Abstract
Nisin A is the most widely characterized lantibiotic investigated to date. It represents one of the many antimicrobial peptides which have been the focus of much interest as potential therapeutic agents. This has resulted in the search for novel lantibiotics and more commonly, the engineering of novel variants from existing peptides with a view to increasing their activity, stability and solubility.
The aim of this study was to compare the activities of nisin A and novel bioengineered hinge derivatives, nisin S, nisin T and nisin V. The microtitre alamar blue assay (MABA) was employed to identify the enhanced activity of these novel variants against M. tuberculosis (H37Ra), M. kansasii (CIT11/06), M. avium subsp. hominissuis (CIT05/03) and M. avium subsp. paratuberculosis (MAP) (ATCC 19698). All variants displayed greater anti-mycobacterial activity than nisin A. Nisin S was the most potent variant against M. tuberculosis, M. kansasii and M. avium subsp. hominissuis, retarding growth by a maximum of 29% when compared with nisin A. Sub-species variations of inhibition were also observed with nisin S reducing growth of Mycobacterium avium subsp. hominissuis by 28% and Mycobacterium avium subsp. paratuberculosis by 19% and nisin T contrastingly reducing growth of MAP by 27% and MAC by 16%.
Nisin S, nisin T and nisin V are potent novel anti-mycobacterial compounds, which have the capacity to be further modified, potentially generating compounds with additional beneficial characteristics. This is the first report to demonstrate an enhancement of efficacy by any bioengineered bacteriocin against mycobacteria.
Key words: mycobacteria, nisin variants, alamar blue, peptide engineering, lantibiotic, bacteriocin
Introduction
Mycobacterium tuberculosis, the etiological agent of tuberculosis (TB), is responsible for approximately 9.27 million incident cases of TB annually, resulting in 2–3 million deaths.1 The recent emergence of drug-resistant mycobacteria, particularly XDR-TB, has prompted the World Health Organisation (WHO) to set clear objectives to control this threat.1 The rising incidence of human and animal infection by non tuberculosis mycobacteria (NTM) has also become a serious public-health concern.2 NTM can cause a broad spectrum of diseases including pulmonary infections resembling tuberculosis3 and extra pulmonary infections affecting lymph nodes, skin and soft tissue.4 Among NTM, Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of Johne's disease in ruminants, has been the focus of much attention in recent years, partly as a consequence of its association with Crohn disease in humans.5–7
Lantibiotics have been suggested as a possible alternative to antibiotics for many drug-resistant infections.8–11 This is due to their multiple mechanisms of action, broad-spectrum activity against a wide variety of Gram-positive targets, gene-encoded nature (making them excellent templates for bioengineering) and their ability to be delivered to the site of infection.12–14 However, although lantibiotics have been extensively researched, little work has been done to investigate their application to treat mycobacterial diseases.15
Nisin A is the prototype lantibiotic and has safely been incorporated into a wide range of commercial products since its acceptance by the Food and Drug Administration (FDA) as a food additive in 1988.16 As with all lantibiotics, synthesis of the mature nisin peptide involves extensive post-translational modification resulting in the formation of the unusual amino acids lanthionine and β-methyllanthionine, as well as dehydrated amino acids.8 Nisin is produced by Lactococcus lactis which is generally regarded as safe (GRAS) for food applications. Nisin is generally used as a food preservative, although it has recently been applied therapeutically in the form of an anti-mastitis product called ‘Wipe-out’.8
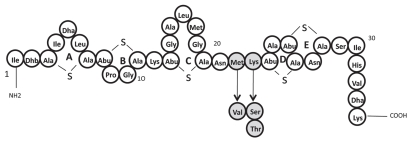
The bio-engineering of lantibiotics has been particularly useful with respect to elucidating the importance of specific residues and domains within the peptides. In particular, the N-terminal region responsible for binding lipid II in the cell wall of target cells was elucidated, as was the C-terminal region which inserts into the cell membrane to cause pore formation. Most note-worthy is the work focusing on the central hinge region which allows the aforementioned domains to move relative to one another.17–19 In contrast however, the generation of lantibiotics with enhanced features has been infrequently reported. Some successes have occurred, such as the generation of nisin variants with enhanced solubility at neutral pH20 or increased antimicrobial activity against non-pathogenic strains.21 Interestingly it has been established that modification of the hinge region in nisin Z (a natural variant of nisin A which differs by only one amino acid) led to the identification of variants (N20K and M21K) with increased efficacy against Gram-negative species, i.e., Shigella, Pseudomonas and Salmonella.18 Perhaps more significantly, variants with enhanced activity against specific Gram-positive pathogens, such as S. aureus, S. agalactiae and L. monocytogenes, have also been generated.19 The enhanced activity of two peptides, i.e., nisinK22T and M21V (hereafter nisin T and V respectively), (Fig. 1) has been confirmed against non mycobacterial targets, in studies with purified peptides.19,22 However, nisinK22S (hereafter nisin S) has not been evaluated in purified form to date. Here we carry out an investigation to determine if any of these three nisin variants display enhanced antimicrobial activity against four representative species of pathogenic mycobacteria.
Figure 1.
Structure of nisin variants Nisin A, Nisin V, Nisin S and Nisin T. Dark circles indicate amino acid differences between Nisin A and its derivatives.
Results
Initial agar-based comparison of the anti-mycobacteria activity of nisin and nisin variants.
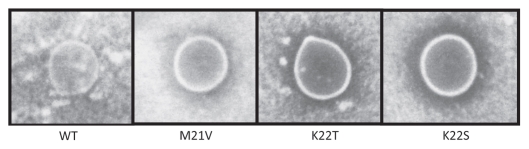
Nisin and nisin variant-producing lactococci were employed for initial agar-based bioactivity studies to assess the anti-mycobacteria activity of the peptides that they produce. Mycobacterium smegmatis was chosen as a target strain due to its fast growing properties. This study indicated that nisin S, T and V all possess greater anti-mycobacteria activity (as evident from zones of activity). These zones were 2.2 mm (nisin V), 2.7 mm (nisin T) and 4 mm (nisin S) compared to their nisin A counterpart (the producer of which failed to generate a zone of clearing) (Fig. 2). To more accurately assess the activity of nisin S, T and V these peptides were purified and equimolar (sub-lethal) concentrations were used to determine their activity against Mycobacterium tuberculosis H37Ra, Mycobacterium kansasii, Mycobacterium avium subsp. hominissuis and Mycobacterium avium subsp. paratuberculosis, using the MABA.
Figure 2.
Agar diffusion assay of Nisin A and its derivatives Nisin V, Nisin T and Nisin S, overlaid with M. smegmatis.
Efficacy of nisin S against four pathogenic mycobacteria.
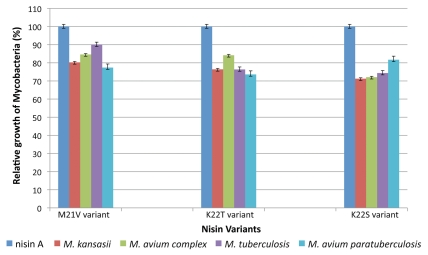
The variant nisin S exhibited the greatest activity of the nisin peptides tested against Mycobacterium tuberculosis H37Ra, Mycobacterium kansasii and Mycobacterium avium subsp. hominissuis (Fig. 3). This bioengineered variant more effectively inhibited all four targets than wild-type nisin A. This impact was most apparent for M. kansasii as relative growth of the target was reduced by 29% compared to that which occurred in the presence of nisin A (Fig. 3). The sensitivity of MAC and M. tuberculosis was comparable as respective 28% and 26% reductions in growth, relative to that brought about by nisin A, were apparent (Fig. 3). MAP was the least susceptible in that there was only a 19% decrease in growth. Thus, the K22S change within nisin S increased the efficacy of nisin A against mycobacteria by an average of 26%.
Figure 3.
Activity of nisin V (M21V), nisin T (K22T), nisin S (K22S) and wild-type nisin A against a cohort of pathogenic mycobacteria. Efficacy was evaluated using alamar blue colorimetric indicator dye. The dye reduction value of nisin A was taken as 100% relative growth, facilitating direct comparison of nisin A to the activity of nisin V, nisin T and nisin S.
Efficacy of nisin T against four pathogenic mycobacteria.
Nisin T also demonstrated enhanced activity, relative to wild-type nisin A, against the four target strains. Mycobacterium avium subsp. paratuberculosis was the most susceptible species in that 27% decrease in growth relative to nisin A was observed, thereby establishing it as the most potent anti-MAP variant (Fig. 3). K22T increased the extent to which M. kansasii and M. tuberculosis were inhibited by 24% (compared to nisin A) while MAC was least susceptible, corresponding to a 16% increase in inhibition (Fig. 3). It is noteworthy that while the activities of nisin T and nisin S against M. tuberculosis and M. kansasii were very similar, their relative activities against Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. hominissuis, both subspecies of M. avium, varied. Overall, relative mycobacterial growth was decreased by 23% when nisin T rather than nisin A was employed.
Efficacy of nisin V against four pathogenic mycobacteria.
The nisin V variant also displayed potent activity against each species of mycobacteria. This was enhanced relative to nisin A but was reduced relative to that of nisin S and T. MAP was the most susceptible species with a 23% decrease in relative growth while efficacy was least enhanced against M. tuberculosis (10% decrease in relative growth). The growth of M. kansasii and MAC was reduced by 20 and 16%, respectively, relative to that which occurred in the presence of nisin A (Fig. 3). Collectively, nisin V brought about an average 17% increase inhibition relative to nisin A.
Discussion
Lantibiotics are well established anti-microbial agents with many attractive attributes. They are gene-encoded, active at low concentrations,15 and do not require access into the mycobacterial cell to exert their effect, unlike conventional anti-mycobacterial drugs like rifampicin.25 While the identification of nisin derivatives with enhanced activity is itself a rare event, this is the first report to demonstrate the increased efficacy of any bioengineered bacteriocins against mycobacteria. Moreover, the further demonstration of the usefulness of MABA indicates that it could be employed on a larger scale to screen banks of bioengineered lantibiotics, and indeed other antimicrobials, to identify anti-mycobacteria compounds with enhanced activity, stability, solubility and/or pH range.
Considering the intentional mutagenesis of lantibiotics was initiated in the early 1990s,26,27 it is somewhat surprising that so few enhanced lantibiotic variants have been identified. Recent work19 employed saturation and site directed approaches in which enhanced variants were identified, producing the first such peptides to display enhanced activity against Gram-positive pathogens.
This study demonstrated the enhanced activity of three purified variants, nisin S, nisin T and nisin V, which retarded relative growth by as much as 29% (Fig. 3). The increased efficacy of nisin S and nisin T is of particular interest in that both variants contain a new incorporated hydroxy-amino acid at the same position but each have differing activities. Interestingly each newly introduced residue, i.e., valine, serine and threonine (in nisin V, S and T, respectively), were much smaller than the residues initially present i.e., methionine (M) and lysine (K). The corollary also seems to be true as the introduction of larger residues such as arginine have previously been found to have a negative impact against other targets.19
Previous observations suggest that the enhanced activities of the nisin variants are strain and species specific.19 Sub-species fluctuations are also apparent in this study i.e., variant nisin S reduced the growth of Mycobacterium avium subsp. hominissuis by 28% and Mycobacterium avium subsp. paratuberculosis by 19% whereas nisin T contrastingly reduced MAP growth by 27% and MAC growth by 16% (Fig. 3). It may be that a detailed understanding of the basis for the enhanced activity of these peptides coupled with the creation of new generations of bioengineered lantibiotics could yield new lantibiotic variants with particular potency against species and sub-species of pathogenic mycobacteria
The generation of species specific lantibiotics with dual modes of action would be an exciting step forward in the development of novel anti-mycobacterial drugs. Moreover, as these variants are the result of just a single nucleotide mutation on the nisin structural gene, it could mean their acceptance may be more readily and rapidly facilitated by food and clinical regulators.19 In conclusion, the creation, identification and evaluation of enhanced variants nisin T, nisin S and nisin V may represent the first step in producing species or even sub-species specific bio-engineered anti-mycobacterial peptides.
Materials and Methods
Bacterial strains and culturing conditions.
Four common infectious species of mycobacteria were chosen as targets for antimicrobial activity studies. These were (period of growth in brackets) Mycobacterium tuberculosis H37Ra (11 days), Mycobacterium kansasii CIT11/06 (10 days), Mycobacterium avium subsp. hominissuis (CIT05/03) (10 days) and Mycobacterium avium paratuberculosis (ATCC 19698) (6–8 weeks). Each was routinely grown in Middlebrook 7H9 broth (MB broth) (Sigma Aldrich), supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Unitech), at 37°C. Culturing of Mycobacterium avium subsp. paratuberculosis also required the addition of 0.2% Mycobactin J (Synbiotics). The rapid growing Mycobacterium smegmatis MC2155, also grown in supplemented MB broth, was used as a mycobacterial indicator strain for initial agar based assays. L. lactis NZ9800 harboring plasmid-associated nisin A and nisin variant genes19 were grown in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) and chloramphenicol (10 µg/ml).
Agar diffusion detection of antimicrobial activity.
To initially determine if the peptide variants showed increased activity compared with wild-type nisin, deferred antagonism assays were performed by spotting 10 µl of nisin A producing strain L. lactis NZ9800 pCI372-nisA and the relevant variant producers L. lactis NZ9800 pCI372nisA-K22S, L. lactis NZ9800 pCI372nisA-K22T and L. lactis NZ9800 pCI372nisA-M21V on GM17 agar plates and allowing them to grow overnight. Each spot was then irradiated under ultra violet light for 10 minutes to kill the nisin producing L. lactis strains and subsequently over-laid with Middle-brook (MB) agar (0.75% W/V agar) seeded with the fast growing indicator strain M. smegmatis MC2155 (10%).
Purification of wild-type nisin A and variants.
To determine if an enhancement of activity was a result of increased production or increased potency by each variant, evaluation of equimolar purified peptides was performed. All nisin peptides were purified using the protocol described previously.19 Briefly, overnight cultures of Lactococcus lactis NZ9800 producing either nisin A or each of the nisin variants, were inoculated into 1 L modified TY broth (1% inoculum) and incubated overnight at 30°C. Cells were harvested by centrifugation at 7,000 rpm for 15 minutes. The cell pellet was re-suspended in 300 ml of 70% propan-2-ol and 0.1% trifluoroacetic acid (TFA) pH 2.0. After stirring at room temperature for 4 hours, the cell debris was removed by centrifugation (7,000 rpm for 15 minutes) and the bacteriocin-containing supernatant reduced to approximately 60 ml by rotary evaporation. The resultant preparation was adjusted to pH4 before applying to a 10 g (60 ml volume) Varian C18 Bond Elut Column (Varian, Harbor City, CA) pre-equilibrated with methanol and water. The column was subsequently washed with 100 ml of 20% ethanol and the active bacteriocin was eluted with 100 ml 70% IPA, 0.1% TFA. Aliquots (15 ml) were concentrated to 2 ml through the removal of propan-2-ol by rotary evaporation. Aliquots (1.5 ml) were applied to a Phenomenex (Phenomenex, Cheshire UK) Jupiter Proteo RP-HPLC column (250 × 10 mm, 90 Å, 4 µm) previously equilibrated with 25% propan-2-ol, 0.1% TFA. The column was subsequently developed in a gradient of 30% propan-2-ol containing 0.1% TFA to 60% propan-2-ol containing 0.1% TFA from 10 to 45 min at a flow rate of 1.2 ml/min.
Analysis of the relative activities of nisin and nisin variants against mycobacteria.
A previously optimized microtitre alamar blue assay (MABA)23 was employed to detect viability of target mycobacterial cells in the presence of each nisin variant. AlamarBlue is an indicator dye which quantitatively measures the proliferation of cells including mycobacteria. It consists of an oxidation-reduction (REDOX) indicator that yields a colorimetric change and a fluorescent signal in a response to metabolic activity.
The mycobacterial strains were grown to late log phase and a 10 ml culture of each target strain was centrifuged at 15,000 RPM for 20 minutes using a bench top centrifuge (Model CR 4–12 Jouan Inc., UK). The supernatant was removed and the pellet was washed in fresh Middlebrook 7H9GC broth. The pellet was then re-suspended in 10 ml of fresh supplemented MB broth (containing 0.2% mycobactin J in the case of MAP). To minimize bacterial clumping, cell suspensions were initially vortexed for 2 minutes and subsequently mixed by pipetting each suspension up and down for 2 minutes. This ensured mycobacterial clumping did not adversely interfere with the assay. Cultures were subsequently adjusted to match McFarland standard no. 1 (3 × 108 CFU/ml), prior to a subsequent 1:20 dilution of the culture in MB broth.
The MABA was performed in triplicate in a sterile 96 well microtitre plate. All outer perimeter wells were filled with sterile water to prevent evaporation of test wells. Positive controls containing no nisin and negative controls containing no mycobacteria were included for each test strain. Sub-lethal concentrations of wild-type nisin A or a variant peptide were added to each well (containing 100 µl of the target strain) at a final concentration of 10 µg/ml (20 µg/ml for MAP). Following 24 hours incubation, 20 µl of alamar blue (AbD Serotech) (10% final volume of the well) was added to each well. Subsequent readings were taken at 570 and 600 nm over a 6 day period. Plates were covered and re-sealed with parafilm and incubated at 37°C following each reading. The time taken for a visual color change in all wells varied from four days (MAC) to thirteen days (MAP), with M. kansasii and M. tuberculosis requiring four and six days incubation respectively. The activity of each nisin variant, relative to the wild-type nisin A, was initially observed visually. Once the positive control produced a strong color change indicating cell viability, quantitative measurements of all wells for each target strain (in the presence of wild-type nisin A and variants) were taken and calculated using the appropriate formula.24 Background values from the negative controls were subtracted from each test well.
In the case of each individual species of mycobacteria, the wild-type nisin dye reduction value was taken as 100% and the relative anti-mycobacterial activity of each nisin variant was directly compared to that. This was carried out separately for each individual test species due to their differing growth rates. The formula used for calculating % reduction of alamar blue dye was as follows;24
Percent reduction of alamarBlue
Where:
O1 = molar extinction coefficient (E) of oxidized alamarBlue (Blue) at 570 nm
O2 = E of oxidized alamarBlue at 600 nm
R1 = E of reduced alamarBlue (Red) at 570 nm
R2 = E of reduced alamarBlue at 600 nm
A1 = absorbance of test wells at 570 nm
A2 = absorbance of test wells at 600 nm
N1 = absorbance of negative control well (media plus alamarBlue but no cells) at 570 nm
N2 = absorbance of negative control well (media plus alamarBlue but no cells) at 600 nm.
Acknowledgements
We would like to thank Cork University Hospital for providing the M. kansasii and MAC strains, Dr. Joe Keane, St. James' Hospital, Dublin for providing Mycobacterium tuberculosis H37Ra and Pierre Douarre for providing the MAP strain.
Abbreviations
- MABA
microtitre alamar blue assay
- MAP
M. avium subsp. paratuberculosis
- MB
middlebrook
- GRAS
generally regarded as safe
- TFA
trifluoroacetic acid
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/13642
Financial Support
This research was funded by a Strand I research grant from the Technological Strand Research Program awarded through the Institute of Technology Ireland initiative. J.O.M., R.P.R., C.H. and P.D.C. are recipients of a Department of Agriculture grant (MAPSAFE).
References
- 1.WHO, author. Global tuberculosis control: epidemiology, strategy, financing: WHO report. 2009. [Google Scholar]
- 2.Wang HX, Yue J, Han M, Yang JH, Gao RL, Jing LJ, et al. Nontuberculous mycobacteria: susceptibility pattern and prevalence rate in Shanghai from 2005 to 2008. Chin Med J (Engl) 2010;2:184–187. [PubMed] [Google Scholar]
- 3.Falkinham JO., 3rd Nontuberculous mycobacteria in the environment. Clin Chest Med. 2002;3:529–551. doi: 10.1016/s0272-5231(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 4.Martin-Casabona N, Bahrmand AR, Bennedsen J, Thomsen VO, Curcio M, Fauville-Dufaux M, et al. Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int J Tuberc Lung Dis. 2004;10:1186–1193. [PubMed] [Google Scholar]
- 5.Hermon-Taylor J, Bull TJ, Sheridan JM, Cheng J, Stellakis ML, Sumar N. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can J Gastroenterol. 2000;6:521–539. doi: 10.1155/2000/798305. [DOI] [PubMed] [Google Scholar]
- 6.Scanu AM, Bull TJ, Cannas S, Sanderson JD, Sechi LA, Dettori G, et al. Mycobacterium avium subspecies paratuberculosis infection in cases of irritable bowel syndrome and comparison with Crohn's disease and Johne's disease: common neural and immune pathogenicities. J Clin Microbiol. 2007;12:3883–3890. doi: 10.1128/JCM.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nacy C, Buckley M. Mycobacterium avium paratuberculosis. Infrequent human pathogen or public health theat. American Academy of Microbiology. 2008 [PubMed] [Google Scholar]
- 8.Cotter PD, Hill C, Ross RP. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci. 2005;1:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 9.Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases and antimicrobial peptides. J Appl Microbiol. 2008;1:1–13. doi: 10.1111/j.1365-2672.2007.03498.x. [DOI] [PubMed] [Google Scholar]
- 10.Russell JB, Mantovani HC. The bacteriocins of ruminal bacteria and their potential as an alternative to antibiotics. J Mol Microb Biotech. 2002;4:347–355. [PubMed] [Google Scholar]
- 11.Gillor O, Nigro LM, Riley MA. Genetically engineered bacteriocins and their potential as the next generation of antimicrobials. Curr Pharm Design. 2005;8:1067–1075. doi: 10.2174/1381612053381666. [DOI] [PubMed] [Google Scholar]
- 12.Sosunov V, Mischenko V, Eruslanov B, Svetoch E, Shakina Y, Stern N, et al. Antimycobacterial activity of bacteriocins and their complexes with liposomes. J Antimicrob Chemoth. 2007;5:919–925. doi: 10.1093/jac/dkm053. [DOI] [PubMed] [Google Scholar]
- 13.Benech RO, Kheadr EE, Laridi R, Lacroix C, Fliss I. Inhibition of Listeria innocua in cheddar cheese by addition of nisin Z in liposomes or by in situ production in mixed culture. Appl Environ Microb. 2002;8:3683–3690. doi: 10.1128/AEM.68.8.3683-3690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malheiros PD, Daroit DJ, da Silveira NP, Brandelli A. Effect of nanovesicle-encapsulated nisin on growth of Listeria monocytogenes in milk. Food Microbiol. 2010;1:175–178. doi: 10.1016/j.fm.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Carroll J, Field D, O'Connor PM, Cotter PD, Coffey A, Hill C, et al. A comparison of the activities of lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int J Antimicrob Agents. 2010;36:132–136. doi: 10.1016/j.ijantimicag.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 16.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek. 1996;2:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 17.Hsu S, Breukink E, Tischenko S, Lutters M, De Kruijff B, Kaptein A, et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol. 2004;11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, Zhang ZZ, Chen XZ, Yang W, Huan LD. Site-directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl Microbiol Biotechnol. 2004;6:806–815. doi: 10.1007/s00253-004-1599-1. [DOI] [PubMed] [Google Scholar]
- 19.Field D, Connor PM, Cotter PD, Hill C, Ross RP. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol. 2008;1:218–230. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- 20.Rollema HS, Kuipers OP, Both P, de Vos WM, Siezen RJ. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol. 1995;8:2873–2878. doi: 10.1128/aem.61.8.2873-2878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJ, Kuipers OP, et al. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl Environ Microbiol. 2007;18:5809–5816. doi: 10.1128/AEM.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field D, Quigley L, O'Connor PM. Studies with bioengineered nisin peptides highlight the broad-spectrum potency of Nisin V. Microb Biotechnol. 2010;3:473–486. doi: 10.1111/j.1751-7915.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll J, Douarre P, Coffey A, Buckley J, Cashman B, O'Farrell K, et al. Optimization of a rapid viability assay for Mycobacterium avium subsp. paratuberculosis by using alamarBlue. Appl Environ Microb. 2009;24:7870–7872. doi: 10.1128/AEM.01203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.alarmBlueTechnical Datasheet. 2008. pp. 1–11. www.biokom.com.pl/files/alamarblue.pdf.
- 25.Hartmann GR, Heinrich P, Kollenda MC, Skrobranek B, Tropschug M, Weiss W. Molecular mechanism of action of the antibiotic rifampicin. Angew Chem Int Edit. 1985;12:1009–1014. [Google Scholar]
- 26.Kuipers OP, Rollema HS, Yap WMGJ, Boot HJ, Siezen RJ, Devos WM. Engineering Dehydrated Amino-Acid-Residues in the Antimicrobial Peptide Nisin. J Biol Chem. 1992;34:24340–24346. [PubMed] [Google Scholar]
- 27.Liu W, Hansen JN. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem. 1992;35:25078–25085. [PubMed] [Google Scholar]