholar]