Abstract
Hepatitis C virus (HCV) is the major etiological agent of blood-borne non–A non–B hepatitis and a leading cause of liver cirrhosis and hepatocellular carcinoma worldwide. HCV core protein is a multifunctional protein with regulatory functions in cellular transcription and virus-induced transformation and pathogenesis. Here we report on the identification of a bZIP nuclear transcription protein as an HCV core cofactor for transformation. This bZIP factor, designated LZIP, activates CRE-dependent transcription and regulates cell proliferation. Loss of LZIP function in NIH 3T3 cells triggers morphological transformation and anchorage-independent growth. We show that HCV core protein aberrantly sequesters LZIP in the cytoplasm, inactivates LZIP function and potentiates cellular transformation. Our findings suggest that LZIP might serve a novel cellular tumor suppressor function that is targeted by the HCV core.
Keywords: bZIP transcription factor/hepatitis C virus/hepatitis C virus core protein/hepatocellular carcinoma/LZIP
Introduction
Hepatitis C virus (HCV) is a parenterally transmitted virus etiologically associated with post-transfusion and community-acquired hepatitis (Purcell, 1994; Houghton, 1996; Rice, 1996; Clarke, 1997). Chronic and persistent infection with HCV is common (up to 85% in some populations) and ultimately leads to liver cirrhosis and hepatocellular carcinoma (HCC) (Saito et al., 1990; Bukh et al., 1993). Globally there are 0.25–1.2 million new cases of HCC annually (Idilman et al., 1998). Thus, HCV is one of the major infectious causes of morbidity and mortality worldwide.
HCV, a member of the Flaviviridae family, contains an ∼10 kb positive stranded RNA genome. The HCV genome encodes a single large polyprotein, which is cleaved by cellular and viral proteases into three structural (core, E1 and E2) and at least six non-structural (NS2, NS3, NS4a, NS4b, NS5a and NS5b) proteins. HCV core protein is derived from the N–terminal 191 amino acids of the large precursor polyprotein and is well conserved among all isolates (Bukh et al., 1994). The core is a highly basic protein with RNA-binding activity (Santolini et al., 1994; Hwang et al., 1995) and has the ability to form homo-multimers (Matsumoto et al., 1996; Nolandt et al., 1997). It is found in the cytoplasm (Cerino et al., 1993; Selby et al., 1993; Moradpour et al., 1996; Kawamura et al., 1997; Yasui et al., 1998), and it has been described variously to associate with endoplasmic reticulum (Santolini et al., 1994; Barba et al., 1997), lipid droplets (Barba et al., 1997), apolipoprotein AII (Sabile et al., 1999), lymphotoxin-β receptor (Chen et al., 1997; Matsumoto et al., 1997) and tumor necrosis factor (TNF) receptor 1 (Zhu et al., 1998). A small amount of core protein has also been seen in the nucleus (Chang et al., 1994; Ravaggi et al., 1994; Lo et al., 1995; Suzuki et al., 1995; Liu et al., 1997; Hsieh et al., 1998; Yasui et al., 1998; Sabile et al., 1999; You et al., 1999a).
Biologically, HCV core protein has been implicated in cellular proliferation. Observations relevant to this include transformation of NIH 3T3 (Ray et al., 1996a) and Rat–1 (Chang et al., 1998) cells by core protein; cooperation of core protein with H–ras to transform BALB/3T3 A31-I-1 cells (Tsuchihara et al., 1999) and primary rat embryo fibroblasts (Ray et al., 1996a); and core protein's modulation of anti-Fas- (Ruggieri et al., 1997; Marusawa et al., 1999), cisplatin plus c-myc- (Ray et al., 1996b) and TNF-α(Ray et al., 1998a; Zhu et al., 1998; Marusawa et al., 1999) induced apoptosis. HCV core protein can also regulate the transcription of a diverse array of viral and cellular genes (Kim et al., 1994; Ray et al., 1995, 1997, 1998b; Srinivas et al., 1996; Wang et al., 1997b; Hsieh et al., 1998; You et al., 1999b). Phenotypically, it induces hepatic steatosis (Moriya et al., 1997), and HCC has been observed in core protein-expressing transgenic mice (Moriya et al., 1998).
Many of the descriptive findings for HCV core protein have been poorly explained at the mechanistic level. To address the functional interactions, we searched for cellular cofactors/effectors of the core protein. Here, we report on the identification of the human bZIP transcription factor LZIP as a core protein-binding and core protein-transforming cofactor. Among extant proteins in databases, human LZIP is highly homologous to the Drosophila BBF2/dCREB-A protein, which has been shown to activate transcription from Drosophila fat body- and mammalian liver-specific enhancer elements (Abel et al., 1992; Smolik et al., 1992). We demonstrate that HCV core protein expression results in a loss of LZIP function, leading in NIH 3T3 cells, to focus formation and anchorage-independent growth. We propose that LZIP defines a new tumor suppression pathway that is potentially relevant to HCV-related HCC.
Results
Identification of LZIP as a core-binding protein
Expression of HCV core protein in cultured mammalian cells affects the expression of several cellular genes (Kim et al., 1994; Ray et al., 1995, 1997, 1998b; Srinivas et al., 1996; Wang et al., 1997b; Hsieh et al., 1998). Because there is no current evidence that HCV core protein interacts directly with RNA polymerase II promoters, its effect on gene expression probably comes indirectly from protein–protein interaction with cofactors. To address this issue, we searched for HCV core-binding proteins in a human cDNA library using yeast two-hybrid analysis.
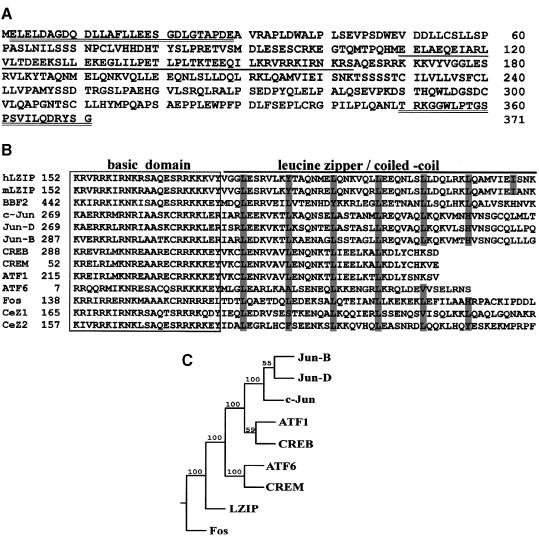
The full-length 191 amino acid core protein from a genotype II/1b HCV isolate (Wang et al., 1997b) was used as bait for screening a human liver cDNA library. From 1.2 × 108 gross transformants, several positive clones were selected for their ability to reconstitute a functional Gal4 transcription complex as measured by induction of a His3p marker and a β–galactosidase reporter. Two overlapping clones described a single, albeit incomplete, open reading frame (ORF). This ORF was repaired to completion by addition to its 5′ end of a 532 bp segment obtained from the IMAGE Consortium (Lawrence Livermore National Laboratory; cDNA clones 38454 and 30070). Thus a 1829 bp cDNA encoding 371 uninterrupted amino acids (Figure 1A) and containing a characteristic bZIP domain was obtained (DDBJ/EMBL/GenBank accession No. U59629). This sequence was reasoned to be complete based on the observation that an in-frame termination codon existed immediately upstream of the putative initiating ATG.
Fig. 1. LZIP represents a new subfamily of bZIP proteins. (A) Amino acid sequence of LZIP. The glutamic acid-rich activation domain is underlined. The sequence of the N– and C–terminal synthetic peptides used to raise antisera in rabbit is doubly underlined. (B) Multiple alignment of bZIP sequences. Periodically occurring leucine or other compatible residues are highlighted by shading. GenBank CDS translation identification numbers (gi) or SWISS-PROT Protein Data Bank accession numbers (sp) of the aligned sequences are: c-Jun, sp/P05412; Jun-D, sp/P17535; Jun-B, sp/P17275; mLZIP, gi/405526; CREB, sp/P16220; CREM, gi/532056; ATF1, sp/P18846; ATF6, sp/P18850; BBF2, sp/P29747; Fos, sp/P01100; CeZ1, gi/1943850; CeZ2, gi/726428. The alignment was generated by the PILEUP program in the Wisconsin package of sequence analysis software (version 8.1, Genetics Computer Group). (C) Consensus phylogenetic tree relating LZIP to other human bZIP proteins. Phylogenies were inferred from protein sequences using parsimony. The parsimony tree was constructed by the PROTPARS program in the PHYLIP package (version 3.5c, J.Felsenstein, University of Washington). SEQBOOT and CONSENSUS programs in the same package were used to perform bootstrap replication and to produce the majority rule consensus tree from 100 replicates. Numbers on the nodes are bootstrap confidence probabilities (%). LZIP does not cluster with Jun, Fos, ATF1, ATF6, CREB or CREM, but forms a separate grouping in this tree.
At the time of initial identification, this core-binding protein had no counterparts in extant databases. Subsequently, several orthologs including mouse LZIP-2/CREB3 (Burbelo et al., 1994; Burbelo and Kozak, 1998) and Drosophila BBF2/dCREB-A (Abel et al., 1992; Smolik et al., 1992) were noted. For simplicity, hereafter we will call this protein human LZIP. The subportion of LZIP that contains its bZIP domain, not surprisingly, is similar to counterparts in other bZIP proteins (e.g. Jun, Fos, CREB, CREM, ATF1, ATF6, Caenorhabditis elegans CeZ1 and CeZ2; Figure 1B). However, the leucine zipper/coiled-coil region of LZIP is particularly long and extends for seven instead of four, five or six heptad repeats. A phylogenetic tree generated by parsimony analysis of protein sequences segregates LZIP into a novel subfamily of bZIP factors distinct from CREB, ATF1, Jun or Fos (Figure 1C).
To characterize LZIP, we raised rabbit antiserum α-ZC to its C–terminal peptide, amino acids 350–371 (Figure 1A). Next, the tissue distribution of LZIP mRNA and protein was assessed using Northern and Western blot analyses. Expression of LZIP is conserved in all human tissues and cell lines tested, including normal liver, hepatoma cell line HepG2, and both the non-tumor and tumor parts of the liver tissue from seven HCV-positive patients (data not shown).
LZIP activates CRE-dependent transcription
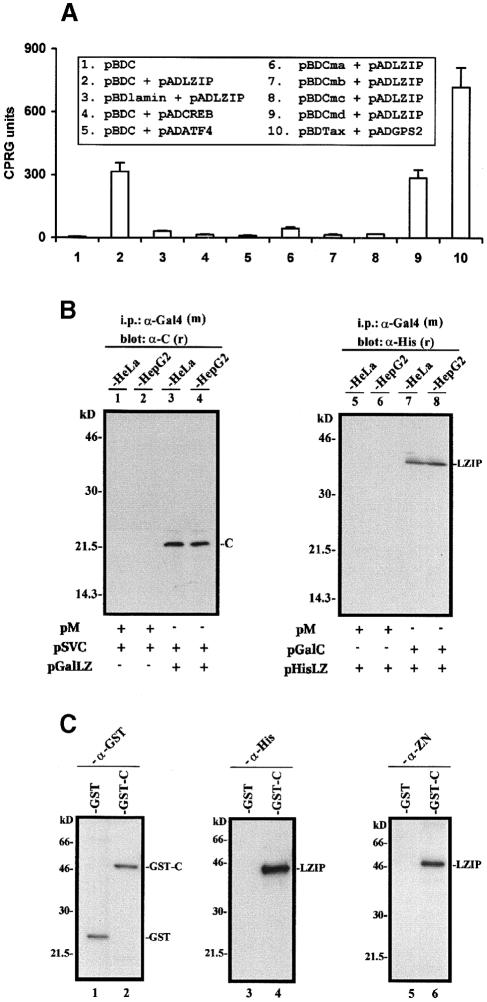
As LZIP contains a bZIP domain, this suggests that it might be a transcription factor. If so, the transcriptional activities previously attributed to HCV core protein (Kim et al., 1994; Ray et al., 1995, 1997, 1998b; Srinivas et al., 1996; Wang et al., 1997b) could be explained through its interaction with LZIP. As a prelude to analyzing transcriptional function, we verified core–LZIP interaction in several ways. First, binding was assessed within yeast cells (Figure 2A). Background activity was seen when Gal4 DNA-binding domain (Gal4BD)–core protein was expressed alone (pBDC; Figure 2A, column 1), or when Gal4 activation domain (Gal4AD)–LZIP and Gal4BD–lamin (pBDlamin + pADLZIP; column 3) were co-expressed. By contrast, co-expression of core (pBDC) with LZIP (pADLZIP) strongly activated the β–galactosidase reporter (Figure 2A, column 2). The HCV core interacts specifically with LZIP and not generally with other bZIP proteins since it did not bind either CREB (Figure 2A, column 4), ATF4 (column 5), c-Jun or c-Fos (data not shown). The C–terminal portion of the core (amino acids 91–191) is sufficient to bind LZIP (Figure 2A, column 9; core mutant pBDCmd), whereas three truncated mutants containing amino acids 1–123 (pBDCma), 1–100 (pBDCmb) or 1–75 (pBDCmc) had marginal or no LZIP-binding ability (columns 6–8).
Fig. 2. Interaction of LZIP with HCV core protein. (A) Yeast two-hybrid assay. Co-expression of pADLZIP and pBDC conferred an ∼80–fold increase in β–galactosidase expression (compare columns 2 and 1). A previously reported (Jin et al., 1997b) interactive pair (pBDTax + pADGPS2) is shown as a positive control (column 10). Core protein (pBDC) interacts specifically with LZIP, since neither CREB, ATF4, c-Jun nor c-Fos could bind the core protein (columns 4 and 5, and data not shown). The LZIP-binding domain in the core protein was mapped to amino acids 91–191 (column 9; core protein mutant pBDCmd). That LZIP does not bind the N–terminus of core protein was shown with pBDCma (column 6), pBDCmb (column 7) and pBDCmc (column 8), which contain, respectively, amino acids 1–123, 1–100 and 1–75 of core protein. Truncated naturally occurring versions of core protein similar to pBDCma have been described elsewhere (Lo et al., 1995; Suzuki et al., 1995). Relative β–galact– osidase activity was expressed in CPRG units (Jin and Jeang, 1997). (B) Co-immunoprecipitation of LZIP and core protein from transfected cells. HeLa and HepG2 cells were transfected individually with the plasmids indicated (5 μg). Immunoprecipitation was performed with cleared cell lysates using a mouse monoclonal anti-Gal4BD antibody [α-Gal4 (m)]. Blots were probed with rabbit polyclonal anti-core [α-C (r); lanes 1–4] or anti-His tag [α-His (r); lanes 5–8]. Bands representing core (C) or His-tagged LZIP (LZIP) co-precipitated by α-Gal4 are indicated. pM is an expression vector for Gal4BD. (C) LZIP binds to immobilized GST–C but not to GST. His-tagged LZIP (5 μg) was loaded onto either GST- or GST–C–bound glutathione–Sepharose (Pharmacia). The resins were washed extensively. Proteins bound to the resins were solubilized in SDS gel loading buffer, resolved by 10% SDS–PAGE, transferred to membrane and probed with anti-GST (lanes 1 and 2; α-GST), anti-His tag (lanes 3 and 4; α-His) or anti-LZIP (lanes 5 and 6; α-ZN). Lanes 1 and 2 show the immobilized GST and GST–C proteins in the resins. His-tagged LZIP was retained in the GST–C resin (lanes 4 and 6) and not in the GST resin (lanes 3 and 5). We verified that ∼1 μg of the His-tagged LZIP loaded onto the GST–C resin (∼20% of the input) bound to GST–C.
Next, the interaction between LZIP and core protein was confirmed by co-immunoprecipitation of either native core protein with Gal4-tagged LZIP (Figure 2B, lanes 3 and 4) or Gal4-tagged core protein with His-tagged LZIP (lanes 7 and 8) from transfected human cells, HeLa and HepG2. Finally, direct binding between purified core and purified LZIP was documented by protein affinity chromatography. In this setting, we observed that His-tagged LZIP was retained by GST–core (GST–C; Figure 2C, lanes 4 and 6), but not by GST alone (lanes 3 and 5) or by GST–Tax (data not shown).
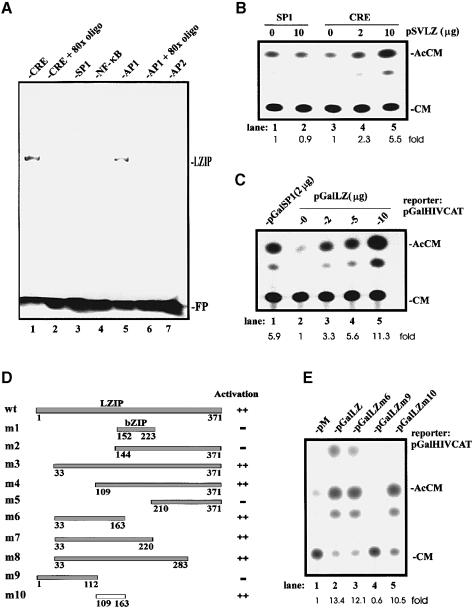
We asked further whether LZIP has a specific transcriptional function. We overexpressed LZIP in Escherichia coli and purified the protein. LZIP's DNA-binding activity was assessed by gel mobility shift analysis. Consistent with its relatedness to the CRE-binding proteins, mouse LZIP-2 (Burbelo et al., 1994) and Drosophila BBF2/dCREB-A (Abel et al., 1992; Smolik et al., 1992), human LZIP bound a consensus CRE (Figure 3A, lane 1) and a related AP1-responsive (lane 5) motif, but it did not bind SP1- (lane 3), NF-κB- (lane 4) or AP2-responsive (lane 7) sequences. An 80-fold excess of either unlabeled CRE (lane 2) or AP1 (lane 6) oligonucleotides effectively eradicated LZIP-specific binding.
Fig. 3. LZIP is a sequence-specific transcription factor. (A) DNA-binding activity of LZIP by gel mobility shift assay. Each reaction contains ∼0.1 μg of purified His-tagged LZIP. The migration position of the free probe (FP) is indicated. An 80-fold excess of unlabeled CRE (lane 2) or AP1 (lane 6) oligonucleotides was used to compete for binding. In parallel assays (data not shown), mutant LZIPm2 (see below) was unable to bind either CRE or AP1. (B) LZIP activates CRE-dependent transcription. HepG2 cells were transiently transfected with reporter plasmid pSP1CAT (2 μg; lanes 1 and 2) or pCRECAT (2 μg; lanes 3–5) and the amounts indicated of pSVLZ. Relative CAT activity was quantitated. Migration positions of acetylated chloramphenicol (AcCM) and unacetylated chloramphenicol (CM) are indicated. Results are representative of triplicate experiments. Parallel experiments in HeLa cells yielded similar results (data not shown). (C) LZIP activates transcription when targeted to a minimal promoter. HepG2 cells were transiently transfected with a reporter plasmid pGalHIVCAT (5 μg) and the amounts indicated of plasmid pGalSP1 (lane 1) or pGalLZ (lanes 2–5). The results represent three independent experiments. (D) Mapping of the trans-activation domain in LZIP by yeast one-hybrid assay. Experiments similar to those described in (C) were performed in budding yeast. Plasmids encoding the indicated LZIP proteins fused to Gal4BD (wt: Gal4BD–LZIP wild-type; m1–m10: Gal4BD–LZIP mutants) were expressed in yeast strain SFY526 (Clontech). The transactivation activity was scored by relative β–galactosidase units (++ >100; – <10). (E) Definition of the LZIP activation domain in mammalian cells. Experiments similar to those described in (C) and (D) were performed in COS7 cells. A 10 μg aliquot of reporter plasmid pGalHIVCAT and 5 μg of the indicated Gal4BD plasmid were used for each transfection. Similar results were also obtained in HeLa and HepG2 cells (data not shown). Plasmid pM expresses Gal4BD alone (lane 1). Plasmids pGalLZ, pGalLZm6, pGalLZm9 and pGalLZm10 produce Gal4BD–LZIP wild-type (lane 2), Gal4BD–LZIPm6 (lane 3), Gal4BD–LZIPm9 (lane 4) and Gal4BD–LZIPm10 (lane 5), respectively.
To characterize its transcriptional activity in mammalian HepG2 cells, we analyzed the effect of an LZIP-expressing plasmid (pSVLZ) on a CRE-dependent chloramphenicol acetyltransferase (CAT) reporter. Figure 3B shows that while LZIP activated CRE-dependent expression in a dose-dependent manner (lanes 3–5), it did not influence SP1-dependent activity (lanes 1 and 2). Consistent with that result, when LZIP was expressed as a Gal4BD–LZIP fusion protein, it activated a Gal4-binding site-dependent promoter with a potency comparable with that of a well-defined transcriptionally active Gal4BD–SP1 fusion (Figure 3C, compare lane 3 with lane 1).
A discrete activation domain in LZIP was mapped to amino acids 109–163 (m10; Figure 3D). This conclusion emerged from the functional profiles revealed by 10 mutant LZIP plasmids in yeast one-hybrid assays (Figure 3D). The validity of these yeast readouts for mammalian cells was verified by transfecting Gal4BD–LZIP expression plasmids into COS7 cells. Thus, in agreement with the yeast results, the 109–163-containing Gal4BD–LZIPm10 (pGalLZm10; Figure 3E, lane 5) protein activated transcription from a Gal4-binding site-fused promoter (pGalHIVCAT). Notably, this LZIP activation domain (Figure 1A) is rich in glutamic acid residues (∼20%).
Mapping of dimerization and core-binding domains in LZIP
bZIP proteins homo- and heterodimerize through their leucine zipper/coiled-coil domains. Selective dimerization is one determinant for diversity and specificity of bZIP protein functions (Ziff, 1990; Vinson et al., 1993; Motohashi et al., 1997). Based on its structural domains, we expected that LZIP would be functionally active as a homodimer. Considered in this way, one could imagine two possible consequences of LZIP's interaction with core protein: the core protein could bind to an active dimerized LZIP or it could abolish activity by preventing LZIP dimerization. Using a modified interactive assay in yeast (Figure 4), we explored LZIP homodimerization and how this might be affected by the core.
Fig. 4. Mapping of the dimerization and core-binding domains in LZIP. A TRP1-marked plasmid expressing Gal4BD–LZIPm2 (amino acids 144–371) was targeted via the Gal1 upstream activating sequence containing Gal4-binding sites to a Gal1 minimal promoter in stably transformed yeast strain Y187 (Clontech). Gal4AD–LZIP (column 1), Gal4AD–CREB (column 3), Gal4AD–ATF4 (column 4), Gal4AD–core (column 5) and Gal4AD–LZIP + core protein (column 6) were then expressed individually from stably transformed LEU2- and/or HIS1-marked vectors.
Various combinations of native or Gal4BD–Gal4AD fusion proteins were targeted in yeast via upstream Gal4-binding sites to a minimal promoter–reporter. We observed that Gal4BD–LZIPm2 (amino acids 144–371) with an intact bZIP region, but deleted for its activation domain, induced LacZ reporter expression poorly (Figure 4, column 2). However, when Gal4AD–LZIP, which otherwise was inactive alone (Figure 2A, column 3), was co-expressed with Gal4BD–LZIPm2, a >100-fold increase in activity was observed (Figure 4, column 1). This indicated that LZIPm2 forms a stable complex with full-length LZIP, a finding consistent with homodimerization through the bZIP domain. This self-interaction is the preferred interaction since neither Gal4AD–CREB (Figure 4, column 3) nor Gal4AD–ATF4 (column 4) showed binding to Gal4BD–LZIPm2. A further verification of this was obtained when we screened a HeLa cDNA library using the yeast two-hybrid assay for potential LZIP partners with Gal4BD–LZIPm2 as bait. This screen yielded only Gal4AD–LZIP (data not shown). Taken together, these results suggest that the observed biological findings for LZIPm2 probably occur through its dimerization with LZIP and are unlikely to occur due to a general interaction of LZIPm2 with other bZIP proteins.
To address the role of core protein in LZIP homodimerization, we expressed Gal4AD–core protein and Gal4BD–LZIPm2 together. This co-expression resulted in substantial (>30–fold) activation of reporter activity (Figure 4, column 5), indicating that the same bZIP sequence in LZIP is shared for core protein binding and homodimerization. We next assessed whether core protein would influence LZIP homodimerization. When native core protein was expressed simultaneously with Gal4BD–LZIPm2 and Gal4AD–LZIP, a significant reduction of LZIP dimerization-dependent activity (>8-fold) was observed (Figure 4, compare columns 1 and 6). One interpretation is that contact of one bZIP domain by core protein can prevent homodimerization of LZIP. However, when purified recombinant GST–core protein and His-tagged LZIP were added simultaneously to a gel mobility shift reaction containing the CRE oligonucleotides (see Figure 3A for a similar assay), the CRE-binding activity of LZIP was not reduced significantly (data not shown). We also noted that LZIPm2 binds more potently to wild-type LZIP than to core protein (Figure 4, compare columns 1 and 5). Taken together, these findings suggest that core protein may inhibit formation of active LZIP complex in the nucleus through mechanisms other than displacement of LZIP dimers.
HCV core sequesters LZIP in the cytoplasm
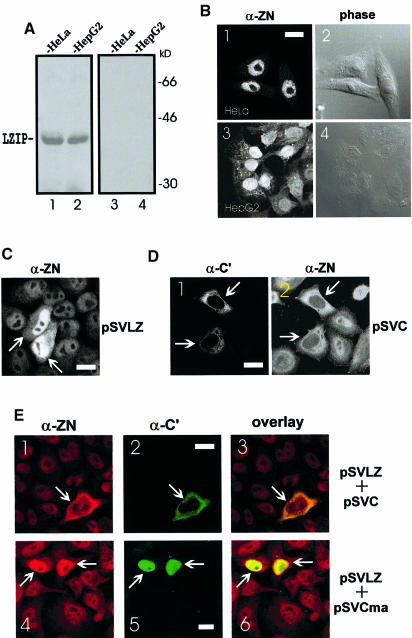
Because the subcellular location of proteins presents an important functional clue, we sought to visualize LZIP and HCV core immunologically in human cells. The specificity of our polyclonal anti-LZIP serum (α-ZN raised to N–terminal amino acids 2–29; Figure 1A) was verified first by Western blotting, carried out in either the absence or presence of a competitive excess of immunizing peptide (Figure 5A). The reactivity of a second monoclonal anti-core antibody has been detailed elsewhere (Cerino et al., 1993; Santolini et al., 1994). Using these specific reagents, endogenous LZIP in HepG2 and HeLa cells was found to be predominantly nuclear (Figure 5B). Similarly, over– expressed LZIP from an exogenously introduced SV40 promoter-driven plasmid (pSVLZ) was also observed in the nucleus (Figure 5C).
Fig. 5. Subcellular localization of LZIP and HCV core protein. (A) Specificity of anti-LZIP antibody α-ZN. HeLa (lanes 1 and 3) and HepG2 (lanes 2 and 4) cell extracts containing equal amounts (20 μg) of protein were separated by 10% SDS–PAGE. Immunoblotting was performed separately with α–ZN (lanes 1 and 2) and α–ZN pre-incubated with 6 μg of immunizing peptide (lanes 3 and 4). (B) Nuclear localization of endogenous LZIP protein in serum-stimulated cells. HeLa (panels 1 and 2) and HepG2 (panels 3 and 4) cells were fixed and monitored by staining with α-ZN (panels 1 and 3) and by phase contrast microscopy (panels 2 and 4). The same fields are shown in panels 1 and 2 and panels 3 and 4. Bar = 20 μm. (C) Nuclear localization of exogenously expressed LZIP in HeLa cells. Cells were transfected with pSVLZ and stained with α-ZN. The arrows indicate transfected cells. Bar = 20 μm. (D) Localization of core and LZIP in core protein-expressing HeLa cells. Cells were transfected with pSVC alone and co-stained with anti-core protein (α-C′; panel 1) and anti-LZIP (α-ZN; panel 2) antibodies. The arrows indicate transfected cells. The same fields are shown in panels 1 and 2. Bar = 20 μm. Parallel experiments in pBDC–transfected NIH 3T3, HepG2 and Chang liver cells yielded similar results (data not shown). (E) Localization of core protein and LZIP in HeLa cells co-expressing core protein and LZIP. Cells were transfected with either pSVLZ plus pSVC (panels 1–3) or pSVLZ plus pSVCma (panels 4–6) and stained simultaneously with rabbit antibody α-ZN (panels 1 and 4) and mouse/human antibodies α-C′ (panels 2 and 5). Red (LZIP) and green (core) fluorescent signals were overlaid by computer assistance (panels 3 and 6). Co-localizations are shown in yellow. The same fields are shown in panels 1–3 and 4–6. The arrows indicate transfected cells. The nuclear localization of the core mutant containing amino acids 1–123 (pSVCma) has been documented elsewhere (Ravaggi et al., 1994; Lo et al., 1995; Liu et al., 1997; Yasui et al., 1998; Sabile et al., 1999). Bar = 20 μm. All examples shown are representative of at least three independent experiments.
We next asked if and how LZIP might be affected by HCV core protein. To address this, HeLa cells were transfected with an LZIP-expressing plasmid (pSVLZ) and a core protein-expressing plasmid (pSVC) either singly or together. Consistent with previous reports (Cerino et al., 1993; Selby et al., 1993; Santolini et al., 1994; Lo et al., 1995; Moradpour et al., 1996; Barba et al., 1997; Chen et al., 1997; Kawamura et al., 1997; Matsumoto et al., 1997; Yasui et al., 1998; Zhu et al., 1998; Sabile et al., 1999), expression of core protein from transfected pSVC was found in the cytoplasm (Figure 5D, panel 1). Interestingly, in marked distinction to untransfected cells (Figure 5B, panels 1 and 3; D, panel 2, cells without arrow) or cells overexpressing LZIP (Figure 5C), the core protein-expressing cells (Figure 5D, panel 1, cells with arrow) showed a dramatic relocalization of LZIP from its normal nuclear location into the cytoplasm (panel 2, cells with arrow). We also examined pSVC and pSVLZ co-transfected cells. Because DNAs co-precipitated using calcium phosphate are taken up together into cells, co-transfection of pSVC and pSVLZ showed that >95% of core-expressing cells (Figure 5E, panel 2) co-stained for LZIP (panel 1). LZIP was found in the nucleus of HeLa cells co-expressing LZIP and a core protein mutant (amino acid 1–123; pSVCma) defective for LZIP binding (Figure 5E, panel 4, cells with arrow). In sharp contrast, LZIP redistributed to the cytoplasm in cells co-expressing core protein and LZIP (Figure 5E, panel 1, cell with arrow). Digital measurements of fluorescent images showed that >80% of the LZIP signal (re)-localized with the core protein (Figure 5E, panel 3). Thus one simple interpretation is that HCV core protein sequesters in the cytoplasm LZIP that would otherwise locate into the nucleus.
HCV core protein inhibits LZIP-dependent transcription in mammalian cells
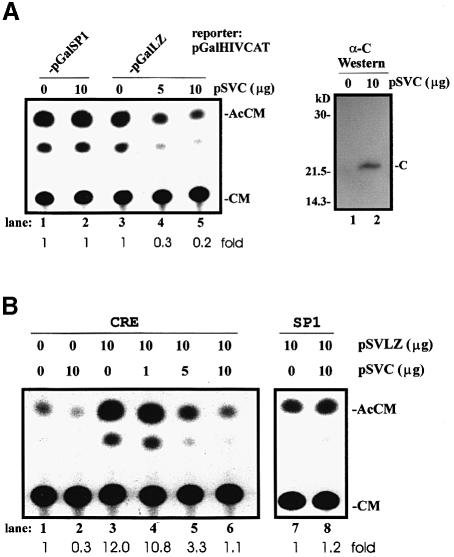
Above, we showed LZIP to be a nuclear CRE-activating factor (Figure 3). The finding that core protein can prevent formation of LZIP dimers in the nucleus (Figure 4) and can sequester LZIP in the cytoplasm (Figure 5) predicts a repressive effect of the former on the transcriptional activity of the latter. To explore this, we co-expressed core protein and Gal4BD–LZIP in HepG2 cells and assessed the effects on reporter expression (Figure 6A). Indeed, Gal4BD–LZIP-dependent transcription decreased progressively when expression of core protein increased (Figure 6A, lanes 3–5). By contrast, Gal4BD–SP1dependent expression was totally unaffected by core protein (Figure 6A, compare lanes 1 and 2). Next, various combinations of core protein-expressing plasmid (pSVC) and/or LZIP-expressing plasmid (pSVLZ) were assayed with either CRE- or SP1-dependent reporters to characterize the effects further. Again, we observed that increased expression of core protein in HepG2 cells reproducibly repressed cell-endogenous (Figure 6B, lanes 1 and 2) and LZIP-stimulated (lanes 3–6) CRE-dependent activity, while cell-endogenous SP1-dependent transcription was not affected (lanes 7 and 8). An interpretation of this result is that core protein physically binds endogenous LZIP in HepG2 cells in a manner similar to its interaction with exogenously overexpressed LZIP (Figure 2).
Fig. 6. HCV core protein inhibits LZIP-dependent transcription. (A) Effects of core expression on Gal4BD–LZIP transactivation. HepG2 cells were transiently transfected with pGalHIVCAT (5 μg) plus pGalSP1 (8 μg; lanes 1 and 2) or pGalLZ (8 μg; lanes 3–5) plus the amounts indicated of pSVC. The results are representative of triplicate experiments. Western blotting was performed to verify the overexpression of HCV core protein (right panel). (B) Effects of core protein expression on activation of CRE by LZIP. HepG2 cells were transiently transfected with pCRECAT (4 μg; lanes 1–6) or pSP1CAT (4 μg; lanes 7 and 8), and the amounts indicated of pSVLZ and pSVC. The results are representative of triplicate experiments.
Loss of LZIP function correlates with cellular transformation
Several viral bZIP proteins such as v-Jun, v-Fos and v-Maf serve transforming functions for retroviruses (Maki et al., 1987; Nishizawa et al., 1987; Motohashi et al., 1997). Cellular bZIPs including avian C/EBP-β/NF-M have also been implicated in limited aspects of leukemogenesis (Katz et al., 1993). Furthermore, chromosomal translocations in acute leukemia and solid tumors have been found to confer transforming roles to various bZIP proteins (Cleary, 1991; Look, 1997). Because HCV is closely associated with HCC (Saito et al., 1990; Bukh et al., 1993; Di Bisceglie, 1995; Wang and Jin, 1997; Idilman et al., 1998) and because the oncogenic mechanism(s) for this virus remains incompletely defined, we wondered whether the HCV core–LZIP interaction might explain the alterations in cell growth control.
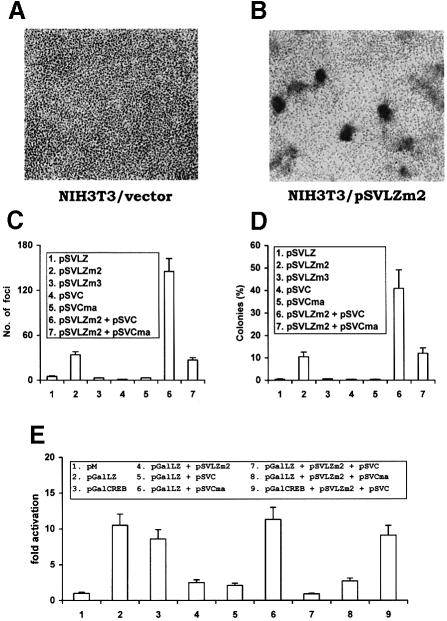
We explored this hypothesis by assessing the growth phenotypes of wild-type or mutant LZIP-expressing NIH 3T3 cells. NIH 3T3 cells were selected separately for stable overexpression of wild-type LZIP, of a transcriptionally competent LZIPm3 mutant or of a transcriptionally incompetent LZIPm2 mutant (Figure 3D). Several independently selected cells were then assayed for focus-forming and anchorage-independent growth. On visual inspection, we noted that cells that expressed activation-incompetent mutant LZIPm2 (Figure 3D) differed morphologically from parental NIH 3T3 cells and from cells that overexpressed either wild-type LZIP or LZIPm3 (data not shown). When considered for growth characteristics, LZIPm2-expressing cells exhibited loss of contact inhibition, which manifested as dense foci of cells. This finding was not observed for either parental NIH 3T3 cells (compare Figures 7A and B) or cells that overexpressed LZIP wild-type or LZIPm3 (Figure 7C), which were cultured in parallel. When compared further with parental NIH 3T3 cells, LZIP wild-type or LZIPm3 cells, LZIPm2 cells were also found to grow easily in soft agar in an anchorage-independent manner (Figure 7D).
Fig. 7. HCV core protein enhances the transforming potential of LZIPm2. NIH 3T3 cells were stably transfected with either empty vector (A) or SV40 promoter-driven plasmid pSVLZm2 expressing LZIPm2 (B). Transfectants were selected for neo expression with G418/geneticin (800 μg/ml). Drug-selected cells were seeded to flasks, and the morphology of the transfected cells was photographed at the same magnification after 12 days. (C) Focus formation in monolayer NIH 3T3 cells. Representative clones of NIH 3T3 cells stably transfected with empty vector, pSVLZ, pSVLZm2, pSVLZm3, pSVC, pSVCma, pSVLZm2 + pSVC or pSVCm2 + pSVCma were grown in monolayer. The numbers of foci were counted 12 days after seeding as in (A) and (B). Expression of LZIP and core protein from exogenously introduced plasmids was confirmed by Western blotting (data not shown). The results represent the average of four dishes of the same clone. Three independent clones were tested and similar results were obtained. (D) Colony formation in soft agar. Representative clones of stably transfected NIH 3T3 cells (1 × 104) were suspended in 0.33% top agar/growth medium and overlaid onto a 0.5% agar/growth medium bottom layer. Colonies were scored after 12 days. The results represent the average of three dishes of the same clone. Three independent clones were tested and similar results were obtained. (E) HCV core enhances the inhibitory effect of LZIPm2 on LZIP-dependent transcription. HeLa cells were transfected with the plasmids indicated (8 μg) plus pGalHIVCAT (5 μg). Relative CAT activity was quantitated and is presented as fold activation.
For these cell growth phenotypes, we noted that this aspect of LZIP function correlated inversely with its transcriptional capacity (Figure 3D). Thus, cells with exogenously added wild-type LZIP or its transcriptionally active mutant, LZIPm3, were unperturbed in normal growth control. By contrast, overexpression of a loss-of-function LZIPm2 mutant produced a new and distinctive proliferative phenotype. These findings suggest that LZIPm2 exerts a transdominant negative effect on endogenous LZIP and that it is this loss of LZIP function that accounts for dysregulated cell growth. In support of this interpretation, it was demonstrated directly that LZIPm2 is a functional transdominant negative form of LZIP. Thus, LZIP-dependent activation was repressed by trans-expression of LZIPm2 (Figure 7E, compare columns 2 and 4).
How would dysregulated cellular proliferation occurring due to a loss of LZIP function be relevant to core protein? We noted that LZIP transcription function was suppressed by both expression of HCV core protein (Figures 6 and 7E, column 5) and co-expression of core protein with LZIPm2 (Figure 7E, column 7). Thus, if loss of LZIP function leads to a transformed cellular phenotype, then it stands to reason that core protein overexpression should increase this effect of LZIPm2. In exploring this, we observed that core protein synergized with LZIPm2 to increase significantly (above that seen with LZIPm2 alone) the number of anchorage-independent NIH 3T3 foci in soft agar (Figure 7C, column 6). Moreover, a C–terminal truncated core mutant containing amino acids 1–123 (pSVCma), which is defective for physical interaction with LZIP (Figure 2A, column 6) and functional suppression of LZIP activity (Figure 7E, compare columns 5 and 2, and columns 8 and 4), did not synergize with LZIPm2 in the transformation assays (Figures 7C and D, compare columns 7 and 2). These results support the hypothesis that LZIP is targeted by core protein in the transforming synergy of core protein with LZIPm2. Curiously, in these assays, simple overexpression of core protein alone was insufficiently transforming (Figure 7C and D, column 4). This is probably due to a failure to achieve sufficient amounts and/or duration of expression, since others have shown that core protein alone can transform NIH 3T3 cells (Ray et al., 1996a). Another possibility is that inhibition of LZIP-dependent transcription is required but not sufficient for transformation. In this scenario, LZIPm2 may act through as yet unknown mechanisms (e.g. induction of a conformational change in wild-type LZIP) to trigger a transforming process that can be potentiated by core protein. LZIPm2 and core protein may represent different steps in a multistage process leading to transformation. HCV-induced hepatocarcinogenesis is complicated and poorly understood. Mice transgenic for HCV core protein did not show inflammatory or neoplastic changes at the age of 3–12 months, but developed HCC after 16 months (Moriya et al., 1998). The development of HCC in human patients is also a chronic process that takes 10–20 years (Di Bisceglie, 1995; Idilman et al., 1998). Presumably, several discrete events must occur within the infected cell prior to fully developed transformation. The dysregulation of LZIP by core protein may represent only one aspect of this pathological course.
Discussion
HCV is an RNA virus that causes hepatitis, cirrhosis, liver failure and HCC (Purcell, 1994; Houghton, 1996). Development of HCC occurs in 10% of HCV-infected individuals (Di Bisceglie, 1995; Idilman et al., 1998). Globally, the seroprevalence of HCV is >100 million (Alter et al., 1995; Idilman et al., 1998); hence, this implies that >10 million individuals are at risk for HCV-associated HCC. The magnitude of this potential cancer burden presents an impetus to understand the transforming mechanism(s) of this virus. Although multiple factors probably contribute to HCV-associated HCCs, currently, the most compelling viral candidate oncoprotein is HCV core protein (Ray et al., 1996a; Chang et al., 1998; Moriya et al., 1998).
There is abundant descriptive evidence that links core protein to cellular transformation (Ray et al., 1996a; Chang et al., 1998; Tsuchihara et al., 1999). Despite the plethora of reports, mechanistic details as to how core protein effects the requisite cell growth dysregulation are, lamentably, sketchy. Ideally, in order to understand processes of viral transformation, it is important to identify cellular effectors of viral oncoproteins and to characterize the functions of these cofactors. Here, for HCV core protein, a cellular bZIP protein, LZIP (Figure 1), was identified as a direct core-binding protein (Figure 2) and a cofactor in cellular transformation (Figure 7). While other as yet uncharacterized activities cannot be excluded, LZIP is shown to be a transcriptional factor in a cAMP-potentiated pathway (Figure 3). In the context of HCV core protein, LZIP presents activities consistent with that of a cellular tumor suppressor (Figure 7).
Several aspects of core protein–LZIP interaction are informative. LZIP is a nuclear CRE-activating factor whose transcriptional activity is repressed by core protein (Figures 6 and 7). Specific interaction with HCV core protein (Figure 2) influences LZIP in several ways. First, core protein prevents formation of active LZIP homodimers in the nucleus (Figure 4). Secondly, a substantial amount of nuclear LZIP was observed to relocate into the cytoplasm in core protein-expressing cells (Figure 5). Finally, the proliferative activity of a transdominant negative form of LZIP (LZIPm2) was potentiated by core protein (Figure 7). Taken together, these findings establish that core protein inactivates LZIP function through subcellular sequestration, and that a loss of endogenous LZIP function correlates with abnormal cellular proliferation. In the absence of a reliable tissue culture system for HCV, it remains to be determined whether the expression of other HCV proteins could affect the ability of core protein to modulate LZIP function. In this regard, we note that core protein was reported to interact with E1 protein (Lo et al., 1996).
Other evidence compatible with the role of LZIP in virus-induced cell proliferation was noted after the completion of our study. Recently, we noticed that LZIP is closely related to, if not an isoform of, another human protein, luman (Freiman and Herr, 1997; Lu et al., 1997, 1998; J.Vogel and T.Kristie, personal communication). Whereas LZIP is targeted directly by HCV core protein, luman is targeted indirectly by herpes simplex virus through its VP16-associated cell proliferation factor, HCF. HCF is evolutionarily conserved from C.elegans to human (Kristie, 1997) and has been clearly shown to be important in regulating cell proliferation (Goto et al., 1997). The as yet to be established possibility that HCF and LZIP operate through a shared pathway would represent a further example of how divergently different viruses conserve related mechanisms in order to disrupt host cell growth control.
Gain of bZIP functions, including those for Jun and Fos, has been implicated clearly in various cellular transformation events (Gao et al., 1996). An unexpected finding from this study is that the loss of an endogenous bZIP function, as defined by LZIP, is correlated with the proliferative effects of a hepatotropic viral oncoprotein, HCV core. Thus, at least in this restricted perspective, LZIP could be defined operationally as a tumor suppressor. Two of the best characterized tumor suppressors are p53 and Rb. Other emerging tumor suppressors continue to be characterized in mice and humans (McClatchey and Jacks, 1998). To our knowledge, as yet, a tumor suppressor protein in a human cAMP signaling pathway has not been described. Since there is an indisputably large body of evidence linking cAMP signaling to cell proliferation and cell death (e.g. Lanotte et al., 1991; McConkey et al., 1992; Aharoni et al., 1995; Lomo et al., 1995; Ruchaud et al., 1997), the existence of a cAMP-responsive/regulated tumor suppressor should not be unexpected. Whether, beyond the context of HCV core protein, LZIP fulfills tumor suppressor functions remains to be determined by future studies.
Materials and methods
Plasmids
Plasmid pBDC expressing Gal4BD–core protein was constructed by in-frame insertion into plasmid pGBT9 (Clontech) of a cDNA (DDBJ/EMBL/GenBank accession No. U63376) encoding the full-length 191 amino acid core protein (Wang et al., 1997b) from a Chinese HCV isolate HUNAN–AB belonging to genotype II/1b (Yang et al., 1994). pBDCma, pBDCmb, pBDCmc and pBDCmd express truncated Gal4BD–core proteins corresponding to amino acids 1–123, 1–100, 1–75 and 91–191, respectively. pADLZIP was obtained by cloning the complete LZIP coding sequence into Gal4AD plasmid pGADGH (Clontech). pADCREB and pADATF4 contain the full-length cDNA for human CREB (IMAGE clone 758241) and ATF4 (IMAGE clone 46141), respectively. pM (Clontech), pGalLZ, pGalLZm6, pGalLZm9, pGalLZm10, pGalC, pSVC, pSVCma, pSVLZ, pSVLZm2 and pSVLZm3 are SV40 promoter-driven vectors expressing Gal4BD, Gal4BD–LZIP, Gal4BD–LZIPm6, Gal4BD–LZIPm9, Gal4BD–LZIPm10, Gal4BD–core, core protein (amino acids 1–191), core mutant Cma (amino acids 1–123), LZIP, LZIPm2 and LZIPm3, respectively. pHisLZ expressing His-tagged LZIP is based on pEBVHis (Invitrogen). pGSTC and pTrcHisLZ were prokaryotic expression vectors derived from pGEX-4T-3 (Pharmacia) and pTrcHis (Invitrogen). pSP1CAT and pCRECAT are CAT reporter plasmids based on pCAT-basic (Promega) with a minimal human immunodeficiency virus type 1 (HIV-1) promoter (Berkhout and Jeang, 1992). SP1 and CRE motifs in these plasmids were derived from the following synthetic oligonucleotides: SP1, 5′–AGCTTGGGGAGTGGCGGATCCGGGGAGTGGCA–3′ and 5′–AGCTTGCCACTCCCCGGATCCGCCACTCCCCA–3′; CRE, 5′–AGCTTGGTGACGCGGATCCGGTGACGCA–3′ and 5′–AGCTTGCGTCACCGGATCCGCGTCACCA–3′. pGalHIVCAT contains four consensus Gal4-binding sites and an HIV minimal promoter upstream of the CAT gene (Berkhout and Jeang, 1992; Xiao et al., 1997). Plasmid expressing Gal4BD–LZIPm2 was derived from TRP1-marked pAS2-1 (Clontech). Plasmids expressing Gal4AD–CREB, Gal4AD–ATF4, Gal4AD–LZIP and Gal4AD–core protein were derived from LEU2-marked pGAD424 (Clontech) or HIS1-marked pGAD424H. Plasmid pGHnfC expressing the core protein was constructed by inserting a full-length HCV core gene into pGHnf (Jin et al., 1997b). Plasmids pGalSP1, pBDlamin, pBDp53, pADLT (SV40), pBDTax and pADGPS2 have been described elsewhere (Jin and Jeang, 1997; Jin et al., 1997a, 1998; Xiao et al., 1997).
Antibodies
Rabbit polyclonal anti-LZIP antibody α-ZC was raised against a C–terminal peptide (amino acids 350–371; Figure 1A) conjugated to keyhole limpet hemocyanin (KLH). Rabbit polyclonal anti-LZIP antibody α-ZN was raised to a KLH-conjugated peptide corresponding to the N–terminus of LZIP (amino acids 2–29; Figure 1A). Rabbit polyclonal anti-core sera α-C are mixed from antibodies (kindly provided by Drs J.-H.Ou and M.Houghton; Selby et al., 1993; Lo et al., 1995) raised to bacterially produced full-length core protein. Monoclonal anti-core antibodies α-C′ are mixed from one human monoclonal (a gift from Dr M.U.Mondelli; Cerino et al., 1993; Santolini et al., 1994) and two mouse monoclonal antibodies (Chemicon). Rabbit polyclonal anti-His tag (H-15) and mouse monoclonal anti-Gal4BD (clone RK5C1) were from Santa Cruz. Rabbit polyclonal anti-GST was from Pharmacia.
Yeast two-hybrid assay
Yeast two-hybrid assays were based on Gal4BD (1–147 amino acids) and Gal4AD (768–881 amino acids) fusions. Library screening was performed in strain CG-1945 according to the Matchmaker Two-hybrid System Protocol (Clontech). CG-1945 was transformed with Gal4BD plasmid pBDC and then screened with a human liver Matchmaker cDNA library constructed in Gal4AD plasmid pACT2 (Clontech). Positive clones were selected for expression of both His3p and β-galactosidase. Plasmid DNA was recovered from CG-1945 and electroporated into E.coli. Reporter assays were performed as described previously (Jin and Jeang, 1997; Jin et al., 1997a, b, 1998, 1999). Additional assays were also performed in yeast strain SFY526.
Molecular sequence analysis
Analyses of DNA and protein sequence data, including database searching, multiple alignment and phylogenetic reconstruction, were performed as described previously (Jin et al., 1996, 1998; Wang et al., 1997a, c).
Immunoprecipitation
Immunoprecipitation was performed as described (Jin et al., 1997a, b, 1998). HeLa or HepG2 cells harvested into buffer [phosphate-buffered saline (PBS), 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 μg/ml antipain, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aminoethyl– benzene sulfonyl fluoride and 1 μg/ml aprotinin] were disrupted by repeated aspiration through a 21 gauge needle.
Protein affinity chromatography
Escherichia coli-expressed proteins, GST, GST–C and His-tagged LZIP, were purified according to the manufacturers' protocols (Pharmacia, Invitrogen and Qiagen). His-LZIP was eluted from Ni-NTA resin (QiagenN) with 50 mM sodium phosphate, pH 6.0, 300 mM NaCl, 0.25 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 M imidazole and 10% glycerol. Desalted His-LZIP was loaded onto either GST- or GST–C–bound glutathione–Sepharose (Pharmacia). The resins were washed extensively with 10 mM HEPES, pH 7.5, 0.15 M NaCl, 2 mM dithiothreitol (DTT), 0.25 mM PMSF and 10% glycerol. Bound proteins were solubilized in SDS–gel loading buffer (50 mM Tris–HCl, pH 6.8, 100 mM DTT, 2% SDS, 0.001% bromophenol blue and 10% glycerol).
Gel mobility shift assay
Probe labeling and gel mobility shift assays were performed as previously described (Jin and Jeang, 1997; Jin et al., 1997a). Each reaction contains ∼0.1 μg of purified His-LZIP. Oligonucleotides used to produce consensus factor-binding motifs (Swiss-Prot Protein Sequence Data Bank, Geneva, Switzerland) are as follows: CRE, 5′–AGCTTGGTGACGCGGATCCGGTGACGCA–3′ and 5′–AGCTTGCGTCACCGGATCCGCGTCACCA–3′; SP1, 5′–AGCTTGGGGAGTGGCGGATCCGGGGAGTGGCA–3′ and 5′–AGCTTGCCACTCCCCGGATCCGCCACTCCCCA–3′; NF-κB, 5′–AGCTTGGGGAATCTCCGGATCCGGGGAATCTCCA–3′ and 5′–AGCTTGGAGATCCGCGTCACCA–3′; AP-1, 5′–AGCTTGACTCAGGATCCGGTCAATGACTCA–3′ and 5′–AGCTTGAGTCATTGACCGGATCCTGAGTCA–3′; and AP2, 5′–AGCTGTCCCCAGGCTCGGATCCCCAGGCTC–3′ and 5′–AGCTGAGCCTGGGGATCCGAGCCTGGGGAC–3′.
Chloramphenicol acetyltransferase (CAT) assay
CAT assay was performed as described previously (Jin et al., 1997a). Relative radioactivity on thin-layer chromatography plates was quantitated with a Fuji FLA-2000 phosphorimager.
Laser scanning confocal microscopy
Transfected or mock-transfected HepG2 and HeLa cells were grown to 75–95% confluence on 18 mm diameter No.1 glass coverslips in 60 mm diameter tissue culture dishes. Cells were washed three times with PBS and fixed with 4% paraformaldehyde in PBS (pH 7.4) for 15 min at room temperature. The cells were washed three times with PBS, permeabilized by absolute methanol for 2 min at room temperature and rehydrated by three washes with PBS. Diluted antibodies were beaded in 60 μl on tissue culture dishes, and coverslips with fixed cells were inverted onto the antibodies. Antibodies typically were incubated for 1 h at room temperature. Excess antibodies were removed by multiple washes with PBS. Coverslips were mounted on slides with Fluormount (Virotech). Double-label immunofluorescence was performed using a confocal microscope (Zeiss Axiophot) with primary antibodies from different species and species-specific secondary antibodies pre-absorbed for dual labeling: Texas red-conjugated goat anti-rabbit immunoglobulin G (ICN), fluorescein-conjugated donkey anti-mouse immunoglobulin G (Chemicon) and fluorescein-conjugated goat anti-human immunoglobulin G (ICN).
Biological assays for transformation
Focus formation and colony formation assays in soft agar were performed as described previously (Clark et al., 1995; Benkirane et al., 1997).
Acknowledgments
Acknowledgements
We gratefully acknowledge Drs J.-H.Ou, M.Houghton, M.U.Mondelli, J.Vogel, T.Kristie, C.D.Southgate, M.R.Green, J.Bukh and R.H.Purcell for providing reagents. We thank K.-T.Chin and O.G.W.Wong for critical reading of the manuscript, and K.H.Kok for assistance in preparation of figures. This work was supported in part by the State Key Laboratory for Molecular Virology and Genetic Engineering, Beijing, China and by the University of Hong Kong.
References
- Abel T., Bhatt, R. and Maniatis, T. (1992) A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev., 6, 466–480. [DOI] [PubMed] [Google Scholar]
- Aharoni D., Dantes, A., Oren, M. and Amsterdam, A. (1995) cAMP-mediated signals as determinants for apoptosis in primary granulosa cells. Exp. Cell Res., 218, 271–282. [DOI] [PubMed] [Google Scholar]
- Alter M.J. (1995) Epidemiology of hepatitis C in the West. Semin. Liver Dis., 15, 5–14. [DOI] [PubMed] [Google Scholar]
- Barba G., et al. (1997)Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl Acad. Sci. USA, 94, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M., Neuveut, C., Chun, R.F., Smith, S.M., Samuel, C.E., Gatignol, A. and Jeang, K.-T. (1997) Oncogenic potential of TAR RNA-binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J., 16, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B. and Jeang, K.-T. (1992) Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J. Virol., 66, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Miller, R.H., Kew, M.C. and Purcell, R.H. (1993) Hepatitis C virus RNA in southern African blacks with hepatocellular carcinoma. Proc. Natl Acad. Sci. USA, 90, 1848–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Purcell, R.H. and Miller, R.H. (1994) Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl Acad. Sci. USA, 91, 8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D. and Kozak, C.A. (1998) Mapping of the murine LZIP gene (Creb3) to chromosome 4. Genomics, 54, 357–358. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Gabriel, G.C., Kibbey, M.C., Yamada, Y., Kleinman, H.K. and Weeks, B.S. (1994) LZIP-1 and LZIP-2: two novel members of the bZIP family. Gene, 139, 241–245. [DOI] [PubMed] [Google Scholar]
- Cerino A., Boender, P., La Monica, N., Rosa, C., Habets, W. and Mondelli, M.U. (1993) A human monoclonal antibody specific for the N terminus of the hepatitis C virus nucleocapsid protein. J. Immunol., 151, 7005–7015. [PubMed] [Google Scholar]
- Chang J., Yang, S.-H., Cho, Y.-G., Hwang, S.B., Hahn, Y.S. and Sung, Y.C. (1998) Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J. Virol., 72, 3060–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.C., Yen, J.H., Kang, H.Y., Jang, M.H. and Chang, M.F. (1994) Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun., 205, 1284–1290. [DOI] [PubMed] [Google Scholar]
- Chen C.-M., You, L.-R., Hwang, L.-H. and Lee, Y.-H.W. (1997) Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-β receptor modulates the signal pathway of the lymphotoxin-β receptor. J. Virol., 71, 9417–9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G.J., Cox, A.D., Graham, S.M. and Der, C.J. (1995) Biological assays for Ras transformation. Methods Enzymol., 255, 395–412. [DOI] [PubMed] [Google Scholar]
- Clarke B. (1997) Molecular virology of hepatitis C virus. J. Gen. Virol., 78, 2397–2410. [DOI] [PubMed] [Google Scholar]
- Cleary M.L. (1991) Oncogenic conversion of transcription factors by chromosomal translocations. Cell, 66, 619–622. [DOI] [PubMed] [Google Scholar]
- Di Bisceglie A.M. (1995) Hepatitis C and hepatocellular carcinoma. Semin. Liver Dis., 15, 64–69. [DOI] [PubMed] [Google Scholar]
- Freiman R.N. and Herr, W. (1997) Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev., 11, 3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Morgan, I. and Vogt, P.K. (1996) Differential and antagonistic effects of v-Jun and c-Jun. Cancer Res., 56, 4229–4235. [PubMed] [Google Scholar]
- Goto H., Motomura, S., Wilson, A.C., Freiman, R.N., Nakabeppu, Y., Fukushima, K., Fujishima, M., Herr, W. and Nishimoto, T. (1997) A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev., 11, 726–737. [DOI] [PubMed] [Google Scholar]
- Houghton M. (1996) Hepatitis C viruses. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds.), Fields Virology. 3rd edn. Lippincott-Raven, Philadelphia, PA, pp. 1035–1058. [Google Scholar]
- Hsieh T.-Y., Matsumoto, M., Chou, H.-C., Schneider, R., Hwang, S.B., Lee, A.S. and Lai, M.M.C. (1998) Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem., 273, 17651–17659. [DOI] [PubMed] [Google Scholar]
- Hwang S.B., Lo, S.-Y., Ou, J.-H. and Lai, M.M.C. (1995) Detection of cellular proteins and viral core protein interacting with the 5′ untranslated region of hepatitis C virus RNA. J. Biomed. Sci., 2, 227–236. [DOI] [PubMed] [Google Scholar]
- Idilman R., De Maria, N., Colantoni, A. and Van Thiel, D.H. (1998) Pathogenesis of hepatitis B and hepatitis C–induced hepatocellular carcinoma. J. Viral Hepatitis, 5, 285–299. [DOI] [PubMed] [Google Scholar]
- Jin D.-Y. and Jeang, K.-T. (1997) HTLV-I Tax self-association in optimal trans-activation function. Nucleic Acids Res., 25, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.-Y., Lyu, M.S., Kozak, C.A. and Jeang, K.-T. (1996) Function of 14-3-3 proteins. Nature, 382, 308. [DOI] [PubMed] [Google Scholar]
- Jin D.-Y., Chae, H.Z., Rhee, S.G. and Jeang, K.-T. (1997a) Regulatory role for a novel human thioredoxin peroxidase in NF-κB activation. J. Biol. Chem., 272, 30952–30961. [DOI] [PubMed] [Google Scholar]
- Jin D.-Y., Teramoto, H., Giam, C.-Z., Chun, R.F., Gutkind, J.S. and Jeang, K.-T. (1997b) A human suppressor of c-Jun N–terminal kinase 1 activation by tumor necrosis factor α. J. Biol. Chem., 272, 25816–25823. [DOI] [PubMed] [Google Scholar]
- Jin D.-Y., Spencer, F. and Jeang, K.-T. (1998) Human T cell leukemia virus type 1 oncoprotein targets the human mitotic checkpoint protein MAD1. Cell, 93, 81–91. [DOI] [PubMed] [Google Scholar]
- Jin D.-Y., Giordano, V., Kibler, K.V., Nakano, H. and Jeang, K.-T. (1999) Role of adapter function in oncoprotein-mediated activation of NF-κB: human T-cell leukemia virus type I Tax interacts directly with IκB kinase γ. J. Biol. Chem., 274, 17402–17405. [DOI] [PubMed] [Google Scholar]
- Katz S., Kowenz-Leutz, E., Muller, C., Meese, K., Ness, S.A. and Leutz, A. (1993) The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J., 12, 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Furusaka, A., Koziel, M.J., Chung, R.T., Wang, T.C., Schmidt, E.V. and Liang, T.J. (1997) Transgenic expression of hepatitis C virus structural proteins in the mouse. Hepatology, 25, 1014–1021. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Suzuki, R., Harada, T., Saito, I. and Miyamura, T. (1994) Trans-suppression of gene expression by hepatitis C viral core protein. Jap. J. Med. Sci. Biol., 47, 211–220. [DOI] [PubMed] [Google Scholar]
- Kristie T.M. (1997) The mouse homologue of the human transcription factor C1 (host cell factor): conservation of forms and function. J. Biol. Chem., 272, 26749–26755. [DOI] [PubMed] [Google Scholar]
- Lanotte M., Riviere, J.B., Hermouet, S., Houge, G., Vintermyr, O.K., Gjertsen, B.T. and Doskeland, S.O. (1991) Programmed cell death (apoptosis) is induced rapidly and with positive cooperativity by activation of cyclic adenosine monophosphate-kinase I in a myeloid leukemia cell line. J. Cell Physiol., 146, 73–80. [DOI] [PubMed] [Google Scholar]
- Liu Q., Tackney, C., Bhat, R.A., Prince, A.M. and Zhang, P. (1997) Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol., 71, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.-Y., Masiarz, F., Hwang, S.B., Lai, M.M.C. and Ou, J.-H. (1995) Differential subcellular localization of hepatitis C virus core gene products. Virology, 213, 455–461. [DOI] [PubMed] [Google Scholar]
- Lo S.-Y., Selby, M.J. and Ou, J.-H. (1996) Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol., 70, 5177–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomo J., Blomhoff, H.K., Beiske, K., Stokke, T. and Smeland, E.B. (1995) TGF-β1 and cyclic AMP promote apoptosis in resting human B lymphocytes. J. Immunol., 154, 1634–1643. [PubMed] [Google Scholar]
- Look A.T. (1997) E2A–HLF chimeric transcription factors in pro-B cell acute lymphoblastic leukemia. Curr. Top. Microbiol. Immunol., 220, 45–53. [DOI] [PubMed] [Google Scholar]
- Lu R., Yang, P., O'Hare, P. and Misra, V. (1997) Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol., 17, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Yang, P., Padmakumar, S. and Misra, V. (1998) The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol., 72, 6291–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki Y., Bos, T.J., Davis, C., Starbuck, M. and Vogt, P.K. (1987) Avian sarcoma virus 17 carries the jun oncogene. Proc. Natl Acad. Sci. USA, 84, 2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusawa H., Hijikata, M., Chiba, T. and Shimotohno, K. (1999) Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor α-mediated apoptosis via NF-κB activation. J. Virol., 73, 4713–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Hwang, S.B., Jeng, K.-S., Zhu, N. and Lai, M.M.C. (1996) Homotypic interaction and multimerization of hepatitis C virus core protein. Virology, 218, 43–51. [DOI] [PubMed] [Google Scholar]
- Matsumoto M., et al. (1997)Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-β receptor. J. Virol., 71, 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey A.I. and Jacks, T. (1998) Tumor suppressor mutations in mice: the next generation. Curr. Opin. Genet. Dev., 8, 304–310. [DOI] [PubMed] [Google Scholar]
- McConkey D.J., Jondal, M. and Orrenius, S. (1992) Cellular signaling in thymocyte apoptosis. Semin. Immunol., 4, 371–377. [PubMed] [Google Scholar]
- Moradpour D., Englert,C., Wakita,T. and Wands,J.R. (1996) Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology, 222, 51–63. [DOI] [PubMed] [Google Scholar]
- Moriya K., Yotsuyanagi, H., Shintani, Y., Fujie, H., Ishibashi, K., Matsuura, Y., Miyamura, T. and Koike, K. (1997) Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol., 78, 1527–1531. [DOI] [PubMed] [Google Scholar]
- Moriya K., et al. (1998)The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nature Med., 4, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Motohashi H., Shavit, J.A., Igarashi, K., Yamamoto, M. and Engel, J.D. (1997) The world according to Maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Goto, N. and Kawai, S. (1987) An avian transforming retrovirus isolated from a nephroblastoma that carries the fos gene as the oncogene. J. Virol., 61, 3733–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolandt O., Kern, V., Muller, H., Pfaff, E., Theilmann, L., Welker, R. and Krausslich, H.G. (1997) Analysis of hepatitis C virus core protein interaction domains. J. Gen. Virol., 78, 1331–1340. [DOI] [PubMed] [Google Scholar]
- Purcell R.H. (1994) Hepatitis viruses: changing patterns of human disease. Proc. Natl Acad. Sci. USA, 91, 2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravaggi A., Natoli, G., Primi, D., Albertini, A., Levrero, M. and Cariani, E. (1994) Intracellular localization of full-length and truncated hepatitis C virus core protein expressed in mammalian cells. J. Hepatol., 20, 833–836. [DOI] [PubMed] [Google Scholar]
- Ray R.B., Lagging, L.M., Meyer, K., Steele, R. and Ray, R. (1995) Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res., 37, 209–220. [DOI] [PubMed] [Google Scholar]
- Ray R.B., Lagging, L.M., Meyer, K. and Ray, R. (1996a) Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J. Virol., 70, 4438–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R.B., Meyer, K. and Ray, R. (1996b) Suppression of apoptotic cell death by hepatitis C virus core protein. Virology, 226, 176–182. [DOI] [PubMed] [Google Scholar]
- Ray R.B., Steele, R., Meyer, K. and Ray, R. (1997) Transcriptional repression of p53 promoter by hepatitis C virus core protein. J. Biol. Chem., 272, 10983–10986. [DOI] [PubMed] [Google Scholar]
- Ray R.B., Meyer, K., Steele, R., Shrivastava, A., Aggarwal, B.B. and Ray, R. (1998a) Inhibition of tumor necrosis factor (TNF-α)-mediated apoptosis by hepatitis C virus core protein. J. Biol. Chem., 273, 2256–2259. [DOI] [PubMed] [Google Scholar]
- Ray R.B., Steele, R., Meyer, K. and Ray, R. (1998b) Hepatitis C virus core protein represses p21WAF1/Cip1/Sid1 promoter activity. Gene, 208, 331–336. [DOI] [PubMed] [Google Scholar]
- Rice C.M. (1996) Flaviviridae: the viruses and their replication. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology. 3rd edn. Lippincott-Raven, Philadelphia, PA, pp. 931–959. [Google Scholar]
- Ruchaud S., Seite, P., Foulkes, N.S., Sassone-Corsi, P. and Lanotte, M. (1997) The transcriptional repressor ICER and cAMP-induced programmed cell death. Oncogene, 15, 827–836. [DOI] [PubMed] [Google Scholar]
- Ruggieri A., Harada, T., Matsuura, Y. and Miyamura, T. (1997) Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology, 229, 68–76. [DOI] [PubMed] [Google Scholar]
- Sabile A., et al. (1999)Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology, 30, 1064–1076. [DOI] [PubMed] [Google Scholar]
- Saito I., et al. (1990)Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl Acad. Sci. USA, 87, 6547–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E., Migliaccio, G. and La Monica, N. (1994) Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol., 68, 3631–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M.J., et al. (1993)Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J. Gen. Virol., 74, 1103–1113. [DOI] [PubMed] [Google Scholar]
- Shrivastava A., Manna, S.K., Ray, R. and Aggarwal, B.B. (1998) Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J. Virol., 72, 9722–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolik S.M., Rose, R.E. and Goodman, R.H. (1992) A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol. Cell. Biol., 12, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas R.V., Ray, R.B., Meyer, K. and Ray, R. (1996) Hepatitis C virus core protein inhibits human immunodeficiency virus type 1 replication. Virus Res., 45, 87–92. [DOI] [PubMed] [Google Scholar]
- Suzuki R., Matsuura, Y., Suzuki, T., Ando, A., Chiba, J., Harada, S., Saito, I. and Miyamura, T. (1995) Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol., 76, 53–61. [DOI] [PubMed] [Google Scholar]
- Tsuchihara K., Hijikata, M., Fukuda, K., Kuroki, T., Yamamoto, N. and Shimotohno, K. (1999) Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology, 258, 100–107. [DOI] [PubMed] [Google Scholar]
- Vinson C.R., Hai, T. and Boyd, S.M. (1993) Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev., 7, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Wang H.-L. and Jin, D.-Y. (1997) Prevalence and genotype of hepatitis G virus in Chinese professional blood donors and hepatitis patients. J. Infect. Dis., 175, 1229–1233. [DOI] [PubMed] [Google Scholar]
- Wang H.-L., Hou, Y.-D. and Jin, D.-Y. (1997a) Identification of a single genotype of hepatitis G virus by comparison of one complete genome from a healthy carrier with eight from patients with hepatitis. J. Gen. Virol., 78, 3247–3253. [DOI] [PubMed] [Google Scholar]
- Wang H.-L., Yan, Z.-Y., Hou, Y.-D. and Jin, D.-Y. (1997b) Molecular characterization of suppression of hepatitis B virus transcription by hepatitis C virus core protein. Sci. China C Life Sci., 40, 648–656. [DOI] [PubMed] [Google Scholar]
- Wang H.-L., Yan, Z.-Y. and Jin, D.-Y. (1997c) Reanalysis of published DNA sequence amplified from cretaceous dinosaur egg fossil. Mol. Biol. Evol., 14, 589–591. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Freiman, R.N., Goto, H., Nishimoto, T. and Herr, W. (1997) VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol., 17, 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Lis, J.T. and Jeang, K.-T. (1997) Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol., 17, 6898–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-P., Liu, C.-B., Jin, D.-Y., Zhan, M.-Y., Tang, Q., Xia, N.-S., Cao, J.-Y. and Li, J.-Y. (1994) cDNA cloning of c33-c antigen gene derived from NS3 region of Chinese HCV genome, expression in Escherichia coli and development of HCV EIA second-generation diagnostic kit. Sci. China B, 37, 190–202. [PubMed] [Google Scholar]
- Yasui K., Wakita, T., Tsukiyama-Kohara, K., Funahashi, S.-I., Ichikawa, M., Kajita, T., Moradpour, D., Wands, J.R. and Kohara, M. (1998) The native form and maturation process of hepatitis C virus core protein. J. Virol., 72, 6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L.-R., Chen, C.-M. and Lee, Y.-H.W. (1999a) Hepatitis C virus core protein enhances NF-κB signal pathways triggering by lymphotoxin-β receptor ligand and tumor necrosis factor α. J. Virol., 73, 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L.-R., Chen, C.-M., Yeh, T.-S., Tsai, T.-Y., Mai, R.-T., Lin, C.-H. and Lee, Y.-H.W. (1999b) Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol., 73, 2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Khoshnan, A., Schneider, R., Matsumoto, M., Dennert, G., Ware, C. and Lai, M.M.C. (1998) Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol., 72, 3691–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E.B. (1990) Transcription factors: a new family gathers at the cAMP response site. Trends. Genet., 6, 69–72. [DOI] [PubMed] [Google Scholar]