Abstract
We are analyzing highly conserved heat shock genes of unknown or unclear function with the aim of determining their cellular role. Hsp15 has previously been shown to be an abundant nucleic acid-binding protein whose synthesis is induced massively at the RNA level upon temperature upshift. We have now identified that the in vivo target of Hsp15 action is the free 50S ribosomal subunit. Hsp15 binds with very high affinity (KD <5 nM) to this subunit, but only when 50S is free, not when it is part of the 70S ribosome. In addition, the binding of Hsp15 appears to correlate with a specific state of the mature, free 50S subunit, which contains bound nascent chain. This provides the first evidence for a so far unrecognized abortive event in translation. Hsp15 is suggested to be involved in the recycling of free 50S subunits that still carry a nascent chain. This gives Hsp15 a very different functional role from all other heat shock proteins and points to a new aspect of translation.
Keywords: heat shock/Hsp15/ribosome/RNA binding/50S subunit
Introduction
The explosion in sequencing has revealed a substantial number of conserved genes without assigned functions. No function can be assigned to 30–60% of the open reading frames (ORFs) identified in each organism (Koonin et al., 1998). Even in Escherichia coli, which is extremely well characterized, >30% of the ORFs have no known function (Blattner et al., 1997; Richmond et al., 1999). One of the major challenges of the post-genomic era is to ascertain what these genes do. We have decided to analyze the function and three-dimensional structure of highly conserved, newly identified heat shock proteins. We were motivated by the possibility of discovering new functions that would help us to understand the role of the heat shock response in the cell.
The previously studied heat shock proteins generally function as molecular chaperones or proteases. They act to ensure that the cellular protein pool is maintained in a native, functional state (Becker and Craig, 1994; Gross, 1996). The role of these evolutionarily ancient proteins is essential to the cell not only after heat shock but also under normal conditions. We decided that the uncharacterized heat shock proteins recently uncovered by genomic expression screening technology represent a promising source of new chaperones, proteases and possibly other important cellular functions as well.
Very recently, a global screening of the 4290 ORFs in E.coli by ‘gene chip’ array technology revealed a total of 77 ORFs that are induced >5–fold at the RNA level upon temperature upshift (Richmond et al., 1999). The function of at least one-third of these proteins is unknown (Gross, 1996; Richmond et al., 1999).
We are investigating the function of three highly conserved, very heat-inducible ORFs: yrfH/hslR, yrfI/hslO and ftsJ. The gene for Hsp15 (yrfH/hslR) shows a 43–fold induction as assayed by radioactivity, making it the fifth most highly heat inducible of the 4290 genes in E.coli (Richmond et al., 1999). Seventy-seven ORFs showed a >5–fold induction with this assay. This makes the gene for Hsp15 more highly heat inducible than nearly all well studied heat shock genes including groEL, groES dnaK, dnaJ, clpA, clpP, rpoD, rpoH and lon. When assayed by using a fluorescent microarray gene chip, Hsp15 RNA showed a similar, 51–fold induction.
In a previous report (Korber et al., 1999), we described the purification of the gene product of yrfH/hslR, Hsp15, and its in vitro characterization as a nucleic acid-binding protein. This activity is different from that of most heat shock proteins, which function at the protein level as chaperones or proteases. We are interested in determining the precise role of this newly isolated heat shock protein as this could illuminate new venues within the heat shock response.
The first step was to identify the in vivo binding site of Hsp15. The high abundance of Hsp15 (∼12 000 molecules per cell at 29°C; Korber et al., 1999) made a specific DNA-binding site unlikely. The identification of a novel sequence motif within Hsp15's sequence implicated in RNA binding suggested an RNA substrate for Hsp15 (Aravind and Koonin, 1999; Korber et al., 1999).
This notion received strong support from the crystal structure of Hsp15, which we have solved to 2.0 Å (Staker et al., 2000). Comparison of Hsp15's structure with other known proteins revealed close similarities to two structures of RNA-binding proteins, ribosomal protein S4 and threonyl-tRNA synthetase. Hsp15 is composed almost entirely of this novel motif that we term the αL motif. The αL motif is present in at least 400 sequenced members that comprise eight different RNA-binding families including the FtsJ RNA methylase family that is also heat inducible (Aravind and Koonin, 1999; Korber et al., 1999; Staker et al., 2000; H.Buegl, E.B.Fauman, B.L.Staker, F.–Z.Zheng, S.R.Kushner, M.A.Saper, J.C.A.Bardwell and U.Jakob, submitted). These structural and sequence hom– ologies strongly implicate the αL motif in RNA binding and inspired us to attempt to determine a specific RNA substrate for Hsp15.
Here, we present evidence that Hsp15 binds with high affinity specifically to the free 50S subunit of the ribosome. The binding site is accessible only in the absence of the 30S ribosomal subunit and appears to correspond to a particular state of the free 50S ribosomal subunit. This state seems to be the product of an abortive event during translation that renders a free 50S subunit still carrying a nascent chain. We propose that Hsp15 takes part in removing the nascent chain and making such 50S subunits available again for translation.
Results
Hsp15 binds to the 50S ribosomal subunit
Hsp15 is an abundant heat shock protein that binds to various types of nucleic acids in vitro (Korber et al., 1999). The dissociation constant for the non-specific interaction with chromosomal DNA or RNA oligomers is in the range of 4–20 μM. To characterize Hsp15's function further, we sought to determine whether a higher affinity binding substrate could be found in cell lysates, one that might correspond to the in vivo target of Hsp15 action. The homology between Hsp15 and a number of RNA-binding proteins suggested that Hsp15 may bind to an RNA target.
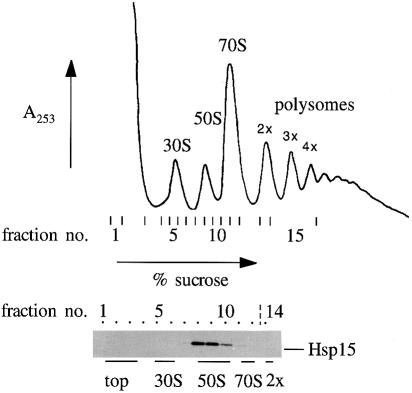
Total cellular RNAs from E.coli cell lysates were fractionated according to their sedimentation coefficient in sucrose gradients (Figure 1). The tRNA and mRNA molecules migrated with cellular protein near the top of the gradient, followed by the major stages of ribosome association: free 30S and 50S subunits, 70S ribosomes and polysomes with two, three, four or more ribosomes stalled on mRNA molecules. The extent of polysomes in the profile was enhanced by the addition of chloramphenicol to the growing cells shortly before harvest. Aliquots of fractions throughout the gradient were assayed for the presence of Hsp15 by Western blot analysis. Hsp15 was found exclusively in fractions of the peak of the free 50S ribosomal subunit, suggesting that Hsp15 associates with the 50S ribosomal subunit and binds strongly enough to remain associated during the centrifugation run. Hsp15 was not found in fractions containing 70S ribosomes or polysomes even though the 50S subunit is a part of these ribosomes. Apparently the binding site for Hsp15 on the 50S subunit is not accessible upon association of 50S with the 30S subunit.
Fig. 1. Hsp15 associates with the free 50S subunit of ribosomes. Polysome profile of wild-type cells that were treated with 100 μg/ml chloramphenicol. A total of 17.5 A260 units of the lysate were loaded onto a sucrose gradient and fractionated as described in Materials and methods. The fractions indicated were analyzed by Western blotting. Hsp15 is present only in the fractions of the 50S subunit peak.
Salt dependence of the binding of Hsp15 to the 50S ribosomal subunit
The interactions of proteins with nucleic acids usually have electrostatic contributions that render the binding affinity sensitive to the salt concentration (Record et al., 1976). The concentration of Mg2+ shows especially large effects on nucleic acid–protein interactions, and Mg2+ plays a particularly important role in the association state of the ribosome (Tissières et al., 1959). Thus, to characterize the interaction of Hsp15 with the 50S ribosomal subunit further, its dependence on concentrations of NH4Cl and Mg2+ was assayed. This was done by loading lysates of chloramphenicol-treated cells onto sucrose gradients with differing buffer composition (Figure 2A and B).
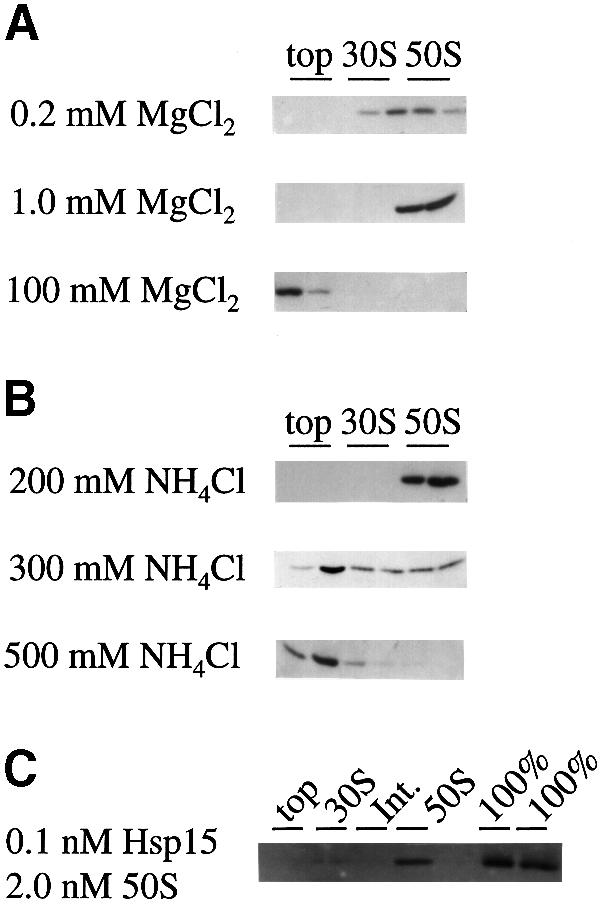
Fig. 2. The influence of salt conditions on the binding of Hsp15 to 50S subunits and estimation of the dissociation constant. (A) Varied MgCl2 concentration at constant 150 mM NH4Cl. (B) Varied NH4Cl concentration at constant 10 mM MgCl2. See Figure 1 for the 10 mM MgCl2, 150 mM NH4Cl condition. The Western blot analyses of top, 30S and 50S peak fractions of the corresponding profiles are shown. Hsp15 is stably bound to the 50S subunit in and beyond the range of physiological ionic strength. (C) For the estimation of the dissociation constant (at 10 mM MgCl2, 150 mM NH4Cl), purified Hsp15 was added at 0.1 nM to various amounts of a chloramphenicol-treated lysate of Hsp15-depleted cells. For calibration, the total amount of Hsp15 in the binding reaction was loaded in parallel on the gel (‘100%’). ‘Int.’ is the fraction between the 30S and 50S subunit peaks. The exogenously added Hsp15 is again found in the 50S peak. A representative example is shown with Hsp15 and 50S concentrations of 0.1 and 2.0 nM, respectively. From five such reconstitution reactions, the KD was estimated to be in the nanomolar range, <5 nM.
At a constant NH4Cl concentration of 150 mM, the Mg2+ concentration was varied as 0.2, 1, 10 and 100 mM (Figure 2A; see Figure 1 for the condition of 10 mM Mg2+ and 150 mM salt). At 1 mM Mg2+, the ribosome is dissociated into the 30S and 50S subunits, and at lower Mg2+ concentrations it begins to fall apart (Tissieres et al., 1959). The binding of Hsp15 to the 50S subunit is not affected by the Mg2+ concentration between 0.2 and 10 mM and can therefore conveniently be studied under conditions of both association and dissociation of the ribosome. At 100 mM Mg2+, the ionic strength is too high to allow stable binding.
Salt concentrations varying between 150 and 500 mM were tested while keeping the Mg2+ concentration constant at 10 mM (Figures 1 and 2B). Hsp15 was stably bound to the 50S subunit in salt concentrations up to 200 mM. At 300 mM salt, Hsp15 began to dissociate; at 500 mM salt no Hsp15 was found to be bound to ribosomal subunits. Hsp15 was present only on the top of the gradient where it migrates according to its own low molecular weight. In summary, Hsp15 is stably bound to the free 50S subunit in and significantly beyond the physiological range of ionic strength.
Estimation of the KD value for the Hsp15–50S subunit association
To estimate the dissociation constant for the Hsp15–50S interaction, purified Hsp15 was added to various amounts of Hsp15-free lysates at 10 mM Mg2+ and 150 mM salt. These lysates were generated from a mutant that carries a kanamycin resistance cassette inserted into the Hsp15 gene hslR. Western blot analysis of these lysates showed no detectable cross-reacting material to Hsp15 (our unpublished data). The exogenously added Hsp15 reconstituted into the 50S peak of polysome profiles, showing that Hsp15 had been purified in an active form. The ratio of Hsp15 present in the 50S peak to Hsp15 on top of the gradient corresponds to the percentage of bound Hsp15. As an additional control, we loaded the same total amount of Hsp15 as was used in the binding reaction in parallel on the gel to obtain a 100% standard on the blot. The concentration of 50S subunits was estimated by measuring the absorbance at 260 nm of the 50S peak fraction and the conversion factor of 1 A260 ≈ 36 pmol 50S subunits (Bommer et al., 1996).
Figure 2C shows an example where Hsp15, added at 0.1 nM, is found almost exclusively in the 50S peak with a 50S concentration in the binding reaction of 2 nM. Comparison of the amount of Hsp15 in the 50S peak with the 100% standards shows that >40% of total Hsp15 is recovered in the 50S fraction. [The missing Hsp15 material is probably lost in the trichloroacetic acid (TCA) precipitation of the highly diluted top fraction, despite the addition of 0.15 mg/ml bovine serum albumin (BSA) as a carrier]. In this concentration range, our assay is at the limit of detection both for Hsp15 on the Western blots and for the 50S peak in the gradient. We therefore could not establish a rigorous binding curve, but we estimate conservatively that the KD value for the Hsp15–50S interaction is <5 nM.
Hsp15 binds exclusively to the 50S subunit of ribosomes in total cell extracts, and its binding affinity is increased by at least three orders of magnitude in comparison with the non-specific nucleic acid binding affinity previously measured as 4–20 μM (Korber et al., 1999). This implies that we have found a specific binding site for Hsp15.
The presence of Hsp15 in the 50S subunit peak is enhanced by chloramphenicol treatment
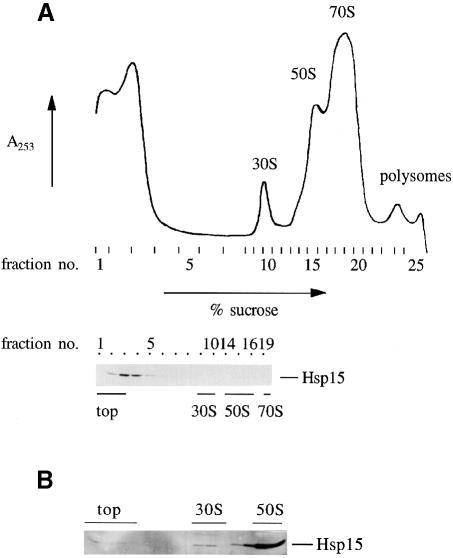
Chloramphenicol treatment increases the amount of polysomes that can be observed in sucrose gradient profiles. Without chloramphenicol treatment, the peak of the 70S monosomes is enlarged at the cost of the polysome peaks, reflecting run-off of the ribosomes during the harvest procedure (Friedman et al., 1971; Figure 3A). To our surprise, Western blot analysis of such gradients showed that Hsp15 was only occasionally detected in the 50S subunit peak. Usually, Hsp15 was not found associated at all with the 50S subunit if no chloramphenicol was added before harvest (Figure 3A; see also Figure 7).
Fig. 3. (A) Polysome profile of wild-type cells that were not treated with chloramphenicol. Hsp15 is no longer bound to the free 50S subunit but is found in the top fractions. The gradient has been centrifuged for a longer time than in Figure 1 (see text). A total of 17.5 A260 units of the lysate were loaded onto the gradient. (B) The same lysate as in (A) was pre-incubated under dissociation conditions (1 mM MgCl2, 150 mM salt). 50S subunits that were generated by the dissociation bind Hsp15. A total of 5 A260 units of the lysate were loaded onto the gradient.
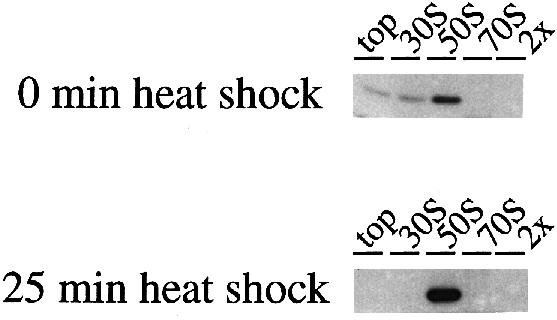
Fig. 7. The high affinity Hsp15-binding state of the 50S subunit is induced under heat shock conditions. Lysates without chloramphenicol treatment were prepared from cells before and during heat shock (30→41°C) and the same amount (17.5 A260 units) was loaded onto the gradients. While only a part of the Hsp15 pool is found in the 50S peak before heat shock (for the extreme example of no binding at all compare Figure 3A), the majority of Hsp15 is bound to 50S during heat shock conditions.
Hsp15 was, however, located in fractions containing higher sucrose concentrations than those where it should be found if it was migrating due to its own hydrodynamic properties. With standard centrifugation conditions (ω2t = 1.2 × 1011 rad2/s), the 30S peak was positioned in these fractions of higher sucrose concentration (Figure 1). However, longer centrifugation runs allowed the 30S peak to ‘out-run’ the Hsp15. For instance, the gradient shown in Figure 3A has been centrifuged for a longer time than usual (ω2t = 4.0 × 1011 rad2/s) to demonstrate that all ribosomal species will out-run Hsp15 migration when using lysates from non-chloramphenicol-treated cells. In contrast, 50S subunits that bind Hsp15 always co-migrate with Hsp15 independently of the centrifugation time.
It appears that the free 50S subunits in lysates of cells without chloramphenicol treatment are mainly in a state where the affinity for Hsp15 is decreased significantly, causing Hsp15 to dissociate during centrifugation. The high affinity binding could not be restored either by adding chloramphenicol to the cells after incubation on ice or by addition to the lysate (our unpublished data), but was found only if chloramphenicol was added to the growing cells (Figure 1).
Hsp15 binds to 50S subunits generated by dissociation of 70S ribosomes
The experiments with lysates without chloramphenicol treatment suggested that the 50S subunit had to be in a specific state that is connected to active metabolism in order to bind Hsp15 with high affinity. We therefore asked whether 50S subunits that were released from 70S ribosomes by incubating under dissociation conditions (1 mM MgCl2, 150 mM NH4Cl), would still be in this high affinity state and bind Hsp15.
The buffer condition of the same wild-type lysate without chloramphenicol treatment, which was analyzed in Figure 3A, was adjusted to dissociation conditions and incubated on ice for 10 min. Binding of Hsp15 under these buffer conditions is not affected (Figure 2B, middle panel), and a 10 min incubation time on ice was sufficient for complete binding of purified Hsp15 to its binding site in lysates of the insertion mutant (see Materials and methods; Figure 2C).
The analysis of such pre-incubated lysates revealed that Hsp15 was again found associated with the 50S subunit (Figure 3B). This experiment rules out the possibility that chloramphenicol itself is necessary for the high affinity binding. It also shows that it is mainly the mature 50S subunit that is recognized by Hsp15, as mainly mature subunits are released from mono- and polysomes.
In this kind of experiment, a small portion of Hsp15 is also associated with the 30S subunit peak (Figure 3B). As Hsp15 has been found in all previous experiments to have a binding site on the 50S subunit, we assume that it is not the 30S subunit but 50S precursors migrating at an approximately 30S position, e.g. p50 S–1 particles with sedimentation constants of 30–36S (Mangiarotti et al., 1968; Lindahl, 1975; for a review, see Nierhaus, 1991), that account for binding this minor fraction of Hsp15.
The high affinity Hsp15-binding 50S subunits can be generated only from polysomes
In order to investigate further where the 50S subunits with high affinity for Hsp15 come from, we divided the wild-type lysate without chloramphenicol treatment into the poly- and the monosome fractions before incubating under dissociation conditions (Figure 4). The monosomes consist mainly of run-off tight coupled ribosomes, whereas the polysomes are active ribosomes stalled in the process of mRNA translation. Endogenous Hsp15 is not present in any of these fractions, so purified Hsp15 was added after dissociation.
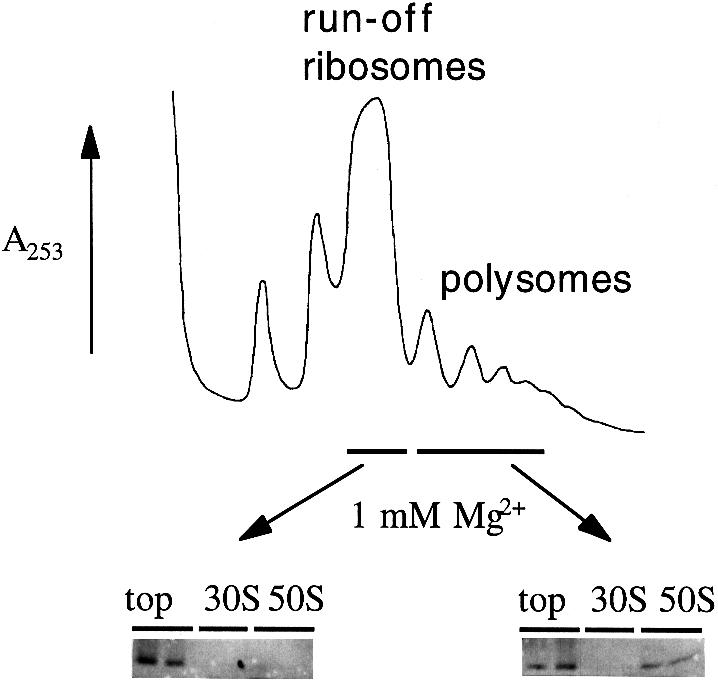
Fig. 4. Only polysomes contain 50S subunits with high affinity for Hsp15. The fractions of run-off ribosomes and of polysomes from a wild-type lysate without chloramphenicol treatment (17.5 A260 units) were subjected to dissociation conditions (1 mM MgCl2, 150 mM salt). The same amounts (24 pmol) of 50S subunits thus generated were tested for their ability to bind purified Hsp15.
The high affinity Hsp15-binding 50S subunits could be generated only from the polysomes (Figure 4). The yield of Hsp15–50S complex in this purified reassociation reaction is low. Hsp15 has been added equimolarly to the subunits, but only a portion of it bound to the 50S subunit. Much Hsp15 remained at the top of the gradient. We consistently found it difficult to isolate the high affinity Hsp15-binding 50S subunits even when starting with the 50S peak of chloramphenicol-treated cells. This implies a labile nature of this high affinity state.
Puromycin abolishes binding of Hsp15 to the 50S subunit
To identify further the nature of the high affinity Hsp15-binding 50S, we employed the antibiotic puromycin which classically has been used to distinguish the pre- and post-translocational states of ribosomes (Rheinberger et al., 1988; Bommer et al., 1996). If puromycin was included into the dissociation experiment with whole lysates, Hsp15 was no longer found in the 50S subunit peak (Figure 5, middle panel). It made no difference whether the puromycin was added before or after the change to the dissociation conditions (our unpublished data). The amount of Hsp15 that was found in the 30S peak (Figures 3B and 5) was not affected by puromycin (Figure 5). Addition of puromycin in the presence of chloramphenicol had no effect; Hsp15 was still associated with the 50S subunit (Figure 5, lower panel).
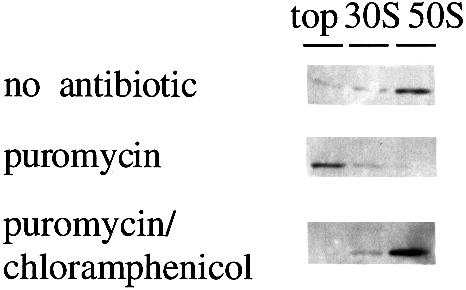
Fig. 5. The reaction with puromycin abolishes the high affinity of 50S subunits for Hsp15. The same experiment as in Figure 3B was conducted additionally in the presence of puromycin (1 mM) and a combination of puromycin and chloramphenicol (1 and 0.3 mM, respectively). Chloramphenicol inhibits the puromycin reaction and preserves the 50S subunits with high affinity for Hsp15.
In vitro translation generates the high affinity Hsp15 binding state of the 50S subunit: Hsp15 co-migrates with the nascent chain
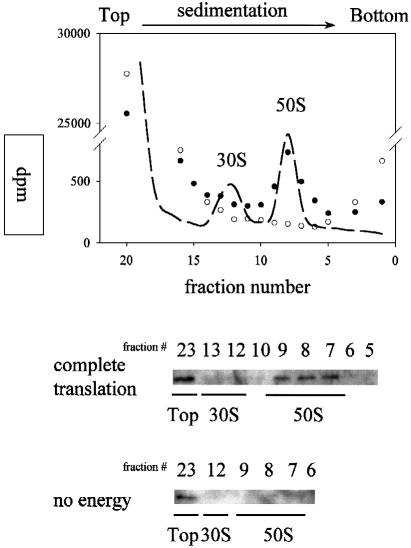
The effect of puromycin on the binding of Hsp15 to the 50S subunit suggested that the presence of a nascent chain may lead to high affinity for Hsp15 of the 50S subunit. In order to test this hypothesis, we generated a nascent poly(phenylalanine) chain by in vitro translations of poly(U) (Bommer et al., 1996). As a negative control, the in vitro translation reaction was performed in parallel without the addition of energy. The tight coupled 70S ribosomes that were used in this system at first did not contain Hsp15-binding 50S subunits (Figures 4 and 6). However, after successful translation (∼30 incorporated phenylalanine residues per ribosome) and subsequent dissociation, addition of Hsp15 and separation on sucrose gradients, Hsp15 was again found in the 50S peak (Figure 6). This is clear evidence that the high affinity Hsp15-binding state can be generated from previously non-binding 50S subunits by going through several rounds of elongation. When the nascent chain was radioactively labeled, it could be found associated with the 50S subunit peak. The fractions with the highest amount of label in the peak also showed the highest amount of Hsp15 in the Western blot (Figure 6). Thus, Hsp15 co-migrates with the nascent chain on the 50S subunit in the sucrose gradient.
Fig. 6. The affinity for Hsp15 of the 50S subunit can be generated by in vitro translation. In vitro translation of poly(U) was performed with 20 pmol of tight coupled 70S ribosomes. The ribosomes were dissociated after the translation reaction and incubated with Hsp15. The subunits thus generated were analyzed on sucrose gradients for the presence of radioactively labeled poly(phenylalanine) and Hsp15. (•), Complete translation reaction; (○), negative control, i.e. translation reaction without addition of energy. The UV absorption profile of the gradients is shown as a dashed line overlay on the radioactivity profile. Only after positive translation is a nascent chain generated that co-migrates with the 50S peak, and only in this case is Hsp15 found associated with the 50S subunit.
Heat shock induces the Hsp15-binding state of the 50S subunit
We asked if the specific 50S subunit state that is recognized by Hsp15 and carries a nascent chain is connected to heat shock conditions. As lysates without chloramphenicol always showed much less, if any, Hsp15 bound to the 50S subunit than lysates with chloramphenicol (compare Figures 1 and 3A), we wanted to test whether heat shock conditions would have a similar effect to the chloramphenicol in generating the high affinity Hsp15-binding state in cell lysates. As shown in Figure 7, Hsp15 was indeed found to a much higher extent in the 50S peak of heat-shocked cell lysates in comparison with the pre-heat shock sample. In contrast to Figure 3A, Figure 7 shows an example where some Hsp15 is located with the 50S peak before heat shock even though no chloramphenicol was added.
Discussion
Previously, we have shown that Hsp15 is distinguished from most characterized heat shock proteins in that it binds to nucleic acids instead of acting at the protein level as a chaperone or protease (Korber et al., 1999). This opens up a whole new functional arena for heat shock proteins. The next step in characterizing the function of Hsp15 was to find its nucleic acid substrate in vivo. The discovery of a sequence and folding motif that Hsp15 has in common with a large number of RNA-binding proteins (Aravind and Koonin, 1999; Korber et al., 1999; Staker et al., 2000) led us to test the notion that Hsp15 interacts with a specific RNA target.
By fractionating different RNA species from lysates, we succeeded in revealing a high affinity interaction of Hsp15 with the free 50S ribosomal subunit (Figure 1). The 50S subunit consists of 60% of RNA (Tissières et al., 1959) and is therefore an appropriate substrate for a nucleic acid-binding protein. The constitutive expression level of Hsp15 was determined previously to be ∼12 000 molecules per cell at 29°C (Korber et al., 1999), which is of the right order of magnitude to postulate that the ribosome is the in vivo substrate for Hsp15. There are ∼70 000 ribosomes per cell under our growth conditions (Bremer and Dennis, 1996). The KD value of the binding reaction between Hsp15 and the 50S ribosomal subunit was estimated to be <5 nM (Figure 2C). This represents an increase in affinity by three orders of magnitude in comparison with Hsp15's non-specific nucleic acid binding affinity, where the KD value was determined to be 4–20 μM (Korber et al., 1999). We infer from this high affinity interaction that we have identified the actual in vivo binding site for Hsp15. Though the binding is sensitive to conditions of very high ionic strength, the specific interaction is stable at salt concentrations even exceeding physiological levels, again implying an in vivo significance of the observed association (Figure 2A and B).
In polysome profiles of cells where translation had been stopped by chloramphenicol, Hsp15 was found exclusively in the peak of the free 50S subunit (Figure 1). Hsp15 was never seen in the peaks of the 70S ribosome or polysomes. This argues that the binding site of Hsp15 on the 50S subunit is accessible only in the absence of the 30S subunit and occluded upon creation of the subunits to the 70S ribosome.
Surprisingly, if the cells had not been stopped by chloramphenicol but just chilled on ice before harvest, Hsp15 was no longer bound quantitatively to the 50S subunit, and in most cases was not bound at all (Figures 3A and 7). The treatment with chloramphenicol yielded free 50S subunits with high affinity for Hsp15, whereas lysates without chloramphenicol treatment contain mostly free 50S subunits of significantly lower binding affinity for Hsp15. This points to the existence of two different states of the free 50S subunit.
Even though the free 50S subunits in lysates of cells without chloramphenicol treatment did not bind Hsp15, it was possible to generate high affinity 50S subunits from the same lysate by pre-incubation under dissociation conditions (1 mM MgCl2, 150 mM salt; Figure 3B). The low magnesium concentration dissociates all 70S ribosomes into the subunits (Tissières et al., 1959). Some of these 70S ribosomes contain 50S subunits that possess a high binding affinity for Hsp15, which is revealed once they become dissociated from the 30S subunit. This finding rules out the possibility that it is the chloramphenicol itself that is responsible for the high affinity binding state. It also implies that it is the mature 50S subunit that is recognized by Hsp15, as mainly mature subunits are generated from 70S ribosomes. In order to understand the nature of the difference between the two states of the free 50S subunit, we wanted to determine the source of 50S subunits with high affinity for Hsp15.
The 70S ribosomes in cell lysates can be divided into two major groups: those that are stalled on the mRNA and constitute the polysomes, and those that have already run off the mRNA, but are not yet dissociated. The fractions containing polysomes and run-off ribosomes, respectively, were isolated from cells that had not been treated with chloramphenicol. After exchanging the buffer background to dissociation conditions, both fractions were tested with purified Hsp15 to see if they contained high affinity Hsp15-binding 50S subunits. Only 50S subunits that were generated from polysomes had a high affinity for Hsp15 (Figure 4). The yield was low, leaving a significant excess of the equimolarly added Hsp15 at the top of the gradient. The difficulties in isolating the high affinity Hsp15 binding state of the 50S subunit and the fact that each handling step seemed to reduce the percentage of binding subunits suggest that the state of 50S that binds Hsp15 is unstable or that the 50S–Hsp15 interaction requires an additional factor that is lost upon handling.
What is the difference between 50S subunits that are generated from polysomes and those from run-off ribosomes? What is the nature of the unstable high affinity state or the additional factor? Experiments involving puromycin pointed to the role of the nascent chain. Puromycin acts as an aminoacyl-tRNA analog and removes the nascent chain from 50S subunits. The peptidyltransferase center of the 50S subunit transfers the nascent chain to the puromycin molecule where the nascent chain–puromycin adduct dissociates due to the low affinity of puromycin for the 50S subunit.
The addition of puromycin abolished the high affinity state of 50S subunits (Figure 5, middle panel). Chloramphenicol inhibits the peptidyltransferase reaction and also the puromycin reaction (Traut and Monro, 1964). Thus it was expected, and observed, that the addition of puromycin in the presence of chloramphenicol had no effect on Hsp15 binding (Figure 5, lower panel).
These results strongly suggest that the loss of the nascent chain leads to the loss of the high affinity state of the 50S subunit. We could also show the converse: generation of a nascent chain by in vitro translation actually leads to the high affinity state (Figure 6). Tight coupled 70S ribosomes, which at first do not contain Hsp15-binding 50S subunits (Figures 4 and 6), were used to translate poly(U) in vitro. The radioactively labeled nascent poly(phenylalanine) chain was found associated with the 50S subunit after dissociation and analysis on sucrose gradients, as has been shown previously (Gilbert, 1963). In addition, we found that Hsp15 co-migrates with the nascent chain on the 50S subunit in the sucrose gradient. This interaction can only be observed under dissociating conditions, which again confirms that the binding site for Hsp15 is accessible only on the free 50S subunit. A negative control where no energy was added to the translation reaction did not generate a nascent chain and showed no Hsp15 associated with the 50S peak. The same negative result is obtained by omitting the poly(U) instead of omitting the energy source (our unpublished data). Therefore, we argue that it is not just the energy-dependent action of a factor from the S100 enzymes that generates the high affinity binding state of the 50S subunit. 50S subunits from ribosomes in a defined pre- and post-translocational state (as in Rheinberger et al., 1988) also did not bind Hsp15 (our unpublished data). Apparently an acetyl-phenylalanine residue is not sufficient to mimic the effect of a nascent chain with regard to the generation of the high affinity Hsp15 binding state.
Taking all these results together, we explain our data as follows: only free 50S subunits that carry a nascent chain have high affinity for Hsp15. 50S subunits that are generated from polysomes by dissociation still carry the nascent chain and therefore bind Hsp15, in contrast to those from run-off ribosomes. The release of the nascent chain is blocked by the addition of chloramphenicol to growing cells before harvest, whereas just chilling the cells on ice leads to a run-off of translation, including the termination event. Therefore, much less or no Hsp15 is found in the 50S peak of lysates without chloramphenicol treatment.
We do not have any evidence that Hsp15 binds directly to the nascent chain. Although most heat shock proteins function as molecular chaperones, Hsp15 has tested negative in chaperone assays (our unpublished data). Rather, Hsp15 is a nucleic acid-binding protein (Korber et al., 1999), making it much more likely that Hsp15 binds to a part of the rRNA of the 50S subunit that changes its conformation and/or accessibility in the presence of the nascent chain.
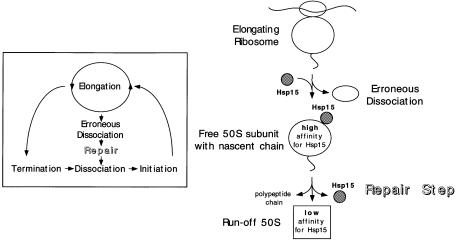
It has been known for a long time that the nascent chain stays associated with the 50S subunit upon dissociation of elongating ribosomes (Gilbert, 1963). Nonetheless, free 50S subunits carrying a nascent chain are considered to be an artifactual state that does not occur in vivo as the nascent chain is released by the action of termination factors before the ribosome is dissociated into its subunits. However, we show that Hsp15 recognizes exclusively the free 50S subunit only when it is not part of the 70S ribosome. Moreover, the present data strongly suggest that only free 50S subunits carrying a nascent chain have a high affinity for Hsp15. This leads us to postulate that a free 50S subunit with a nascent chain does indeed occur in vivo. Figure 8 outlines a model that accounts for this state. The free 50S subunit with high affinity for Hsp15 is thought to be the product of an erroneous dissociation of elongating ribosomes prior to the usual termination step. Such a ‘loaded’ 50S subunit needs to be relieved of its nascent chain in order to re-enter the translational cycle. We suggest that Hsp15 is involved in the recognition and repair of such 50S particles. This erroneous dissociation may be especially prevalent under heat shock conditions, explaining why the Hsp15 mRNA is 40- to 50–fold induced upon heat shock. Consistent with this, we find that the specific state of the 50S subunit that is recognized by Hsp15 is more abundant under heat shock conditions (Figure 7). The recently published structure of the 70S ribosome revealed a surprisingly small area of interface contacts between the 50S and 30S subunits of the ribosome, which supports the idea of a delicate balance between association and dissociation of the ribosome (Cate et al., 1999). Erroneous dissociation of the elongating ribosome under normal and especially under heat shock conditions may indeed be an as yet unrecognized problem in vivo.
Fig. 8. A model explaining the nature of the 50S subunits that have high affinity for Hsp15. Hsp15 recognizes an off-pathway 50S subunit that still carries the nascent chain. It is the product of an erroneous dissociation of elongating ribosomes and has to be repaired for re-use in the translational cycle. For details see text.
It is premature to give definite solutions to the detailed cellular machinery that deals with this problem and the exact role that Hsp15 plays here. Nonetheless, the specific interaction of Hsp15 with free 50S subunits points to a surprising aspect of the translational cycle that has not been seen so far.
Materials and methods
Buffers
All buffers contained 20 mM HEPES–KOH pH 7.5 and 4 mM β-mercaptoethanol. The concentrations of magnesium and ammonium salts were varied as indicated. The association buffer used was 10 mM MgCl2, 150 mM NH4Cl, while 1 mM MgCl2, 150 mM NH4Cl was used for dissociation of the ribosome into its subunits.
Bacterial strains
Escherichia coli MC4100 (Silhavy et al., 1984) was used as wild type. JCB1184 is MC4100 hslR::kan1. It contains the kanR cassette from pUC4–K inserted into hslR, the gene for Hsp15, at the BsmI site that is located at residue Glu50 of Hsp15. This mutant was generated in vitro and then recombined into the chromosome by the process of linear transformation. First, a 7 kb SalI fragment from Kohara clone 620 was ligated into the SalI site of pBluescriptSK(+) (Stratagene; Kohara et al., 1987). This fragment contains the Hsp15 gene and ample flanking DNA. A 1.1 kb PstI fragment from pWW97 that contains the kanamycin-resistant cartridge (Muller et al., 1986) was then blunt ended with T4 polymerase and ligated into the BsmI site in the hslR gene. This plasmid, which contains a kanR disruption in the gene for Hsp15, was linearized and crossed into the chromosome of JCB1185 by selection for kanamycin resistance. JCB1185 is MC1000 recD::Tn10, zxx665::specR. The recD mutation permits high efficiency linear transformation (Bardwell and Craig, 1988). The absence of Hsp15 was confirmed by Western blot.
Preparation of cell lysates
Bacterial strains were grown in LB in baffled Erlenmeyer flasks by shaking at 150 r.p.m. at 37°C. The volume of the culture was maximally a quarter of the flask capacity. Doubling times were typically between 20 and 30 min. If required, chloramphenicol (ICN) was added to a final concentration of 100 μg/ml 3 min before harvest. Cells were harvested when the OD600 reached 0.5. The cells were poured onto an approximately equal volume of chipped ice and harvested by centrifugation. The cell pellet was resuspended in 1 ml of cold buffer (20 mM HEPES–KOH pH 7.5, 4 mM β–mercaptoethanol, 6 mM MgCl2, 30 mM NH4Cl) per 100 ml of culture, and lysozyme was added to a final concentration of 0.75 mg/ml. After 3 min incubation on ice, the cells were frozen for at least 1 h at –80°C and slowly thawed in an ice–water bath. The lysate was clarified by 20 min centrifugation at 2°C, 22 000 r.p.m. in an RP80AT rotor (Sorvall) and either used immediately or shock-frozen in dry ice–ethanol in 1 ml aliquots and stored at –80°C. Before use, the thawed lysates were clarifed by a 5 min centrifugation at 13 500 g.
Sucrose gradients and preparation of ribosomal particles
Sucrose gradients were formed by layering of different concentrations of sucrose followed by diffusion. A 1.8 ml aliquot of a 50% sucrose solution (Fisher ultracentrifugation grade) and 2.5 ml each of 40/30/20 and 10% sucrose solutions in either association or dissociation buffer were layered into Beckman polyallomer SW40 tubes and diffusion was allowed to proceed overnight in the cold or for at least 3 h at 37°C. Cell lysate was applied on the top of the gradient and the gradients were centrifuged in an SW40Ti rotor in a Beckman L8–70 preparative ultracentrifuge at 2°C with a centrifugal effect ω2t of 1.2–4.0 × 1011 rad2/s.
Gradients were analyzed using a model SYS-101 Brandel syringe pump connected to a Brandel model 184 fractionator and an ISCO gradient flow cell (5 mm path length, model 647–030) in an ISCO UV-detector (253 nm, model UA–5). The syringe was filled with 50% sucrose in water and the pump flow rate set to 0.75 ml/min. Peak fractions were collected by hand. For preparation of ribosomal particles, sucrose was removed from the appropriate gradient fractions using Amicon centricon–30 ultrafiltration units.
SDS–PAGE and Western blots
Aliquots of the gradient fractions were analyzed on 14% Tris–glycine NOVEX SDS–PAGE run at 200 V for 47 min, according to the directions of the manufacturer (Novagen). The gels were blotted onto Millipore Immobilon-P PVDF membranes with a Fisher-Biotech semi-dry electroblotting apparatus. The Western blots were treated and developed essentially as described for the ECL system by the manufacturer (Amersham). Polyclonal antiserum against Hsp15 was a gift from Michael Ehrmann (Universität Konstanz, Germany). Due to the 1 ng per lane detection limit of the Western blot, occasionally volumes larger than could be loaded into one well were concentrated by TCA (12% final concentration) precipitation (Figure 2C). To enhance the precipitation, 0.15 mg/ml BSA was added as a carrier.
Binding reactions with purified Hsp15 for estimation of the KD value
Hsp15 was purified as described (Korber et al., 1999). Protein dilutions for binding experiments were done in association buffer. The mutant lysates were prepared in parallel to lysates of the isogenic wild type. Hsp15 was added to varying amounts of lysate in a final volume of 1 ml and the binding reaction was incubated on ice for 10 min and loaded directly onto the sucrose gradient. Protein concentrations were determined by extinction as in Korber et al. (1999). The concentration of 50S subunits in the fraction was determined by measuring the absorbance at 260 nm and the conversion factor of 1 A260 ≈ 36 pmol 50S subunits (Bommer et al., 1996).
Puromycin reaction
Puromycin (Sigma) was added at a concentration of 1 mM and the reaction incubated on ice for 45 min (Rheinberger et al., 1988).
In vitro translations
In vitro translations were done essentially as in Bommer et al. (1996) with tight coupled 70S ribosomes and poly(U) (Amersham Pharmacia) as mRNA. The magnesium concentration was lowered to 3 mM. Instead of the bulk tRNA, purified tRNAPhe was used and the S100 enzymes were made tRNA free as in Rheinberger et al. (1988). For the negative control, ATP, GTP, phosphoenolpyruvate and pyruvate kinase were replaced by water. After translation for 20 min at 37°C, the reaction was cooled on ice, diluted 3–fold with 20 mM HEPES pH 7.5, 150 mM NH4Cl, 4 mM β–mercaptoethanol to yield 1 mM Mg2+ dissociation conditions, and Hsp15 was added at a molar ratio of 0.5:1 to the ribosomes. The specific radioactivity was 40 d.p.m./pmol phenylalanine.
Heat shock induction
MG1655 cultures were grown at 30°C as described above. At an OD600 of 0.5, a 100 ml aliquot was withdrawn, incubated on ice for 50 min to allow run-off of the ribosomes, and harvested and lysed as above. The rest of the culture was transferred to a 41°C water bath with continuous shaking. At various time points, aliquots were withdrawn and treated as above. The lysates thus generated were loaded on sucrose gradients with association conditions.
Acknowledgments
Acknowledgements
We thank Bart Staker and Mark Saper for communication of the three-dimensional structure of Hsp15 prior to publication. We also thank Rachel Green and Gloria Culver in the laboratory of Harry Noller, UC Santa Cruz, as well as Greg Johannes and Ken Kuhn in the laboratory of Peter Sarnow, Stanford University, for technical advice and help with the analysis of sucrose gradients. Dan Herschlag, Stanford University, is gratefully acknowledged for setting up the ‘ribosome connection’ and for his advice and encouragement. We appreciate the support of Rainer Jaenicke, Universität Regensburg, Germany, for his continuous interest in the analysis of new heat shock proteins.
References
- Aravind L. and Koonin, E.V. (1999) Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol., 48, 291–302. [DOI] [PubMed] [Google Scholar]
- Bardwell J.C.A. and Craig, E.A. (1988) Ancient heat shock gene is dispensable. J. Bacteriol., 170, 2977–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. and Craig, E.A. (1994) Heat-shock proteins as molecular chaperones. Eur. J. Biochem., 219, 11–23. [DOI] [PubMed] [Google Scholar]
- Blattner F.R., et al. (1997)The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Bommer U.A., Burkhardt,N., Jünemann,R., Spahn,C.M.T., Triana-Alonso,F.J. and Nierhaus,K.H. (1996) Ribosomes and polysomes. In Graham,J. and Rickwoods,D. (eds), Subcellular Fractionation. A Practical Approach. IRL Press, Oxford, UK, pp. 271–301. [Google Scholar]
- Bremer H. and Dennis,P.P. (1996) Modulation of chemical composition and other parameters of the cell by growth rate. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1553–1569. [Google Scholar]
- Cate J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N. and Noller, H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Friedman H., Lu, P. and Rich, A. (1971) Temperature control of initiation of protein synthesis in Escherichia coli.J. Mol. Biol., 61, 105–121. [DOI] [PubMed] [Google Scholar]
- Gilbert W. (1963) Polypeptide synthesis in Escherichia coli. II. The polypeptide chain and S-RNA. J. Mol. Biol., 6, 389–403. [DOI] [PubMed] [Google Scholar]
- Gross C.A. (1996) Function and regulation of the heat shock response. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1382–1399. [Google Scholar]
- Kohara Y., Akiyama, K. and Isono, K. (1987) The physical map of the whole E.coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell, 50, 495–508. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Tatusov, R.L. and Galperin, M.Y. (1998) Beyond complete genomes: from sequence to structure and function. Curr. Opin. Struct. Biol., 8, 355–363. [DOI] [PubMed] [Google Scholar]
- Korber P., Zander, T., Herschlag, D. and Bardwell, J.C.A. (1999) A new heat shock protein that binds nucleic acids. J. Biol. Chem., 274, 249–256. [DOI] [PubMed] [Google Scholar]
- Lindahl L. (1975) Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol., 92, 15–37. [DOI] [PubMed] [Google Scholar]
- Mangiarotti G., Apirion, D., Schlessinger, D. and Silengo, L. (1968) Biosynthetic precursors of 30S and 50S ribosomal particles in Escherichia coli.Biochemistry, 7, 456–472. [DOI] [PubMed] [Google Scholar]
- Muller W., Keppner, W. and Rasched, I. (1986) Versatile kanamycin-resistance cartridges for vector construction in Escherichia coli.Gene, 46, 131–133. [DOI] [PubMed] [Google Scholar]
- Nierhaus K.H. (1991) The assembly of prokaryotic ribosomes. Biochimie, 73, 739–755. [DOI] [PubMed] [Google Scholar]
- Record M.T. Jr, Lohman, T.M. and de Haseth, P. (1976) Ion effects on ligand–nucleic acid interactions. J. Mol. Biol., 107, 145–158. [DOI] [PubMed] [Google Scholar]
- Rheinberger H.-J., Geigenmüller, U., Wedde, M. and Nierhaus, K.H. (1988) Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol., 164, 658–670. [DOI] [PubMed] [Google Scholar]
- Richmond C.S., Glasner, J.D., Mau, R., Jin, H. and Blattner, F.R. (1999) Genome-wide expression profiling in E.coli K-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T.J., Berman,M.L. and Enquist,L.W. (1984) Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Staker B.L., Korber, P., Bardwell, J.C.A. and Saper, M.A. (2000) Structure of Hsp15 reveals a nove RNA-binding motif. EMBO J., 19, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissières A., Watson, J.D., Schlessinger, D. and Hollingworth, B.R. (1959) Ribonucleoprotein particles from Escherichia coli.J. Mol. Biol., 1, 221–233. [Google Scholar]
- Traut R.R. and Monro, R.E. (1964) The puromycin reaction and its relation to protein synthesis. J. Mol. Biol., 10, 63–72. [DOI] [PubMed] [Google Scholar]