Abstract
Recently, our laboratory demonstrated that Paneth cell defensins, innate antimicrobial peptides that contribute to mucosal host defense, are able to regulate the composition of the intestinal bacterial microbiome. Using complementary mouse models of defensin deficiency (MMP 7−/−) and surplus (HD 5+/+), we noted defensin-dependent reciprocal shifts in the dominant bacterial species of the small intestine, without changes in total bacterial numbers. In addition, mice that expressed HD 5 showed a significant loss of segemented filamentous bacteria (SF B), resulting in reduced numbers of Th17 cells in the lamina propria. This data showed a novel role for PC defensins in intestinal homeostasis, by regulation of the small intestinal microbiome. The microbiome plays an essential role in mediating host physiology, metabolism and immune response. The ability of PC defensins to regulate the composition of the biome suggests a much broader importance of these innate immune effectors than previously considered. In this addendum, the role of PC defensins in the regulation of the intestinal microbiome is reviewed, and discussed in the context of recent evidence that highlights the important role of PC s and defensins in the pathophysiology of inflammatory bowel disease.
Key words: Paneth cell, defensin, microbiome, Th17, intestinal homeostasis, inflammatory bowel disease
Introduction
The intestinal mucosal surface is a dynamic interface linking the host and its external environment. Its role is multifunctional, allowing nutrient and water absorption and sampling of luminal antigens, while excluding potentially harmful microbes, toxins and environmental antigens. These activities are carried out with a minimum of inflammation, and contribute to intestinal homeostasis. Disruption of homeostasis, such as that caused by bacterial infection, results in host inflammatory responses.
The innate mucosal barrier, primarily elaborated by the single layer of epithelial cells that separate the host from the intestinal luminal contents, is composed of mucins, IgA and antimicrobial effector molecules. The external environment contains digestible and non-digestible food, as well as a vast, diverse, complex microbial ecosystem. Recent advances have provided significant insight into the composition and functional capacity of this ecosystem by the application of high throughput 16S ribosomal RNA gene sequencing and metagenomic sequencing approaches.1–3
The intestinal microbiota is Janus-faced. In an intact host, the microbiota are critical to host protection, through their induction of mucosal immune development4,5 and by forming a mucosal barrier that prevents colonization and/or invasion of microbial species that may cause damage to the host. This function, referred to as “colonization resistance”, is thought to arise through effective competition by the indigenous microbiota for nutrients, iron, and available attachment sites in the gut. However, in the absence of the host structural and immune barrier, the microbes that comprise this ecosytem have the potential to invade the host and cause disease. Although it isn't yet entirely clear what comprises a “normal” microbiota, alterations in the bacterial composition of the intestinal microbiota have been implicated in the pathophysiology of several diseases, including obesity,6 diabetes,7 irritable bowel syndrome8 and inflammatory bowel disease.9
Interactions between the colonizing microbiota and the mucosal immune system have come under intense study recently. Studies evaluating this using monoassociation of germ-free animals clearly demonstrated the importance of the commensal biota in the development of acquired immunity (reviewed in ref. 5), and the induction of IgA.10 Several studies have demonstrated the ability of intestinal commensals to induce mucosal innate immune effectors, including RegIIIγ,11 and Angiogenins.12 The induction of these innate antimicrobials acts to contain the microbiota within the intestinal lumen and prevent translocation.13 However, our understanding of the immune factors that shape the composition of the microbiota is more limited. Studies by Rawls et al. demonstrated that the host plays an important role in selecting its particular biota,14 but did not define the specific host factors involved. Early work, investigating the role of immune factors in shaping the microbiota, focused on the role of IgA. Suzuki et al. found that IgA deficient mice had significant alterations in composition of their microbiota.15 We hypothesized that other components of the innate mucosal barrier, would also be involved in shaping and regulating the composition of the intestinal microbial ecosystem, and focused our investigation on the role of defensins.
Defensins are amphipathic cationic peptides that demonstrate broad-spectrum antibiotic activity against bacteria, fungi and viruses.16,17 They make up one of the classes of human cationic antimicrobial peptides, and are characterized by an invariant 6-cysteine array, each cysteine involved in intramolecular disulfide bonding. There are three subclasses of defensins in mammals (α, β, θ), but only α and β have been identified in humans and mice. In humans, α defensins are found primarily in neutrophils and Paneth cells (PCs), while β defensins are much more broadly distributed at mucosal surfaces. In mice, the α defensin distribution is restricted even further, solely to PCs. Defensins are synthesized as propeptides and enzymatically cleaved into their active forms. In humans, PC defensins are stored as inactive propeptides and processed upon secretion by PC trypsin.18 In mice, PC defensins are processed by MMP7 and stored in their processed form in the PC granule.19
PCs, highly secretory cells located at the bases of the crypts of Lieberkuhn, in the small intestine, derive from the crypt stem cell.20 Wnt4 signaling drives PC differentiation, maturation and positioning via β-catenin/Tcf-4.21,22 Paneth cells express and secrete a number of AMPs and inflammatory mediators in addition to defensins, including secretory phospholipase A 2 (sPLA2), TNFα, RegIIIγ and Angiogenins. PC differentiation occurs independent of microbial stimuli, as does expression of several PC products including lysozyme, sPLA2 and defensins.19,23 However several PC products, including RegIIIγ and Angiogenins are induced by bacterial colonization via TLR stimulation.
PC granule secretion is triggered by a variety of stimuli, including cholinergic agonists, bacteria and bacterial products (LPS, LTA).24 After secretion, the concentration of PC defensins in the small intestinal crypt has been calculated to reach 15–100 mg/ml.24 In vitro, defensins have broad-spectrum anti-gram positive and anti-gram negative activity,25 while RegIIIγ,11 sPLA2,26 and lysozyme are most effective against gram-positive bacteria. Earlier work from our laboratory and others demonstrated that PC defensins are important for host defense against enteric pathogens.27,28 However, because of their constitutive expression and secretion, we hypothesized that PC defensins were likely to have an equally critical role in intestinal homoestasis, that of regulating the composition of the colonizing biota.
Mouse Models for the Study of in vivo Defensin Function
To address this question, we used two mouse models that have been crucial to understanding in vivo PC defensin biology, the HD5 transgenic mouse and the MMP7 KO mouse. The HD5 TG mouse expresses the human PC defensin (DEFA5) specifically in the mouse PC.28 HD5 is expressed at physiologic levels, similar to the expression level of a single endogenous mouse PC defensin (cryptdin), and is processed and secreted similarly to the mouse cryptdins. Initial studies using this model demonstrated that the addition of HD5 to the mouse PC defensin “armamentarium” conferred significant resistance to enteric Salmonella infection. The complementary model is one of PC defensin deficiency.27 MMP7, a matrix metalloproteinase, is localized to PC in the mouse small intestine, where it is responsible for processing cryptdins from their proform into active peptides within the PC granules. MMP7 KO mice lack this enzyme and are deficient in active defensins in the small intestine. In vivo, these mice have reduced capacity to clear enteric pathogens and exogenously delivered bacteria. While MMP7 may have other extra-intestinal roles in vivo, notably in models of inflammation and wound healing,29–31 in unchallenged animals it is a useful model for small intestinal defensin deficiency.
PC Defensins Influence the Composition of the Bacterial Microbiome
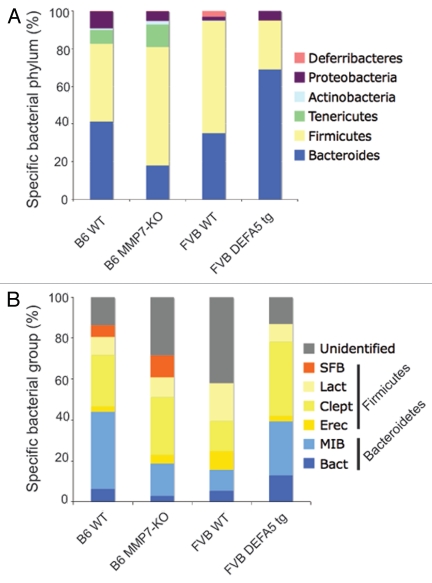
Using a combination of methodologies based on analysis of 16S rRNA gene sequences, including high through put 16S rRNA Sanger gene sequencing, quantitative PCR and fluorescence in situ hybridization (FISH) on terminal ileum tissue, we compared small intestinal colonization of these animal models.32 We generated litters of offspring from either hemizygous HD5 parents (HD5+/−) or partial MMP7 KO mice (MMP7+/−). This resulted in litters containing WT, hemizygous and homozygous offspring. We used this approach to control for parental effects and environmental effects. We found evidence of significant shifts in the dominant bacterial species present in the small intestines. The two dominant bacterial phyla present in the intestinal tract of mice and humans are the Bacteriodetes (gram negative) and the Firmicutes (gram positive), which together comprise about 70–80% of the total bacteria present. In HD5 mice, the percentage of Firmicutes decreased, while the Bacteriodes increased proportionally, when compared to WT littermate controls. In the MMP7 KO litters, we found reciprocal shifts. With loss of active defensins, the abundance of Firmicutes increased, while the Bacteriodes decreased proportionally (Fig. 1). While metabolic effects, primarily obesity, have been ascribed to shifts in the dominant bacterial groups, we did not observe similar effects in our mice. However, our study was focused on the biome of the small intestines and the metabolic effects may be more related to shifts in the large intestine. Nevetheless, it is evident that alterations in PC defensins, innate antibiotic effector peptides, result in alterations in the composition of the small intestinal microbiome.32 Interestingly, the presence, absence or composition of active defensins in the PC had no impact on the total numbers of bacteria colonizing the GI tract. This suggests that other factors are responsible for keeping bacterial numbers in check.
Figure 1.
Comparison of microbial communities of distal small intestines from DEFA5 TG and MMP7−/− mice. Genomic DNA was isolated from the distal small intestines of age matched DEFA5 TG mice and their wild type littermates, and from MMP7−/− mice and their wild type littermates. Microbial community analysis was done based on 16S rRNA gene sequence, using high through put Sanger sequencing (A) and quantitative PCR (B). The stacked graphs show the relative percentages of the bacterial groups identified by each method. Percentages were obtained for Sanger subclone sequencing using the total number of sequences obtained as the denominator. Percentages were obtained for qPCR by using total bacterial 16S gene copies as the denominator, determined by amplifying each sample with universal bacterial primers. Figure reproduced from original publication by Salzman et al.32
PC Defensins Regulate the Abundance of Small Intestinal Segmented Filamentous Bacteria
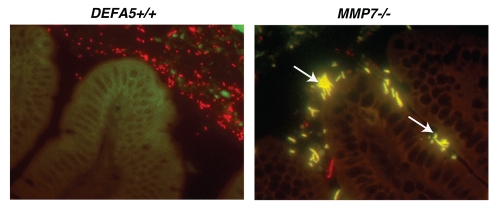
Our study design resulted in an unexpected finding. The litters from the HD5 hemizygous breeding pairs all lacked the presence of one distinctive bacterium, segmented filamentous bacteria (SFB), while MMP7−/− litters retained these bacteria32 (Fig. 2). Interestingly, increased presence of SFB was recently noted in a mouse model of Cd1d deficiency, a model also characterized by PC abnormalities;33 providing additional evidence to support a role for PC antimicrobial effectors in the regulation of SFB abundance. SFB, formally known as Candidatus arthromitis, is an uncultivable, spore forming grampositive member of the Firmicute phylum, and is one of the only members of the commensal biota that directly contacts intestinal epithelial cells.34 It is highly sensitive to exogenous antibiotics,35 and also regulated by IgA.15 A potent stimulator of acquired immune responses and development, several groups have recently characterized this organism's ability to stimulate the expression of IL17A in lamina propria CD4+ T cells (Th17 cells).36,37 Concurrently, we determined that our HD5 homozygous mice also lacked LPL Th17 differentiation, in association with their lack of SFB.32 This offers evidence that PC defensins are able to regulate acquired immune responses via their ability to regulate the commensal microbiota. Interestingly, Th17 cells in the lamina propria have been shown to induce mucosal antimicrobial peptide expression, notably RegIIIγ,38,39 via IL22 secretion-bringing the regulatory pathway to almost a full circle. At the same time, inversion of the Th17/Treg balance has been associated with skewing of the immune response towards proinflammatory responses. Concurrently, the commensal bacterium Bacteroides fragilis has been recently shown to induce Treg development,40 demonstrating that distinct commensal bacteria can direct either iTreg or Th17 differentiation. Therefore, the composition of the microbiome, as determined by antimicrobial peptides, may be a fundamental mediator of iTreg/Th17 balance. We find these results particularly intriguing when we consider the role of PC's and defensins in the pathophysiology of inflammatory bowel disease.
Figure 2.
Segmented filamentous bacteria (SFB) is lost in DEFA5 TG mice. Fluorescence in situ hybridization was performed on the terminal 1.5 cm section of distal small intestine from DEFA5 TG and MMP7−/− mice, for the detection of total bacteria and for SFB. Tissue sections were hybridized with a combination of oligonucleotide probes for all bacteria (Texas red labeled) and SFB (6Fam labeled) and examined by fluorescence microscopy. Images were overlaid in Adobe Photoshop. Bacteria that hybridize with the universal bacterial probe fluoresce red. Bacteria that co-hybrizide with the universal bacterial probe and the specific SFB probe appear yellow. In the small intestine, segmented filamentous bacteria (yellow) directly contact the intestinal epithelium (green), unlike the rest of the commensal microbiota (red), which are located in the intestinal mucus. Arrows point to SFB bacteria. Figure is a composite of unpublished and published data by Salzman et al.32
PCs, Defensins and Crohn Disease (CD)
Crohn disease (CD) is a chronic inflammatory disease of the GI tract. Current theory suggests that CD is caused by an abnormal immune response to colonizing biota in a genetically susceptible host. One of the hallmarks associated with CD is intestinal dysbiosis or abnormal bacterial growth at the intestinal mucosal surface, with evidence of increased numbers of mucosa-adherent bacteria, shifts in bacterial colonization and reduced bacterial diversity.3,9,41 One critical and currently unanswered question is whether the abnormal bacterial colonization is driving chronic immune responses or whether chronic inflammation is triggering the dysbiosis. Recent accumulating evidence supports the hypothesis that an innate immune defect can explain the cluster of observations surrounding CD ileal pathophysiology, and much of this evidence points to the essential role of PCs and PC effectors.42–45
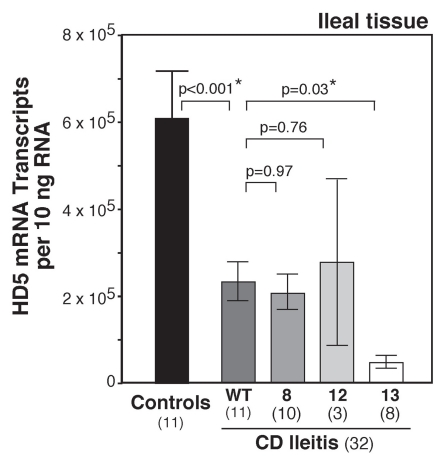
Several genetic defects associated with ileal CD are involved in Paneth cell function. Mutations in the NOD2 gene were among the earliest genetic associations with the development of severe ileitis in CD.46 The NOD2 gene encodes for the nod2 protein, an abbreviation for nuclear oligomerization domain 2. This protein product is a pattern recognition receptor that is important in intracellular bacterial sensing, binding MDP moieties found on the bacterial cell wall. Exactly how this defect may lead to CD is controversial, but a study by Kobayashi et al.47 demonstrated that nod2 deficient mice had reduced expression of PC defensins. Work by Wehkamp et al.44 clearly demonstrated that patients with CD had reduced expression of HD5, and those with the 1007fs (SNP13) NOD2 defect had even greater reductions in their HD5 mRNA and peptide expression (Fig. 3). This specific SNP has been associated with greater ileal involvement and disease severity.48,49 Reductions in HD5 were not related to intestinal inflammation. This suggests that PC deficiency is not the result of chronic inflammation but rather an inciting cause of CD pathophysiology. However, NOD2 mutations are only found in a small subset of patients with CD, while defensin deficiency is associated with CD even in the absence of NOD2 mutations. Further work by Wehkamp et al.45 demonstrated that patients with ileal CD also had deficiencies in Tcf-4, the Wnt signaling transcription factor that is directly associated with PC differentiation and α-defensin expression. Tcf-4 expression levels were not related to treatment, extent of inflammation or NOD2 genotype, delineating a separate mechanism leading to PC defensin deficiency in CD pathogenesis. Additional genetic mutations that result in altered PC function have been associated with CD. One example is the mutation of ATG16L1, a gene that is involved in autophagy.42,50 Examination of small intestinal tissue from both patients and mice harboring this mutation shows severe defects in PC function. A second example is the XBP1 gene. XBP1 is a critical transcription factor induced during the endoplasmic reticulum stress response. Work by Kaser et al.43 showed that XBP1 variants were associated with CD susceptibility. XBP1 deletion in mice resulted in PC dysfunction and loss and increased susceptibility to colitis. Again, these finding support the hypothesis that PC dysfunction is intimately associated with the development and progression of CD.
Figure 3.
HD5 expression related to NOD2 genotype in patients with ileal CD. HD5 mRNA expression was determined by RT -qPCR of RNA isolated from ileal biopsies from patients with CD and controls. Samples were grouped by NOD2 genotype. All patients with CD had significant reductions in HD5 mRNA expression, irrespective of NOD2 genotype. However, there were significantly greater reductions in HD5 expression in patients with the SNP13 NOD2 mutation. The SNP13 mutation has shown increased association with severe ileal disease.48,49 Composite figure and data from Wehkamp et al.44 redrawn by Dr. Charles Bevins, UC Davis School of Medicine, Davis, CA , Copyright (2005) National Academy of Sciences USA.
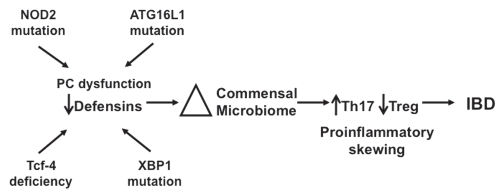
We propose the following provisional model (Fig. 4), which, although speculative, is suggested by the findings of our group and several others, and may offer insight into the dysbiosis and proinflammatory skewing of mucosal immune responses in CD. Antimicrobial effectors secreted by PCs and epithelial cells combine to form a barrier against intruding intestinal microbes. We hypothesize that the concentration of effector molecules diffuses from the point of secretion, setting up a gradient, allowing intestinal microbes to encroach on the epithelial surface until the levels of AMPs are too high for their survival. Genetic defects in innate mucosal defense of the small intestine, such as those currently described regarding PCs and PC defensins result in a “fading back” of the mucosal antimicrobial barrier. This could result in the noted alterations in the composition of the microbiota,3,9,41 which would likely be most pronounced closest to the mucosal surface. In addition, the reduced concentration of antimicrobial effectors could allow bacterial species to approach much closer to the epithelial surface, allowing their interaction with intestinal epithelium and mucosal immune cells (reviewed in ref. 9). The shift in the bacterial composition of the small intestine could then influence mucosal associated T cell differentiation and response, leading to skewing of mucosal T cell responses towards a proinflammatory phenotype.
Figure 4.
An exploratory model for the role of defensin deficiency in CD. Several genetic mutations (NOD2, AT G16L1, XBP1) lead to either global PC dysfunction or specific defensin deficiency (NOD2, Tcf-4). Reduction in PC defensin expression results in alterations in the intestinal bacterial microbiome. Shifts in the biome lead to induction of Th17 cells and proinflammatory skewing of the Th17:Treg balance, resulting in predisposition to the persistent and chronic inflammation associated with CD.
While this highly exploratory model offers an explanation for both dysbiosis and skewing of mucosal T cell responses, it is likely that additional “hits” are required for the complete development of CD, although whether these hits are environmental or immune is yet to be determined. Nevertheless, it is clear that PC defensins have multiple roles that are essential for establishing intestinal homeostasis. Appropriate expression of PC defensins function by directly killing bacterial pathogens, and by regulating the composition of the bacterial microbiome. The ability of PC defensins to support the correct bacterial balance in the small intestine is critical for limiting mucosal inflammatory response, effecting a reduction of hostilities or détente at the intestinal host-microbe interface.
Acknowledgements
Research in our laboratory is supported by the National Institutes of Health, NIAID AI057757. I would like to thank Dr. Charles Bevins and Dr. Jan Wehkamp for providing a composite figure from their published work, and the permission for its use.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/14076
References
- 1.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 5.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkes GC, Brostoff J, Whelan K, Sanderson JD. Gastrointestinal microbiota in irritable bowel syndrome: their role in its pathogenesis and treatment. Am J Gastroenterol. 2008;103:1557–1567. doi: 10.1111/j.1572-0241.2008.01869.x. [DOI] [PubMed] [Google Scholar]
- 9.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 10.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: an new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 13.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 17.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh D, Porter E, Shen B, Lee SK, Drazba WD, Yadav SP, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 19.Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, et al. Activation of paneth cell α-defensins in mouse small intestine. J Biol Chem. 2002;277:5219–5228. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- 20.Porter EM, Bevins CL, Ghosh D, Ganz T. The multifaceted Paneth cell. Cell Mol Life Sci. 2002;59:156–170. doi: 10.1007/s00018-002-8412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batlle E, Henderson JT, Beghtel H, van den Born MMW, Sancho E, Huls G, et al. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 22.van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, et al. Wnt signaling induces maturation of Panteh cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 23.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275:40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 24.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 25.Ouellette AJ, Hsieh MM, Nosek MT, Cano-Gauci DF, Huttner KM, Buick RN, et al. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62:5040–5047. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koduri RS, Gronroos JO, Laine VJ, Le Calvez C, Lambeau G, Nevalainen TJ, et al. Bactericidal properties of human and murine groups I, II V, X and XII secreted phospholipases A(2) J Biol Chem. 2002;277:5849–5857. doi: 10.1074/jbc.M109699200. [DOI] [PubMed] [Google Scholar]
- 27.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, et al. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 28.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 29.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. International J Biochem Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 31.Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y. Role of matrix metalloproteinase-7 (matrilysin) in human cancer invasion, apoptosis, growth and angiogenesis. Exp Biol Med (Maywood) 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- 32.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–82. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, et al. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats and chickens and proposal of “Candidatus arthromitus”. Int J Syst Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 35.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 38.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16L1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wehkamp J, Wang G, Kubler I, Nuding S, Gregorieff A, Schnabel A, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 46.Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 48.Hampe J, Grebe J, Nikolaus S, Solberg C, Croucher PJ, Mascharetti S, et al. Association of NOD2 (CARD15) genotype with clinical course of Crohn's disease: a cohort study. Lancet. 2002;359:1661–1665. doi: 10.1016/S0140-6736(02)08590-2. [DOI] [PubMed] [Google Scholar]
- 49.Seiderer J, Schnitzler F, Brand S, Staudinger T, Pfennig S, Herrmann K, et al. Homozygosity for the CARD15 frameshift mutation 1007fs is predictive of early onset of Crohn's disease with ileal stenosis, entero-enteral fistulas and frequent need for surgical intervention with high risk of re-stenosis. Scand J Gastroenterol. 2006;41:1421–1432. doi: 10.1080/00365520600703900. [DOI] [PubMed] [Google Scholar]
- 50.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]