Abstract
Electron transfer is central to a wide range of essential metabolic pathways, from photosynthesis to fermentation. The evolutionary diversity and conservation of proteins that transfer electrons makes these pathways a valuable platform for engineered metabolic circuits in synthetic biology. Rational engineering of electron transfer pathways containing hydrogenases has the potential to lead to industrial scale production of hydrogen as an alternative source of clean fuel and experimental assays for understanding the complex interactions of multiple electron transfer proteins in vivo. We designed and implemented a synthetic hydrogen metabolism circuit in Escherichia coli that creates an electron transfer pathway both orthogonal to and integrated within existing metabolism. The design of such modular electron transfer circuits allows for facile characterization of in vivo system parameters with applications toward further engineering for alternative energy production.
Key words: synthetic biology, electron transfer, ferredoxin, hydrogen, modularity, iron-sulfur cluster
Synthetic biology uses concepts and language from computer science and electrical engineering to enable control of cellular behavior in a predictable, useful manner.1 Standardized, modular biological components have been isolated and recombined in cellular “chassis,” typically E. coli, creating novel circuits that often have functions reminiscent of electrical engineering. In particular, biological circuits have been built to behave like toggle switches,2 oscillators,3 memory loops4 and logic gates.5 This type of engineering has potentially powerful applications in basic science,6 biotechnology,7 medicine8 and alternative energy.9
Aside from their technological applications, synthetic biological circuits can test basic science hypotheses by mimicking natural genetic networks in a simplified context.10 These pathways are often intended to function orthogonally to existing cellular behaviors, creating a framework for studying in vivo behaviors with the isolated simplicity of in vitro studies.11 Much of the work in synthetic biology has focused on understanding and recombining cellular information processing in this way, integrating environmental signals into a novel, coordinated biological behavior. Here, the indirect analogy to computational information processing serves as a simplifying and abstracting framework for understanding and engineering complex systems. Recently, synthetic biology and biological engineering have begun to address increasingly complex circuits composed of numerous biological parts from many organisms12 as well as the electronic properties of cells and membrane ion channels.13 However, such efforts remain focused primarily on the control of genetic transcription and translation rather than on the unique electronic abilities of living cells.
The engineering of cellular components that manage and transfer electrical charge can lead to novel methods of integrating electronic devices and biological systems,14 as well as the design of complex redox pathways within cells for the production of reduced molecules such as fuels. Physical interfaces between electronic and biological devices can be used as biosensors, combining the strengths of biological environmental sensing with the speed and existing infrastructure of computational processing. Such sensors can be used to measure environmental pollution or as medical devices for monitoring and maintaining health. Connections between electronic and biological systems can also be used as sources of energy, as in microbial fuel cells, while engineering of electron-transfer redox pathways within cells is crucial for rerouting reducing power from natural energy metabolism into useful molecules, including hydrogen. As components of synthetic biological devices, biological electron transfer proteins not only more closely resemble the electronic components in the analogy to electrical engineering but provide a common currency for biological devices integrated into metabolism and producing valuable outputs.
Electron transfer pathways in biology are crucial for the reduction of inorganic chemicals into biologically functional molecules and the metabolic breakdown of organic compounds. Electrons readily tunnel quantum-mechanically between two protein-bound iron-sulfur clusters only when these clusters are in close contact, with an optimal tunneling distance of 14 Å.15 Because they are bound to proteins rather than freely diffusing small molecules, electrons held by reduced ironsulfur clusters are unique metabolites with fascinating opportunities for engineering. Many natural electron transfer proteins have already been studied in detail and described in terms of engineering principles15 and have been recombined both in vitro16,17 and in vivo,18 making them ideal parts for synthetic biology.
Electrons typically tunnel from lower to higher potential iron-sulfur clusters in what can loosely be thought of as an amorphous electronic circuit. While the detailed interaction between iron-sulfur proteins is often poorly characterized, as a common currency of biological electron metabolism, iron-sulfur proteins represent uniquely useful parts for the design of diverse synthetic biological systems. The photosystems of plants and the catabolic enzymes of oxidative respiration or fermentation introduce high-energy electrons into cellular metabolism, and can therefore be thought of as “batteries.” These low potential electrons are then passed to “wires,” proteins or cofactors that can transfer electrons to other oxidoreductase enzymes that then generate a measurable output, such as reduction of protons, sulfur, nitrogen, oxygen and carbon or the hydroxylation of steroids.19
As oxidoreductase enzymes are reversible and these circuits are constructed from cellular components free to move throughout the cell, the electrical potential of the protein bound iron-sulfur cluster and the nature of the protein-protein interactions will determine the direction and the rate of the electron “current” traveling through the pathway. The transcriptional control of genes, surface co-evolution of interacting partners and scaffolding of certain crucial pathways has led to precise control over natural, evolved electron transfer circuits, and these become valuable in synthetic biology as methods of circuit insulation in the recombinant design of novel biological circuits.
Many E. coli redox enzymes are electronically coupled to the chemical “wire” cofactor NADH, with a reducing potential of −320 mV. Numerous enzymes from plants and anaerobic bacteria, however, are partnered with the protein-based electron carrier ferredoxin, which can have a wide range of reducing potentials, typically close to that of the H2/H+ pair (−420 mV). As a class of proteins with different regulatory and structural properties that bind iron-sulfur clusters of variable potential, ferredoxins represent a tremendous toolbox for synthetic biology applications. Furthermore, while ferredoxin activity has been well characterized in many photosynthetic pathways, the function of ferredoxin in E. coli,20 though essential,21 remains poorly understood. Here, synthetic biology, through engineering and functional testing of ferredoxin-based electron transfer pathways and insulation strategies, may be useful in elucidating the natural biology of bacterial iron-sulfur cluster protein systems in a simplified in vivo system.
Ferredoxins are small (∼11 kD) soluble proteins that coordinate at least one iron sulfur cluster through a conserved cysteine motif. They have been proposed to be among one of the earliest proteins in evolution due to their simple structures, basic functions and limited amino acid content.22 The simplest ferredoxins are found in anaerobic bacteria and coordinate two [4Fe-4S] clusters. These short bacterial proteins (∼55 aa) with two cysteine-based iron-sulfur cluster binding motifs likely resulted from the duplication of a still smaller ancestral gene.23 Ferredoxins from higher plants, typically coordinating only one [2Fe-2S] cluster, likely branched off in evolution from the bacterial ferredoxins through a second duplication event.24 Such gene duplication, fusion, horizontal gene transfer and drift has led to the existence of ferredoxin-like domains in many other iron-sulfur oxidoreductases. Indeed, the X-ray structure of the [FeFe]-hydrogenase from Clostridium pasteurianum shows that the hydrogenase electron transfer domain is made up of three smaller iron-sulfur cluster binding domains, one with homology to bacterial ferredoxins, one to plant-type ferredoxins and one a unique linker domain.25 As a common component of electron transfer proteins, ferredoxins are excellent parts for the design of novel electron transfer pathways and novel modular electron transfer proteins.
Ferredoxins are characteristically acidic and electrostatic forces have been shown to be important for the interactions between ferredoxins and some oxidoreductases, in particular the hydrogenase from Chlamydomonas reinhardtii, which does not contain the ferredoxin-like domain of the clostridial hydrogenases.26 Mutational analyses of acidic surface residues in ferredoxin show that different residues are important for binding to different oxidoreductases.27–29 However, electrostatic forces have also been shown to not be important in other ferredoxin-paired reactions, notably that of bacterial ferredoxin and the clostridial hydrogenase.17 In these cases, random transient interactions between proteins may be sufficient to initiate the extremely fast electron tunneling between iron-sulfur proteins and dynamic interactions must be regulated in a different way.
As they are common throughout evolutionary history, ferredoxins can be functionally expressed heterologously in a wide host range,30,31 and have been shown to be amenable to functional gene fusion.32 The genomes of many organisms include several putative ferredoxin proteins, with one ferredoxin particularly suitable for a given oxidoreduction reaction.33 In higher plants, a range of ferredoxins at different electronic potential are typically expressed in leaves to account for the complex range of electron transfer reactions occurring in these organisms.34 The specification of each ferredoxin to its particular application may be due to its electronic potential, determined by the amino acid composition surrounding the iron-sulfur cluster or to the surface complementarity between ferredoxin and the cognate oxidoreductase.35 A synthetic metagenomics36 or directed evolution37 approach to the development of engineered electron-transfer pathways in synthetic biology could identify the ideal ferredoxin for each desired application.
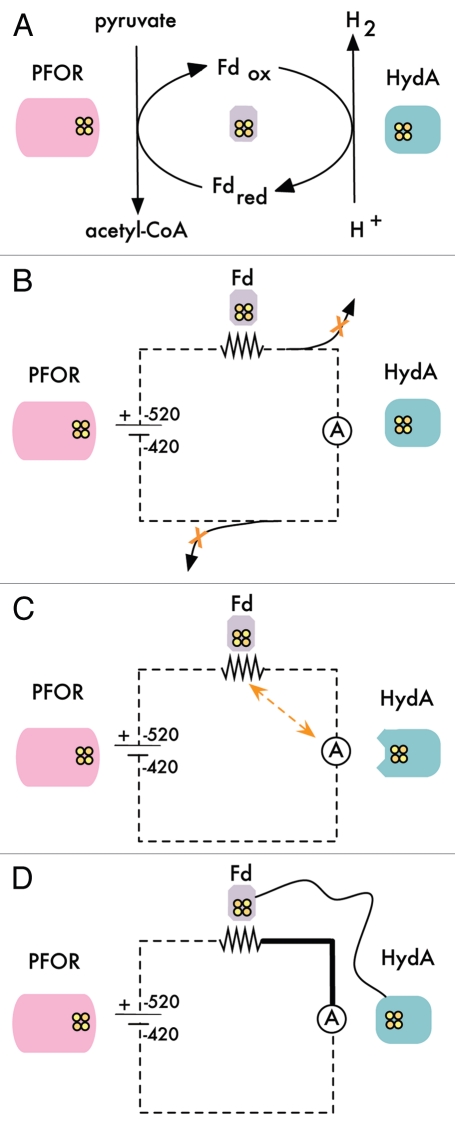
We chose the ferredoxin-paired [FeFe]-hydrogenases as the read-out of “current” through our iron-sulfur protein circuit in E. coli, as these enzymes thermodynamically favor hydrogen production at a high rate, and have been shown to be functionally expressed in E. coli.38 For our “battery” we desired an oxidoreductase enzyme that would be able to generate electrons through the breakdown of a common metabolite, integrating our system into native E. coli metabolism, as well as donate those electrons to our ferredoxin wires. We chose pyruvate-ferredoxin oxidoreductase (PFOR), an enzyme involved in energy metabolism through the hydrogen production circuit of the hydrogenosome,39 and shown to generate reduced ferredoxin recombinantly.17,40 PFOR, with a midpoint reducing potential of −520 mV, breaks down pyruvate, a central molecule of carbon metabolism, into acetyl-CoA, and in the process reduces ferredoxin. Ferredoxin then donates the electron to the hydrogenase, completing the circuit and re-setting cellular redox balance while leading to the production of high levels of hydrogen gas, ranging from 0.005% to nearly 3% of the theoretical maximum level of hydrogen production from pyruvate (Fig. 1A).
Figure 1.
(A) Schematic of the synthetic hydrogen production pathway. The circuit is analogous to an amorphous electronic circuit. Electrons enter the circuit at −520 mV through the function of pyruvate-ferredoxin oxidoreductase (PFOR), which thus behaves as a “battery.” Electrons are transferred to ferredoxin, an electron carrier that behaves as a “resistive wire.” Ferredoxin passes electrons to the hydrogenase, which in turn produces hydrogen at −420 mV that can be measured as as the “current” through the circuit. The circuit can be insulated from competing cellular electron metabolism through: (B) deletion of reactions that can interact with any of the circuit components leading to “short circuits”, (C) improvement of the ferredoxin-hydrogenase binding surface and (D) through direct protein fusion of the ferredoxin and hydrogenase. This fusion provides physical “circuit board” structure that increases the local concentration of electrons available to the hydrogenase.
Rethinking electron transfer-based metabolism as a simple electronic circuit is certainly an oversimplification of an intricate, highly regulated system, but a particularly useful one for the level at which we are currently able to engineer metabolic pathways. There is much that is still unknown about the dynamic regulation of coordinated metabolic processes, but it is possible to quickly combine groups of iron sulfur proteins inside cells, thereby creating novel hydrogen metabolism circuits. As a modular circuit, it is then possible to vary components in order to query different system parameters. These circuits must be integrated into cellular metabolism in order to function, but must be isolated from competing electron-transfer reactions in order to optimize function. We engineered the circuit for preferential transfer through the desired pathway, thus insulating the circuit, through the deletion of competing genes, the modification of protein-protein binding surfaces and the fusion of components into an integrated circuit design (Fig. 1B–D).
We tested three ferredoxins as our circuit's “wires” that represent a wide range of natural structure and function: the bacterial 2[4Fe-4S] ferredoxin from Clostridium acetobutylicum with a midpoint potential of −420 mV, leaf-type [2Fe-2S] ferredoxin I from spinach (−420 mV) and root-type [2Fe-2S] ferredoxin from corn (−345 mV). These three ferredoxins serve as models for ferredoxins of their type, and have been shown to interact specifically with several types of [FeFe]-hydrogenase.33,38
In addition, we tested five [FeFe]-hydrogenases and three PFORs from different species with different properties and behaviors in the synthetic circuit—hydrogenase from Clostridium acetobutylicum, Clostridium saccharobutylicum, Chlamydomonas reinhardtii,38 Shewanella oneidensis and Thremotoga maritima and PFOR from C. acetobutylicum,41 Desulfovibrio africanus,42 and the E. coli homolog YdbK.40 The heterologous expression of functional hydrogenases requires the co-expression of two maturation factors, HydEF and HydG; in our circuit, we used commercially synthesized HydEF and HydG sequences from Chlamydomonas reinhardtii. While the high GC content of the native algal genes made the maturation factors unstable when expressed heterologously in E. coli,38 our codon optimized sequences were stable in our expression system and able to mature hydrogenase from all of the species tested.
All combinations of PFOR-ferredoxinhydrogenase circuits produced measurable amounts of hydrogen over strains expressing the hydrogenase or hydrogenase and ferredoxin alone. Notably, the combination with the highest activity was not that where all the components came from the same organism, but instead when the PFOR came from D. africanus and the ferredoxin and hydrogenase came from C. acetobutylicum. The ability of PFOR, ferredoxin and hydrogenase from diverse organisms to interact when coexpressed in E. coli reflects the generality of iron-sulfur cluster proteins through evolution, as well as the need for regulation of synthetic pathways to ensure proper function, insulating synthetic circuits from natural electron transfer pathways and preventing cross-talk between multiple engineered pathways in more complex designs.
We compared four methods that would “insulate” the circuit from native E. coli electron metabolism, ensuring proper flow of electrons through the circuit and thus increasing the amount of hydrogen produced. These methods are based on how natural electron transfer systems are controlled, simplified to develop generally applicable techniques for pathway insulation in synthetic biology. The temporal control of gene and protein expression is known to be involved in the control of many metabolic pathways, maintaining expression of genes only when needed. Different electron transfer proteins are expressed at different times and in different environmental conditions, ensuring that hydrogenases are expressed only in the absence of oxygen, and when the electrons are not needed in other pathways. 43 We simplified and modeled this kind of transcriptional control through the deletion of reactions that compete for electrons and effectively “short-circuit” the pathway, testing single knockouts of genes with homology to ferredoxins and ferredoxin oxidoreductases: Δfpr, ΔydbK, Δhcr, ΔyeaX, ΔhcaD and ΔfrdB. Of these mutations, only deletion of YdbK, the E. coli PFOR homolog, affected hydrogen production, decreasing background levels when hydrogenase and ferredoxin were expressed alone and increasing production from the whole pathway. Ferredoxin-based electron metabolism in E. coli is still poorly understood, with the function of many iron-sulfur proteins and their interactions still uncharacterized. Further combinatorial knockout analysis and other synthetic pathway design may point to physiological roles for the other ferredoxin-based proteins as well as improve hydrogen production through the synthetic pathway. Deletion of metabolic competitors can help to direct the flux of metabolites through a desired pathway,44 and can be used to make specifically tailored “minimal chassis” strains for any desired application.
Co-evolution of interacting pairs of proteins is crucial to the evolution of biological systems that evolved through the duplication and drift of a small number functional enzymes.45 We synthetically modeled this co-evolution by mutating the surface of the C. reinhardtii hydrogenase with amino acid changes computationally predicted to improve electrostatic interactions with the plant-type ferredoxins.26 Two of these mutations, E5K and D126K, produced a small measurable increase in hydrogen production, indicating that surface mutations can impact the function of electron transfer circuits, as well as shedding light on the computational analysis of electrostatic proteinprotein interactions. Deeper mutational analysis and design of complementary mutations in interacting partners will further impact our ability to design tightly regulated electron transfer systems, as well as likely improve our understanding of the co-evolution of iron-sulfur protein binding surfaces.
The most versatile and easily portable method for pathway insulation does not require deep understanding of protein structures or competing metabolic pathways, but simply requires localizing two or more interacting components in the same place, thereby increasing the likelihood of collision and electron transfer. Natural electron transfer pathways are often regulated through the physical scaffolding of protein components in the membrane, for example in the electron transport chain of mitochondria. Of course, scaffolding of electrical components in a physical circuit defines circuit function, and there is a long history of engineering fusions between electron transfer proteins for dual function, primarily with P450 monooxygenase and/or ferredoxin-NADP+ reductase,32,46 as well as the [NiFe]-hydrogenase.47 By genetically fusing the hydrogenase and ferredoxin or targeting both to a tertiary scaffold48 we were able to increase the local concentration of ferredoxin available to the hydrogenase in our hydrogen production circuit. This effectively insulates the circuits from outside electron competition since electrons will preferentially be transferred between the fused components rather than with outside enzymes.
Because electron transfer proteins must be in brief physical contact and in the proper orientation for quantum tunneling between iron sulfur proteins, the function of the fused or scaffolded proteins exhibit strong dependence on the length of the flexible protein linker between them,49 and on the relative position of each on the scaffold domains. The proteins must be able to easily interact with each other as well as with other electron transfer components, as in the case of the ferredoxin, which both receives and donates an electron through quantum tunneling. In the case of the genetically fused proteins, very short linker lengths (two amino acids) restricted the ability of the ferredoxin and hydrogenase to interact, decreasing hydrogen production, while very long linker lengths (>45 amino acids) were indistinguishable from pathways with no fusion. The optimal linker length of fourteen amino acids increased hydrogen production by nearly five-fold. Genetic or scaffold fusion for physically isolating electron transfer partners can be generally applied to the design of electron transfer circuits, with care needed in determining the positioning and linker lengths for optimal collision rates and electron transfer.
In our system, hydrogenases provided the readout of circuit current, producing varying levels of hydrogen depending on the input parameters. Hydrogenases, however, are reversible, able to consume hydrogen as a source of reducing power. Hydrogenases are “batteries” in several species of bacteria, notably the hydrogen oxidizing [NiFe]-hydrogenases of the chemolithoautotroph Ralstonia eutropha50 and as the source of reducing power to fix sulfur in Desulfovibrio vulgaris.51 Pairing of hydrogenases with the root-type ferredoxins, which have a higher reducing potential, will favor these hydrogen consumption reactions and can aid in the creation of many synthetic metabolic pathways.
The application of engineering principles such as modularity, abstraction and insulation to the design of biological systems has the potential to lead to facile development of biological pathways for diverse applications. The assumptions, abstractions and simplifications of synthetic biology are valuable for developing novel biological technologies and studying biological complexity, as long as such assumptions do not cause researchers to lose sight of important biological dynamics. In developing new parts and platforms for synthetic biology, it is important to look to natural modularity in gene and protein structure as starting points for the design of novel circuits. Through such simplifications we can build networks from isolated genetic and enzymatic elements and begin to characterize and understand how such parts function in novel contexts. Electron transfer circuits built of modular iron-sulfur protein parts can be applied in the design of countless such biological pathways, with potential for integration of electronic and biological systems, industrial-scale production of renewable fuels and importantly, biological assays and tools for better understanding the principles underlying complex biological systems for further engineering.
Acknowledgements
The authors would like to thank the Danny Ducat, Patrick Boyle, Jake Wintermute and Jeff Way for their work on the original paper and Devin Burrill for helpful advice on this manuscript. C.M.A. is supported by an NSF graduate research fellowship and the work described here was funded by Army Research Office Award No. W911NF-09-1-0226.
References
- 1.Andrianantoandro E, Basu S, Karig D, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2:28. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 4.Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EPS, Landgraf D, Phillips I, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin CH, Nielsen DR, Solomon KV, Prather KLJ. Synthetic metabolism: engineering biology at the protein and pathway scales. Chem Biol. 2009;16:277–286. doi: 10.1016/j.chembiol.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 9.Savage DF, Way J, Silver PA. Defossiling Fuel: How Synthetic Biology Can Transform Biofuel Production. Metab Eng. 2007 doi: 10.1021/cb700259j. [DOI] [PubMed] [Google Scholar]
- 10.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filipovska A, Rackham O. Building a parallel metabolism within the cell. ACS Chem Biol. 2008;3:51–63. doi: 10.1021/cb700185e. [DOI] [PubMed] [Google Scholar]
- 12.Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 13.Weber W, Luzi S, Karlsson M, Sanchez-Bustamante CD, Frey U, Hierlemann A, et al. A synthetic mammalian electro-genetic transcription circuit. Nucleic Acids Res. 2009;37:e33. doi: 10.1093/nar/gkp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong TS, Schwaneberg U. Protein engineering in bioelectrocatalysis. Current Opinion in Biotechnology. 2003;14:590–596. doi: 10.1016/j.copbio.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 16.Sadeghi SJ, Meharenna YT, Fantuzzi A, Valetti F, Gilardi G. Engineering artificial redox chains by molecular ‘Lego’. Faraday Disc. 2000;116:135–153. doi: 10.1039/b003180l. [DOI] [PubMed] [Google Scholar]
- 17.Moulis JM, Davasse V. Probing the role of electrostatic forces in the interaction of Clostridium pasteurianum ferredoxin with its redox partners. Biochemistry. 1995;34:16781–16788. doi: 10.1021/bi00051a028. [DOI] [PubMed] [Google Scholar]
- 18.Veit A, Akhtar M, Mizutani T, Jones PR. Constructing and testing the thermodynamic limits of synthetic NAD(P)H:H 2pathways. Microbial Biotechnology. 2008;1:382–394. doi: 10.1111/j.1751-7915.2008.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coghlan VM, Vickery LE. Expression of human ferredoxin and assembly of the [2Fe-2S] center in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:835–839. doi: 10.1073/pnas.86.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoell HE, Knappe J. Escherichia coli ferredoxin, an iron-sulfur protein of the adrenodoxin type. Eur J Biochem. 1974;50:245–252. doi: 10.1111/j.1432-1033.1974.tb03893.x. [DOI] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall DO, Cammack R, Rao KK. Role for ferredoxins in the origin of life and biological evolution. Nature. 1971;233:136–138. doi: 10.1038/233136a0. [DOI] [PubMed] [Google Scholar]
- 23.Otaka E, Ooi T. Examination of protein sequence homologies: IV. Twenty-seven bacterial ferredoxins. J Mol Evol. 1987;26:257–267. doi: 10.1007/BF02099857. [DOI] [PubMed] [Google Scholar]
- 24.Otaka E, Ooi T. Examination of protein sequence homologies: V. New perspectives on evolution between bacterial and chloroplast-type ferredoxins inferred from sequence evidence. J Mol Evol. 1989;29:246–254. doi: 10.1007/BF02100208. [DOI] [PubMed] [Google Scholar]
- 25.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 26.Long H, King P, Ghirardi M, Kim K. Hydrogenase/ferredoxin charge-transfer complexes: effect of hydrogenase mutations on the complex association. J Phys Chem. 2009;113:4060–4067. doi: 10.1021/jp810409z. [DOI] [PubMed] [Google Scholar]
- 27.Coghlan VM, Vickery LE. Site-specific mutations in human ferredoxin that affect binding to ferredoxin reductase and cytochrome P450scc. J Biol Chem. 1991;266:18606–18612. [PubMed] [Google Scholar]
- 28.Lelong C, Sétif P, Lagoutte B, Bottin H. Identification of the amino acids involved in the functional interaction between photosystem I and ferredoxin from Synechocystis sp. PCC 6803 by chemical cross-linking. J Biol Chem. 1994;269:10034–10039. [PubMed] [Google Scholar]
- 29.Hurley JK, Hazzard JT, Martínez-Júlvez M, Medina M, Gómez-Moreno C, Tollin G. Electrostatic forces involved in orienting Anabaena ferredoxin during binding to Anabaena ferredoxin:NADP+ reductase: site-specific mutagenesis, transient kinetic measurements and electrostatic surface potentials. Protein Sci. 1999;8:1614–1622. doi: 10.1110/ps.8.8.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaton BL, Vickery LE. Expression of human ferredoxin in Saccharomyces cerevisiae: mitochondrial import of the protein and assembly of the [2Fe-2S] center. Arch Biochem Biophys. 1992;294:603–608. doi: 10.1016/0003-9861(92)90731-b. [DOI] [PubMed] [Google Scholar]
- 31.Davasse V, Moulis JM. Design and functional expression in Escherichia coli of a synthetic gene encoding Clostridium pasteurianum 2[4Fe-4S] ferredoxin. Biochem Biophys Res Commun. 1992;185:341–349. doi: 10.1016/s0006-291x(05)80991-x. [DOI] [PubMed] [Google Scholar]
- 32.Aliverti A, Zanetti G. A three-domain iron-sulfur flavoprotein obtained through gene fusion of ferredoxin and ferredoxin-NADP+ reductase from spinach leaves. Biochemistry. 1997;36:14771–14777. doi: 10.1021/bi971791t. [DOI] [PubMed] [Google Scholar]
- 33.Guerrini O, Burlat B, Léger C, Guigliarelli B, Soucaille P, Girbal L. Characterization of two 2[4Fe4S] ferredoxins from clostridium acetobutylicum. Curr Microbiol. 2008;56:261–267. doi: 10.1007/s00284-007-9072-x. [DOI] [PubMed] [Google Scholar]
- 34.Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004;134:255–264. doi: 10.1104/pp.103.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuyama K. Structure and function of plant-type ferredoxins. Photosyn Res. 2004;81:289–301. doi: 10.1023/B:PRES.0000036882.19322.0a. [DOI] [PubMed] [Google Scholar]
- 36.Bayer T, Widmaier D, Temme K, Mirsky E, Santi D, Voigt C. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 37.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 38.King PW, Posewitz MC, Ghirardi ML, Seibert M. Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J Bacteriol. 2006;188:2163–2172. doi: 10.1128/JB.188.6.2163-2172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams K, Lowe PN, Leadlay PF. Purification and characterization of pyruvate: ferredoxin oxidoreductase from the anaerobic protozoon Trichomonas vaginalis. Biochem J. 1987;246:529–536. doi: 10.1042/bj2460529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhtar M, Jones PR. Construction of a synthetic YdbK-dependent pyruvate:H2 pathway in Escherichia coli BL21(DE3) Metab Eng. 2009;11:139–147. doi: 10.1016/j.ymben.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Meinecke B, Bertram J, Gottschalk G. Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch Microbiol. 1989;152:244–250. doi: 10.1007/BF00409658. [DOI] [PubMed] [Google Scholar]
- 42.Pieulle L, Guigliarelli B, Asso M, Dole F, Bernadac A, Hatchikian EC. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim Biophys Acta. 1995;1250:49–59. doi: 10.1016/0167-4838(95)00029-t. [DOI] [PubMed] [Google Scholar]
- 43.Richard DJ, Sawers G, Sargent F, McWalter L, Boxer DH. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology. 1999;145:2903–2912. doi: 10.1099/00221287-145-10-2903. [DOI] [PubMed] [Google Scholar]
- 44.Kennedy CJ, Boyle PM, Waks Z, Silver PA. Systems-level engineering of nonfermentative metabolism in yeast. Genetics. 2009;183:385–397. doi: 10.1534/genetics.109.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skerker J, Perchuk B, Siryaporn A, Lubin E, Ashenberg O, Goulian M, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacour T, Ohkawa H. Engineering and biochemical characterization of the rat microsomal cytochrome P4501A1 fused to ferredoxin and ferredoxin-NADP(+) reductase from plant chloroplasts. Biochim Biophys Acta. 1999;1433:87–102. doi: 10.1016/s0167-4838(99)00154-5. [DOI] [PubMed] [Google Scholar]
- 47.Ihara M, Nishihara H, Yoon KS, Lenz O, Friedrich B, Nakamoto H, et al. Light-driven hydrogen production by a hybrid complex of a [NiFe]-hydrogenase and the cyanobacterial photosystem I. Photochem Photobiol. 2006;82:676–682. doi: 10.1562/2006-01-16-RA-778. [DOI] [PubMed] [Google Scholar]
- 48.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 49.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J Am Chem Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cramm R. Genomic view of energy metabolism in Ralstonia eutropha H16. J Mol Microbiol Biotechnol. 2009;16:38–52. doi: 10.1159/000142893. [DOI] [PubMed] [Google Scholar]
- 51.Pereira P, He Q, Valente F, Xavier A, Zhou J, Pereira I, et al. Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Van Leeuwenhoek. 2007;92:347–362. doi: 10.1007/s10482-007-9212-0. [DOI] [PubMed] [Google Scholar]