Abstract
We have solved the crystal structure of the heat shock protein Hsp15, a newly isolated and very highly inducible heat shock protein that binds the ribosome. Comparison of its structure with those of two RNA-binding proteins, ribosomal protein S4 and threonyl-tRNA synthetase, reveals a novel RNA-binding motif. This newly recognized motif is remarkably common, present in at least eight different protein families that bind RNA. The motif's surface is populated by conserved, charged residues that define a likely RNA-binding site. An intriguing pattern emerges: stress proteins, ribosomal proteins and tRNA synthetases repeatedly share a conserved motif. This may imply a hitherto unrecognized functional similarity between these three protein classes.
Keywords: heat shock proteins/protein–RNA interactions/protein structure/ribosome/X-ray crystallography
Introduction
The availability of complete genome sequences has ushered in the era of structural genomics: the solving of three-dimensional structures of proteins derived from open reading frames of unknown function (Sali, 1998). We focus on newly identified Escherichia coli heat shock proteins, attempting simultaneously to determine their function and tertiary structure.
Richmond et al. (1999) recently identified 77 heat shock loci by a sensitive genomic expression technique. We are studying the function and determining the three-dimensional structure of three of these newly identified heat shock proteins: Hsp15, Hsp33 and FtsJ (Jakob et al., 1999; Korber et al., 1999; H.Buegl, E.B.Fauman, B.L.Staker, F.-Z.Zheng, S.R.Kusher, M.A.Saper, J.C.A.Bardwell and U.Jakob, submitted). Here we present the X–ray crystal structure of the heat shock protein Hsp15. Hsp15 is an abundant, heat-inducible protein that binds nucleic acids in vitro. Hsp15 has been determined to be the fifth most highly induced heat shock protein on a genome-wide expression profiling of E.coli that identified 77 heat-inducible genes (Richmond et al., 1999). This makes it more highly induced than most of the well-studied heat shock genes including groEL, groES, dnaK, dnaJ, clpA, clpP, rpoD, rpoH and lon. It is the most highly induced of the genes that lack an assigned function. The dissociation constant for the non-specific binding of Hsp15 to nucleic acids is 4–20 μM, as determined by filter binding assays and quantitative zonal affinity chromatography (Korber et al., 1999). Hsp15 binds specifically and tightly to the free 50S ribosomal subunit with a sub-nanomolar dissociation constant (Korber et al., 2000). While most other heat shock proteins are molecular chaperones or proteases (for a review, see Gross, 1996), Hsp15 appears to be involved in ribosome recycling (see Korber et al., 2000). Our finding of a heat shock protein that functions at the RNA level, instead of at the protein level, opens up a new perspective on the heat shock response.
Hsp15 is highly conserved among eubacteria (Korber et al., 1999). Sensitive homology search programs allowed us to declare that Hsp15 defines a previously unrecognized but very widespread and ancient RNA-binding motif. This motif is present in at least 500 sequenced proteins, including the ribosomal protein S4 family, the 16S rRNA pseudouridine synthase family, the tyrosyl-tRNA synthetase family and an RNA methylase family that also includes heat-inducible members (Korber et al., 1999; H.Buegl, E.B.Fauman, B.L.Staker, F.-Z.Zheng, S.R.Kusher, M.A.Saper, J.C.A.Bardwell and U.Jakob, submitted). Similar conclusions were obtained independently by sequence analysis by Aravind and Koonin (1999). This RNA-binding motif usually occurs in a modular way in combination with other functional domains.
The 133-residue Hsp15 is the smallest member of this newly discovered RNA-binding superfamily and consists almost entirely of the RNA-binding motif. The Hsp15 crystal structure presented here and the comparison with the recently determined structure of the ribosomal S4 protein highlights the unique fold of this motif and the residues likely to be involved in RNA binding. Interestingly, a highly divergent but structurally homologous domain is also found in the structure of threonyl-tRNA synthetase.
Results
Structure determination
Hsp15 readily crystallized in solutions of ammonium sulfate, allowing its structure to be determined to 2.0 Å resolution (see Materials and methods and Table I). The Hsp15 structure was solved using standard multiple isomorphous replacement (MIR) techniques. Five different heavy atom combinations were used to make initial phase estimates and calculate experimental MIR electron density maps. Two heavy atom derivatives of the native protein were found using uranyl acetate and potassium platinate chloride (IV). The native Hsp15 protein contains no cysteine residues, a common binding site for heavy atoms. Thus, we introduced a cysteine residue at position 43, replacing a serine residue. This substitution was chosen because serine to cysteine replacements are rather conservative in nature and because this substitution is actually observed to occur at this position in the Hsp15 homologs present in Pseudomonas aeruginosa and Helicobacter pylori. These facts make it unlikely that this substitution will interfere substantially with the folding or stability of Hsp15. The S43C Hsp15 protein was expressed, purified and derivatized successfully by mercury chloride (II). A selenomethionine-derivatized Hsp15 protein was also expressed and crystallized in a manner similar to the native Hsp15. A fifth heavy atom derivative was generated by soaking a selenomethionine-derivatized crystal with uranyl acetate. The data collection and phasing statistics are listed in Table I. MIR electron density maps calculated to 2.2 Å with the five derivatives provided readily interpretable electron density. Although multiwavelength anomalous dispersion (MAD) experiments were attempted using the selenomethionine derivative, the electron density maps were of lesser quality than maps calculated using MIR techniques. The complete main chain from residue 4 to 110 was traced and side chains located. The final Rcryst and Rfree of the refined model were 22.6 and 28.6%, respectively, using all data in the resolution range 20.0–2.0 Å. Mass spectrometry indicates that crystals contain the complete intact protein of 133 residues (data not shown). The C-terminal 23 residues were never visible in electron density maps despite map improvement and refinement, and are presumably disordered, which may contribute to the observed R–factors. The statistics of the final model are shown in Table II.
Table I. Hsp15 data statisticsa.
| Derivative | Native | Uranyl acetate | HgCl2-S43C | Se-Met peak | PtCl4 | Se/U |
|---|---|---|---|---|---|---|
| Heavy atom | U | Hg | Se | Pt | Se/U | |
| Molarity (mM) | 1 | 5 | 2 | 1 | ||
| Length of soak (h) | 8 | 8 | 24 | 8 | ||
| No. of sites per asymmetric unitb | 6 | 4 | 4 | 7 | 9 | |
| Wavelength (Å)c | 1.5418 | 1.5418 | 1.5418 | 0.979314 | 1.5418 | 1.5418 |
| Unique reflections | 21 673 (1911) | 10 535 (999) | 15 032 (1258) | 39 889 (3424) | 12 291 (1121) | 7172 (640) |
| Resolution (Å) | 2.0 (2.07–2.00) | 2.5 (2.59–2.50) | 2.2 (2.28–2.50) | 2.0 (2.07–2.00) | 2.4 (2.49–2.40) | 2.8 (2.90–2.80) |
| Completeness (%) | 98.4 (88.9) | 92.3 (89.8) | 89.7 (76.4) | 96.1 (82.1) | 95.5 (91.4) | 87.6 (81.5) |
| Rsymd | 5.1 (35.3) | 8.9 (38.0) | 7.0 (37.1) | 9.6 (35.0) | 5.0 (30.7) | 8.3 (38.4) |
| Rmergee | 21.3 | 26.2 | 11.8 | 22.0 | 12.8 | |
| Rcullisf | 0.84 | 0.80 | 0.86 | 0.87 | 0.85 | |
| Phasing powerg | 1.22 | 1.35 | 0.78 | 1.03 | 1.00 |
aNumbers in parentheses represent the highest resolution shell of the data.
bEach asymmetric unit contains two Hsp15 molecules.
cData collected on RAXIS-IV detector. Se-Met data were collected at IMCA-CAT, Advanced Photon Source, Argonne National Laboratory.
dRsym = Σ|Ii – Im |/ΣIm where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
eRmerge = Σ|FPH – FP|/Σ|FPH| where FPH is the derivative observed structure factor and FP is the native structure factor.
fRcullis = Σ|(FPH ± FP) – FH(calc)|/Σ|FPH – FP|
gPhasing power = FH/ERMS where ERMS is the residual lack of closure.
Table II. Refinement statisticsa.
| Resolution (Å) | 20.0–2.0 (2.07–2.00) |
| No. of reflections | 20 776 (1729) |
| Reflections used in Rfree | 2062 (178) |
| No. of protein atoms | 1637 |
| No. of solvent atoms | 137 |
| No. of sulfate ions | 5 |
| R-factor | 22.6 (29.4) |
| Rfree | 28.6 (31.9) |
| R.m.s.d. from ideal stereochemistry | |
| bond lengths (Å) | 0.016 |
| bond angles (°) | 1.608 |
| dihedrals (°) | 23.98 |
| impropers (°) | 1.025 |
| Mean B-factor—all atoms (Å3) | 40.9 |
| Ramachandran plot | |
| residues in most favored regions (%) | 89.7 |
| residues in additionally allowed regions (%) | 10.3 |
aNumbers in parentheses represent the highest resolution shell of the data.
Structure description
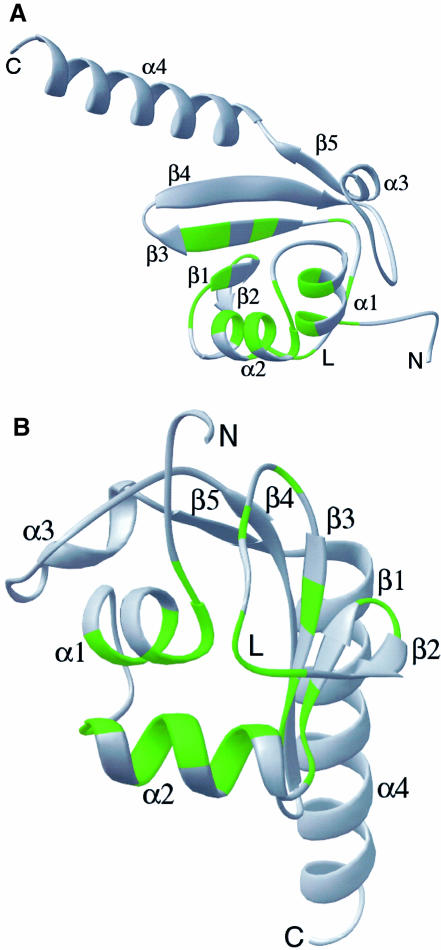
The three-dimensional structure of Hsp15 is comprised of an α+β domain (residues 1–88) followed by a 22-residue α–helix (α4) that projects into the solvent (Figure 1A). The N–terminal helices α1 and α2 are connected by a five-residue loop and are antiparallel (Figure 1B). The polypeptide continues into the first four strands of an antiparallel β-sheet that lies below and approximately perpendicular to α1. Loops between strands β1 and β2, and between β3 and β4, are β–hairpins, while the nine residue loop (residues 43–52) between β2 and β3 is a distinctive L-shaped meander. Strand β4 is followed by helix α3, the short β5 strand and the α4 helix. The final residues of this helix have high temperature factors, suggesting a mobile structure.
Fig. 1. Ribbon representation of Hsp15 colored by conservation. (A) Forty-three eubacterial members of the Hsp15 family were identified by a BLAST search of the non-redundant database and the unfinished microbial databases in July, 1999 (Altschul et al., 1997). Green residues are identical or contain conservative substitutions in at least 50% of the sequences. Residues 4–110 are shown. The portion of the αL loop is labeled with an L. (B) As (A), but oriented with the proposed RNA-binding αL motif in front. This is the orientation of Hsp15 used in all subsequent figures.
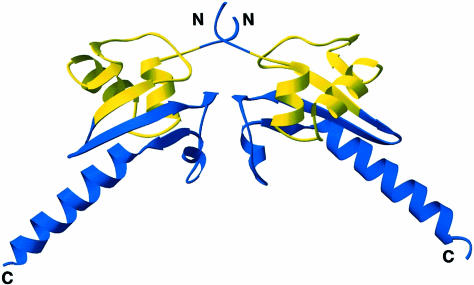
In the crystal's asymmetric unit, two Hsp15 molecules contact each other and bury ∼790 Å2 of solvent-accessible surface (Figure 2). The loop following β4 forms an antiparallel β–bridge with the same loop of the dyad-related molecule. The remainder of the interface is comprised of van der Waals contacts between the non-polar side chains of residues V7, V9, L51, A70, T72 and L83. Helix α4 also makes contacts with the α4 helices from other non-crystallographic and crystallographic symmetry-related molecules. The apparent dimer may be an artifact of crystallization. The non-polar residues at the primary interface are only conserved in a subset of the eubacterial Hsp15 homologs. Furthermore, gel filtration suggests that Hsp15 is a monomer in solution, and binding studies to ribosomal subunits suggest a stoichiometry of 0.7:1 (Korber et al., 1999, 2000).
Fig. 2. Ribbon diagram of non-crystallographic symmetry mates. Residues 9–57 are colored yellow and represent the proposed αL RNA-binding motif, the remaining residues are in blue. The secondary structure was identified and the drawing rendered using RIBBONS (Carson, 1997).
Location of conserved residues: delineation of the αL motif
Figure 1 highlights in green the locations of Hsp15 residues that are identical or structurally similar in 50% or more of the 39 sequenced Hsp15 homologs. These residues are especially abundant in the region from residue 9 to 57. This region, extending from α1 through β3, forms a distinct structural motif defined here as the αL motif, because of the two α-helices and the L-shaped loop located between β2 and β3 (Figure 1B). Buried and conserved residues populate the interface between α1, α2 and the L loop in the vicinity of residue 45 and probably influence the folding and structural integrity of the αL motif (Figure 1A and B). These residues include L11, L15, I31, V36, V49, L55 and L57. Other conserved residues form turns between secondary structure elements: G34 forms a turn between α2 and β1, N39 and G40 form a tight reverse turn between β1 and β2. P45 and S46 are located at the base of the L-shaped loop and form part of the interface with α1 and α2.
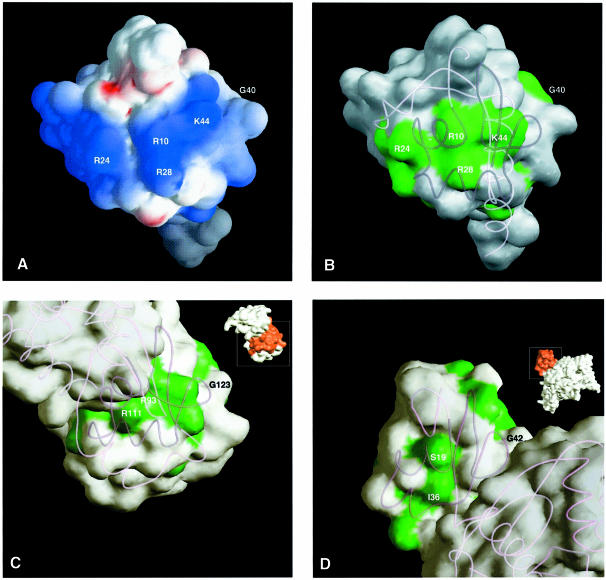
Conserved and solvent-accessible protein residues are often involved in interactions with ligands. Figure 3 highlights in green those residues that are conserved in all Hsp15 homologs and that are also surface exposed. Side chains from five of these residues cluster to form a positively charged surface on the αL domain. K22, R24 and R28 are on adjacent turns of the α2 helix (see Figure 4). Nearby is the side chain of R10 on α1, which is positioned by a salt bridge to the conserved D12. K44 is on the L-shaped loop near the two helices. That positively charged side chains are accessible and clustered near each other is consistent with the hypothesis that the αL motif is directly involved in nucleic acid binding.
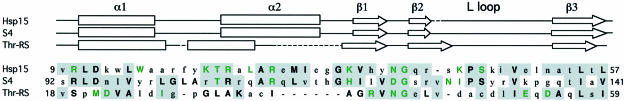
Fig. 3. Structure-based sequence alignment of the proposed αL RNA-binding motif in heat shock protein 15 kDa (Hsp15), ribosomal protein S4 (S4) and threonyl-tRNA synthetase (Thr-RS). Only the portion of each protein sequence that is structurally similar is shown. The secondary structures as identified by RIBBONS (Carson, 1997) are shown above the sequence alignments. Residues that are >50% conserved or identical within each family are shown in bold upper case letters. Residues in bold green upper case letters are both conserved in at least 50% of the members of each family, and are solvent accessible as determined by XPLOR (Brünger, 1997). Residues that are similar between at least two of the three structures are shaded gray.
Fig. 4. Conservation of surface-exposed residues in the αL motif. (A) Surface electrostatic potential of Hsp15 orientated to show the proposed RNA-binding αL motif facing outwards. This is the orientation identical to that in Figure 1B. The extreme ranges of red (negative) and blue (positive) represent electrostatic potentials of less than –9 to greater than +9 kbT, where kb is the Boltzmann constant and T is the temperature. The figure was calculated with GRASP (Nicholls et al., 1991) showing the accessible surface. (B) Surface-exposed residues of Hsp15 that are conserved within the Hsp15 family are shown in green. (C) Surface-exposed residues of ribosomal protein S4 that are conserved within the S4 family are shown in green. Inset is a diagram of the complete S4 protein with the domain homologous to Hsp15 highlighted in red. (D) Surface-exposed residues of threonyl-tRNA synthetase that are conserved within the threonyl-tRNA synthetase family are shown in green. Inset is a diagram of the complete threonyl-tRNA synthetase protein with the domain homologous to Hsp15 highlighted in red.
There are three additional highly conserved residues in Hsp15: L26, M30 and K35. These lie on the opposite face of α2, in comparison with the conserved arginines that are presumed to be involved in RNA binding. These additional residues point out from the interface of α1, α2 and the L-shaped loop and are primarily solvent exposed in Hsp15. Equivalent residues in the S4 structure are involved in protein–protein interactions between the two domains of S4. These residues in Hsp15 may be involved in interactions with protein components of the ribosome.
Structural homology to ribosomal protein S4
Use of the PSI BLAST program (Altschul et al., 1997) reveals that >500 proteins contain sequences similar to the αL motif of Hsp15. These proteins are found in both prokaryotes and eukaryotes. They include the ribosomal protein S4 and eukaryotic homolog S9, 16S rRNA pseudouridine synthase, tyrosyl-tRNA synthetase and an RNA methylase family, a member of which, like Hsp15, is heat inducible (Aravind and Koonin, 1999; Korber et al., 1999; H.Buegl, E.B.Fauman, B.L.Staker, F.-Z.Zheng, S.R.Kusher, M.A.Saper, J.C.A.Bardwell and U.Jakob, submitted). All of these protein families bind RNA. To assess whether the αL motif is folded similarly in these proteins, we compared the structure of Hsp15 with the structure of ribosomal protein S4 from Bacillus stearothermophilus (Davies et al., 1998). S4 is a two-domain protein that binds to 16S rRNA.
Main chain atoms of the first 50 residues of the S4 second domain (residues 92–141) superpose on Hsp15 (residues 9–57) with a root mean square deviation (r.m.s.d.) of 1.8 Å (Figure 5). Of the 24 positions with conserved residues in Hsp15 family members, 20 of the structurally equivalent residues are also conserved within the S4 family (Figure 3). Most of these residues are also similar between the two families. S4 conserved residues that are solvent accessible include R93, T106, R111, H118, D122 and G123 (Figure 4). These are likely candidates for forming an RNA-binding surface. Of these, R93 and R111 in S4 (R10 and R28 in Hsp15) are absolutely conserved in all members of the Hsp15 and S4 families (Figure 4A and B). In the S4 crystal structure, these two residues coordinate a sulfate that may mimic a phosphate group on the backbone of a bound RNA (Davies et al., 1998). Hsp15 also has a sulfate bound to the structurally equivalent residues.
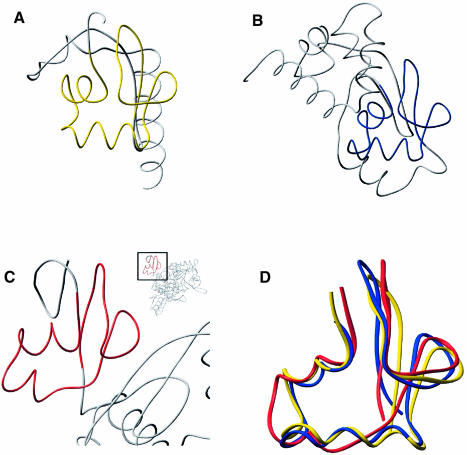
Fig. 5. The αL motif in three different protein structures. The peptide backbones of three structures, ribosomal protein S4, Hsp15 and threonyl–tRNA, are compared. The region highlighted in color is the αL motif that is shared by all three proteins. (A) Hsp15 with its αL motif highlighted in yellow. (B) Ribosomal protein S4 with its αL motif highlighted in blue. (C) Threonyl-tRNA synthetase with its αL motif highlighted in red. (D) Overlay of residues 9–57 of Hsp15 (yellow), 92–141 of ribosomal protein S4 (blue) and 18–59 of threonyl-tRNA synthetase (red).
In summary, Hsp15 and S4 bind RNA but have different biological functions. Each has a similarly folded αL motif with a comparable patch of surface-exposed residues. The same residues in S4 have been implicated in RNA binding. These observations suggest that the αL motif provides a module for RNA binding in a large number of protein families.
Structural homology to threonyl-tRNA synthetase
We were interested in whether the αL motif of Hsp15 is structurally homologous to other proteins in addition to the >500 proteins shown to be homologous at the sequence level. We searched a non-redundant subset of structures from the Protein Data Bank with residues 9–57 of the Hsp15 structure corresponding to the αL motif (DALI server; Holm and Sander, 1993). The only statistically significant fold detected (Z = 2.5) was from the N1 domain of E.coli threonyl-tRNA synthetase (PDB code 1qf6; Sankaranarayanan et al., 1999). Residues 18–59 of threonyl-tRNA synthetase superposed on residues 9–57 of Hsp15 (Figure 5D).
This structural similarity is remarkable in light of the extremely limited sequence homology between threonyl-tRNA synthetase and Hsp15 or S4. A structure-based sequence alignment of the αL regions of Hsp15, S4 and threonyl-tRNA synthetase revealed only two identities between structurally equivalent residues (Figure 3). N56 and G57 of threonyl-tRNA synthetase line up precisely with N39 and G40 of Hsp15. In structures of Hsp15, the S4 protein and threonyl-tRNA synthetase, these surface-exposed residues form a reverse turn between β1 and β2 of the αL motif. Furthermore, these two residues are highly conserved between all seven families that contain the αL motif (Aravind and Koonin, 1999; Korber et al., 1999).
Despite low sequence identity, 18 other residues of the 42 residue αL region in the threonyl-tRNA synthetase N1 domain are similar to the equivalent residues in either Hsp15 or S4. Many of these residues are buried: V18, V23, I27, V40, I50, I51, L57 and I59. This limited conservation is apparently sufficient to maintain the αL motif in threonyl-tRNA synthetase.
Discussion
The structure of a new RNA-binding motif: αL
Recently, two groups have independently identified a new RNA-binding sequence motif that is shared by >500 proteins, from at least seven protein families (Aravind and Koonin, 1999; Korber et al., 1999). In six of these families, the motif is combined with other functional domains, e.g. a methylase domain, and appears to provide the RNA-binding site. The seventh family is the heat shock protein Hsp15 (Korber et al., 1999) and consists almost entirely of just the domain containing the motif. Thus, Hsp15 provides a clearly defined model system for the study of this widespread RNA-binding motif.
We have solved the crystal structure of Hsp15 to 2.0 Å and identified the conserved motif in structural terms. We call it the αL motif as it is composed of an α–helical part in conjunction with a distinctively L-shaped loop. Comparison of the Hsp15 structure with the two recently solved structures of the ribosomal protein S4 and the threonyl-tRNA synthetase (Davies et al., 1998; Sankaranarayanan et al., 1999) permitted us to identify the potential RNA-binding surface shared by these three protein families.
The RNA-binding surface of the αL motif
Hsp15 binds non-specifically with micromolar dissociation constants to different kinds of nucleic acids (Korber et al., 1999) and specifically with a nanomolar dissociation constant to the free 50S subunit of the ribosome (see Korber et al., 2000). Phylogenetic analysis of Hsp15 open reading frames found in the sequence databases (Korber et al., 1999) has revealed a number of highly conserved residues in the Hsp15 family. The conserved residues from Hsp15 cluster in the interface of α1 and α2 and the L loop, forming the αL motif (Figures 1 and 3). This motif is found in other RNA-binding proteins including two whose structure is known (Figure 5). Surface-exposed residues conserved in each family cluster in the same way for all three proteins to form a similar patch (Figure 4). This patch is probably a binding site for the RNA substrate.
Role of the αL motif in ribosomal protein S4
The ribosomal protein S4 belongs to the group of 16S rRNA-binding proteins that act to initiate the assembly of the 30S ribosomal subunit (Mizushima and Nomura, 1970). The S4 protein also autoregulates its own expression by binding to the mRNA of the α–operon (Dean and Nomura, 1980). Extensive studies have tried to elucidate how and what parts of the S4 protein specifically recognize these two different RNA substrates, but the region involved has remained imprecisely defined and spans large portions of the protein (Baker and Draper, 1995). Even determination of the structure for the S4 protein (Davies et al., 1998; Markus et al., 1998) did not fully resolve the long-standing question of how the S4 protein interacts with RNA. It appeared that the RNA-interacting residues are distributed over two domains, roughly centered around the domain interface, confined to one face of the elongated molecule (Davies et al., 1998). Each domain was suggested to have homology to DNA-binding motifs; domain 1 showed weak homology to the helix–turn–helix (HTH) motif of the Tet repressor and domain 2 showed some similarity to the ETS DNA-binding domain. Closer analysis, however, discouraged the use of these homologies to model the S4 interaction with RNA (Davies et al., 1998).
The identification of the αL motif along with its conserved surface-exposed regions provides a clearer model to delineate the RNA-binding region of domain 2 of the ribosomal protein S4. As expected from the sequence alignments, the structure of residues 9–57 of Hsp15, defining the αL motif, is structurally homologous to the region 92–141 of the S4 protein (Figures 3–5). This region is at the beginning of the second domain of the S4 protein and near the domain interface. It contains the two highly conserved arginine residues R93 and R111 which Davies et al. (1998) proposed as the putative RNA-binding site. Markus et al. (1998) already noted the highest degree of conservation within the S4 protein family in this region, and proposed that the RNA-binding site may be located here. Deletion studies on the S4 protein have shown that residues 48–104 contain the core of the RNA-binding site and are responsible for specific recognition of 16S rRNA (Conrad and Craven, 1987). Residues 48–177 are necessary to provide specificity for binding mRNA and to promote proper assembly of 30S subunits (Conrad and Craven, 1987; Baker and Draper, 1995). It was speculated that a C–terminal region after residue 104 provides important RNA contacts. This supports our proposal that the contacts to RNA are made by the αL motif of the second domain of S4 protein (92–141), which would have been disrupted if S4 were truncated at position 104. It seems very likely that the αL motif forms the core of the RNA-binding site of domain 2.
A function for the N1 domain of threonyl-tRNA synthetase
The αL motif of Hsp15 is also structurally homologous to the N1 or first N-terminal domain of the threonyl-tRNA synthetase (Figure 5). This structural similarity is surprisingly high given the limited sequence similarity (Figure 3). Determinants for the αL motif fold have been conserved, but surface features have diverged greatly from the αL motifs in proteins identified by sequence homology to Hsp15. This suggests that additional proteins which contain an αL motif await discovery. The very distant relationship of the primary structures is reflected in the increased deviation of the tertiary structures; see, for instance, the region of helix α2 (Figure 5D).
The N-terminal extension is a common feature of threonyl-tRNA synthetases, although its function remains unclear (Freist and Gauss, 1995). Threonyl-tRNA synthetase binds to tRNAThr and couples it to a threonyl residue, but also binds to its own mRNA to autoregulate its own expression (Moine et al., 1990). Deletion of both N-terminal domains N1 and N2 has no effect on tRNA or mRNA binding but does reduce the efficiency of threonyl-tRNA synthetase in regulating protein synthesis. Sankaranarayanan et al. (1999) have hypothesized that the N-terminal domain may bind to the 30S ribosomal subunit and hinder it from binding to the mRNA initiation site. Such repression of translation, by the so-called entrapment model (Draper, 1987), is supported further by our identification of the αL motif in the N1 domain of threonyl-tRNA synthetase. Ribosomal protein S4 and Hsp15 bind with very high affinity to rRNA in ribosomal subunits (Draper and Reynaldo, 1999; Korber et al., 2000). By analogy, the N1 domain of threonyl-tRNA synthetase may interact with the 16S rRNA on the ribosomal subunit. However, since the N1 domain of threonyl-tRNA synthetase lacks many of the positively charged surface residues conserved in S4 and Hsp15, it might not ligate the rRNA directly, but instead interact with nearby protein subunits of the ribosome. Alternatively, the αL motif of threonyl-tRNA synthetase may bind to rRNA but in a different manner, without ionic interactions to phosphate groups. These models remain to be tested experimentally.
A preview of the C-terminal domain of tyrosyl-tRNA synthetase
Hsp15 is also homologous to a portion of another tRNA synthetase, tyrosyl-tRNA synthetase. The αL motif sequence homology is located in the C-terminal domain of tyrosyl-tRNA synthetase (Markus et al., 1998; Aravind and Koonin, 1999; Korber et al., 1999). R351 of the B.stearothermophilus tyrosyl-tRNA synthetase is in this domain and was directly implicated in tRNA binding (Bedoulle and Winter, 1986). It aligns with the highly conserved and surface-exposed R28 of Hsp15, which we suggest forms part of Hsp15's RNA-binding surface. The crystal structure of tyrosyl-tRNA synthetase was solved but, unfortunately, the C-terminal domain was disordered (Brick et al., 1989). We propose, based upon the clear sequence homology to Hsp15, that this C–terminal region contains an αL motif. The αL motif therefore presents a preview of the not yet crystallographically resolved portion of the tyrosyl-tRNA synthetase structure, as well as the homologous portion of the structures of pseudouridine tRNA synthase and all the other ∼500 proteins related in sequence to Hsp15.
Comparison with other RNA- and ribosome-binding proteins
RNA-binding proteins are a diverse group of proteins whose structures vary widely from each other. Proteins have clearly evolved multiple strategies to interact with RNA. However, there are some generalizations that can be made to help to understand the RNA-binding mechanisms of proteins. Many RNA-binding proteins use independent globular domains of 60–90 residues to interact with RNA. One common property of these small RNA-binding domains is that they are often α+β proteins composed of a β–sheet on one face of the protein supporting a network of α–helices on the other face (Varani, 1997). The most prevalent of these are the ribonucleoprotein, K–homology and double-stranded RNA-binding domains. Additionally, a concentration of conserved basic amino acids is usually present on the surface of the RNA interaction face (Varani, 1997; Patel, 1999).
Hsp15 has a number of properties in common with these proteins. The primary globular domain of Hsp15 is a 90 residue α+β structure that contains a highly conserved RNA-binding motif of 49 amino acids rich in arginines at the proposed RNA-binding surface. However, the topology of Hsp15 is different from other families of RNA-binding proteins in that the β–sheet is composed of parallel β–strands, in contrast to antiparallel strands found in other RNA-binding proteins (Cusack, 1999; Draper and Reynaldo, 1999). The two helices implicated in RNA binding on Hsp15 are reminiscent of DNA-binding homeodomain proteins, a characteristic also noticed for another RNA-binding protein, ribosomal protein L11 (Xing and Draper, 1996; Draper and Reynaldo, 1999). However, the highly conserved L–shaped loop positioned near the interface of these two homeodomain-like helices in Hsp15 is unique. The structure of Hsp15 or homolog in a complex with RNA substrate will illuminate the precise RNA-binding mechanism utilized by these proteins and whether the L–loop interacts directly with RNA or is necessary for stabilizing the RNA-binding surface.
Extended αhelix
The native Hsp15 structure is primarily a single compact globular domain with a very long α–helix extending out from the protein (Figure 1A). The electron density ends at approximately residue 110, leaving 23 residues unaccounted for in the electron density. Mass spectrometry of the crystals reveals completely intact protein of ∼133 amino acids, suggesting that the unobserved residues are present in the crystal lattice but not visible in the electron density.
Isolated long helices such as this are uncommon in solved structures; however, comparison with other ribosomal proteins shows a high percentage of long finger-like exten– sions pointing away from the main globular domain (Ramakrishnan and White, 1998). We speculate that the C-terminal 23 residues of Hsp15 become ordered upon binding to the RNA substrate and may confer additional contacts in addition to the specific αL motif. Wang and Schimmel (1999) have shown that addition of a non-specific RNA-binding domain can enhance the binding affinity of a specific RNA-binding interaction. We note that some members of the Hsp15 family lack this C–terminal domain (Korber et al., 1999).
Common threads of ancient RNA-binding folds
We have shown that the αL motif identified in a stress protein (Hsp15) is present in a ribosomal protein (S4) and two tRNA synthetases. Interestingly, two other ribosomal proteins have now been shown to have an RNA-binding motif that is also present in a stress protein and a tRNA synthetase. The ribosomal protein L25 shares an RNA-binding motif unrelated to the αL motif with the general stress protein CTC and with the glutaminyl-tRNA synthetase (Stoldt et al., 1998; and references therein). This led Stoldt et al. (1998) to hypothesize an evolutionary relationship between these three protein families. Still another unrelated RNA-binding fold, the OB-fold, is present in the ribosomal proteins S1 and S17, in the aspartyl-, lysyl- and phenylalaninyl-tRNA synthetases, and in the cold shock proteins (Murzin, 1993; Bycroft et al., 1997; and references therein).
Our discovery of a homologous structural motif present in the ribosomal protein S4, the heat shock protein Hsp15 and the threonyl- and tyrosyl-tRNA synthetases represents the third such linking of ribosomal proteins, stress proteins and tRNA synthetases. This unexpected finding provides further evidence for an ancient relationship between these three classes of proteins. RNA was thought to be one of the first biological macromolecules. In an RNA world, RNA binding would clearly be an essential feature for early proteins, and RNA-binding motifs would be one of the first building blocks. Today, members of ancient protein families still share these building blocks as structural scaffolds. The substrate specificity has changed somewhat; nonetheless, it is noteworthy that the interaction with the ribosome is a common thread. The structural or sequence features of the RNA substrates recognized by these proteins are not sufficiently well characterized to determine what they may have in common.
We have discovered an evolutionarily ancient motif that binds RNA in a heat shock protein. This implies that the heat shock response acts not only at the protein level, as exemplified so far by molecular chaperones and proteases, but also at the RNA level (Korber et al., 1999, 2000). It may be that RNA-binding stress proteins represent a more ancient mechanism of cell machinery protection.
In conclusion, the structure of Hsp15 reveals a new RNA-binding fold, termed the αL motif. This motif is remarkably common, present in at least eight different families of RNA-binding proteins including ribosomal protein S4 and threonyl- and tyrosyl-tRNA synthetases. Comparison of these structurally diverse proteins provides further insight into what regions are involved in binding RNA. Stress proteins, ribosomal proteins and tRNA synthetases often share a conserved motif. This is further evidence that RNA-binding motifs are ancient, and may imply a hitherto unrecognized functional similarity between these three protein classes.
Materials and methods
Crystallization and data collection
Hsp15 was purified as described previously (Korber et al., 1999). Crystallization conditions were screened with Crystal Screens I and II (Hampton Research; Jancarik and Kim, 1991). Diffraction quality crystals grew in hanging drops that contained equal volumes of Hsp15 (11 mg/ml in 20 mM NaCl, 1 mM EDTA, 50 mM HEPES pH 7.0) and precipitant (2.0 M ammonium sulfate, 0.1 M sodium acetate pH 4.6, 10% glycerol). Drops were equilibrated by microvapor diffusion against 1 ml of the same precipitant. Crystals appeared in 2–3 days and typically grew to a size of 0.6 mm × 0.4 mm × 0.2 mm after 1 week. Crystals belong to space group P21212, have cell dimensions a = 113.1 Å, b = 61.9 Å and c = 41.1 Å, and diffract to at least 2.0 Å resolution.
Prior to data collection, crystals were transferred to precipitant solution containing 25% glycerol for 5 min. Crystals were frozen directly in a –180°C cold nitrogen stream (X-Stream; Molecular Structure Corp.). X–ray diffraction intensities were measured on a Rigaku RAXIS–IV phosphoimaging plate detector with a rotating anode X–ray source (Rigaku RUH3R) operating at 50 kV and 100 mA and equipped with focusing mirrors (Molecular Structure Corp). Diffraction images (20 min exposure, 1° crystal rotation, crystal–detector distance = 120 mm) were indexed and reflections integrated with the HKL package (Otwinowski, 1993). Data collection and phasing statistics are summarized in Table I.
The selenomethionine derivative of Hsp15 was prepared by growing the overexpression strain B834(DE3) pTHZ25 in M63 minimal media supplemented with 300 μg/ml ampicillin and all amino acids at 40 μg/ml except that 50 μg/ml dl–selenomethionione was used instead of methionine. Cells were grown to an OD600 of 0.6 and Hsp15 expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to 1 mM. Cells were harvested 3 h later and the Hsp15 protein was purified as previously described (Korber et al., 1999). Diffraction data from selenomethionine-containing crystals were collected on a Bruker 2K CCD installed on the 17–1D beamline IMCA at the Advanced Photon Source, Argonne National Laboratories. Images were processed with the HKL2000 package.
Structure determination
Observed structure factors from five heavy atom derivatives were included in MIR calculations using SCALEIT and MLPHARE (CCP4, 1994; Dodson et al., 1997). A mercury chloride derivative was prepared by substituting a cysteine for serine at residue 43 by oligo-directed mutagenisis. This conservative substitution is actually observed in the Hsp15 homologs present in P.aeruginosa and H.pylori. Electron density maps were solvent flattened with DM assuming a solvent content of 40%. The overall figure of merit of the experimental phases calculated to 2.2 Å was 0.54 and 0.65 after solvent flattening to 2.2 Å. Phasing statistics are summarized in Table I.
Residues 4–110 were traced in the electron density maps and modeled with O (Jones et al., 1991). Two Hsp15 molecules in the asymmetric unit, 137 water molecules, five sulfate ions and a bulk solvent contribution were refined in XPLOR (Brünger and Rice, 1997) and CNS (Brünger et al., 1998). The two non-crystallographic symmetry (NCS)-related molecules were restrained to each other throughout the refinement. During the final stages of refinement, the NCS restraints were removed. The angle of the final helix, residues 93–110, with respect to the rest of the structure varies by ∼2° between the two molecules in the asymmetric unit. The final Rcryst and Rfree of the refined model were 22.6 and 28.6%, respectively, for all data in the resolution range 20.0–2.0 Å. Coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb/), PDB code 1dm9.
Acknowledgments
Acknowledgements
We thank Hans Buegl for help with protein purification, D.Draper for providing the coordinates of the ribosomal protein S4 crystal structure, and Andy Howard and colleagues for their generous help collecting data at the Industrial Macromolecular Crystallography Association Collaborative Access Team facility, beamline 17-ID, Advanced Photon Source. This work was supported, in part, by NIH Grants to M.A.S. and J.C.A.B. Additional support came from a BMBF (Bundesministerium für Bildung und Forschung, Germany) grant to J.C.A.B., a scholarship of the Studienstiftung des deutschen Volkes to P.K., Pew Scholar in Biomedical Sciences awards to M.A.S. and J.C.A.B. and an NIH training grant to B.L.S. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
References
- Altschul S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. (1999) Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol., 48, 291–302. [DOI] [PubMed] [Google Scholar]
- Baker A.-M. and Draper, D.E. (1995) Messenger RNA recognition by fragments of ribosomal protein S4. J. Biol. Chem., 270, 22939–22945. [DOI] [PubMed] [Google Scholar]
- Bedouelle H. and Winter, G. (1986) A model of synthetase/transfer RNA interaction as deduced by protein engineering. Nature, 320, 371–372. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat, T.N. and Blow, D.M. (1989) Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J. Mol. Biol., 208, 83–98. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1997) Free R value: cross-validation in crystallography. Methods Enzymol., 277, 366–396. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. and Rice, L.M. (1997) Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol., 277, 243–269. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998)Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Hubbard, T.J.P., Proctor, M., Freund, S.M.V. and Murzin, A.G. (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Conrad R.C. and Craven, G.R. (1987) A cyanogen bromide fragment of S4 that specifically rebinds 16S RNA. Nucleic Acids Res., 15, 10331–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S. (1999) RNA–protein complexes. Curr. Opin. Struct. Biol., 9, 66–73. [DOI] [PubMed] [Google Scholar]
- Davies C., Gerstner, R.B., Draper, D.E., Ramakrishnan, V. and White, S.W. (1998) The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA binding motif. EMBO J., 17, 4545–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. and Nomura, M. (1980) Feedback regulation of ribosomal protein gene expression in Escherichia coli.Proc. Natl Acad. Sci. USA, 77, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson E.J., Winn, M. and Ralph, A. (1997) Collaborative Computation Project, Number 4: providing programs for protein crystallography. Methods Enzymol., 277, 621–633. [DOI] [PubMed] [Google Scholar]
- Draper D.E. (1987) Translational regulation of ribosomal proteins in Escherichia coli. In Ilan, J. (ed.), Translational Regulation of Gene Expression. Plenum Press, New York, NY, pp. 1–26. [Google Scholar]
- Draper D.E. and Reynaldo, L.P. (1999) RNA binding strategies of ribosomal proteins. Nucleic Acids Res., 27, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freist W. and Gauss, D.H. (1995) Threonyl-tRNA synthetase. Biol. Chem., 376, 213–224. [PubMed] [Google Scholar]
- Gross C.A. (1996) Function and regulation of the heat shock response. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1382–1399. [Google Scholar]
- Holm L. and Sander, C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jakob U., Muse, W., Eser, M. and Bardwell, J.C.A. (1999) Chaperone activity with a redox switch. Cell, 96, 341–352. [DOI] [PubMed] [Google Scholar]
- Jancarik J. and Kim, S.H. (1991) Sparse matrix sampling: a screen method for crystallization of proteins. J. Appl. Crystallogr., 24, 409–411. [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Korber P., Zander, T., Herschlag, D. and Bardwell, J.C.A. (1999) A new heat shock protein that binds nucleic acids. J. Biol. Chem., 274, 249–256. [DOI] [PubMed] [Google Scholar]
- Korber P., Stahl, J.M., Nierhaus, K.H. and Bardwell, J.C.A. (2000) Hsp15: a ribosome-associated heat shock protein. EMBO J., 19, 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus M.A., Gerstner, R.B., Draper, D.E. and Torchia, D.A. (1998) The solution structure of ribosomal protein S4 D41 reveals two subdomains and a positively charged surface that may interact with RNA. EMBO J., 17, 4559–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S. and Nomura, M. (1970) Assembly mapping of 30S ribosomal proteins from E.coli.Nature, 226, 1214. [DOI] [PubMed] [Google Scholar]
- Moine H., Romby, P., Springer, M., Grunberg-Manago, M., Ebel, J.-P., Ehresmann, B. and Ehresmann, C. (1990) Escherichia coli threonyl-tRNA synthetase and tRNAThr modulate the binding of the ribosome to the translational initiation site of the ThrS mRNA. J. Mol. Biol., 216, 299–310. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Data collection and processing. In Sawyer,L., Isaacs,N. and Bailey,S. (eds), Proceedings of the CCP4 Study Weekend. SERC Daresbury Laboratory, Warrington, UK, pp. 56–62. [Google Scholar]
- Patel D.J. (1999) Adaptive recognition in RNA complexes with peptides and protein modules. Curr. Opin. Struct. Biol., 9, 74–87. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. and White, S.W. (1998) Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biol. Sci., 23, 208–212. [DOI] [PubMed] [Google Scholar]
- Richmond C.S., Glasner, J.D., Mau, R., Jin, H. and Blattner, F.R. (1999) Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A. (1998) 100,000 protein structures for the biologist. Nature Sruct. Biol., 5, 1029–1032. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon, A.-C., Romby, P., Caillet, J., Springer, M., Rees, B., Ehresmann, C., Ehresmann, B. and Moras, D. (1999) The structure of threonyl-tRNA synthetase–tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell, 97, 371–381. [DOI] [PubMed] [Google Scholar]
- Stoldt M., Wöhnert, J., Görlach, M. and Brown, L.R. (1998) The NMR structure of Escherichia coli ribosomal protein L25 shows homology to general stress proteins and glutaminyl-tRNA synthetases. EMBO J., 17, 6377–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G. (1997) RNA–protein intermolecular recognition. Acc. Chem. Res., 30, 189–195. [Google Scholar]
- Wang C.-C. and Schimmel, P. (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Xing Y. and Draper, D.E. (1996) The RNA binding domain of ribosomal protein L11 is structurally similar to homeodomains. Biochemistry, 35, 1581–1588. [DOI] [PubMed] [Google Scholar]