Abstract
The genetic heterogeneity of the nitrite reductase gene (nirK and nirS) fragments from denitrifying prokaryotes in a non-agricultural forest soil in Thailand was investigated using soil samples from the Plant Germplasm-Royal Initiation Project area in Kanchanaburi Province, Thailand. Soil bacteria were screened for denitrification activity and 13 (from 211) positive isolates were obtained and further evaluated for their ability to reduce nitrate and to accumulate or reduce nitrite. Three species with potentially previously unreported denitrifying activities were recorded. Analysis of the partial nirK and nirS sequences of these 13 strains revealed a diverse sequence heterogeneity in these two genes within the same environment and even potentially within the same host species, the potential existence of lateral gene transfer and the first record of both nirK and nirS homologues in one bacterial species. Finally, isolates of two species of bacteria (Corynebacterium propinquum and Micrococcus lylae) are recorded as denitrifiers for the first time.
Keywords: Denitrifying bacteria, Genetic diversity, Nitrite reductase gene, Nitrite reductase ability
Introduction
Denitrification is an anaerobic process, occuring only in low-oxygen or anoxic environments and consists of four reaction steps by which nitrate is reduced to dinitrogen gas sequentially by the four metalloenzyme classes of nitrate reductases, nitrite reductases, nitric oxide reductases and nitrous oxide reductases. The nitrite reductase is the key enzyme of this respiratory process since it catalyses the reduction of soluble nitrite into gas (Zumft 1997). Two types of nitrite reductase, that differ in terms of their structure and prosthetic metal, have been characterized: a copper nitrite reductase (EC 1.7.99.3) encoded by the nirK gene and a cytochrome cd 1-nitite reductase (EC 1.7.2.1) encoded by the nirS gene that is the key enzyme and forms the distinguishing feature between denitrifiers and nitrate reducers (Henry et al. 2004).
Denitrifying microorganisms span a wide range of taxonomic groups including over 50 different genera and are not defined by a close phylogenetic relationship. Thus a phylogenetic approach involving simply the species identification, such as by the 16S rRNA gene, to detect denitrifying microorganisms in the environment is not suitable (Santoro et al. 2006; Heylen et al. 2006) Therefore, an alternative approach has been to amplify denitrifying bacteria nir gene fragments by PCR amplification. To this end, degenerate nirK and nirS primers have been applied to identify denitrifiers, and to study the denitrification diversity and community structures in various environments (Braker et al. 2000; Priemé et al. 2002; Rich et al. 2003; Wang and Skipper 2004; Santoro et al. 2006; Oakley et al. 2007).
However, the research into variation and genetic analysis of the nitrite reductase genes of denitrifying bacteria in Thailand is limited, despite the diverse geochemical soil arrays and generally high but fragmented biodiversity in this region. In this study, the relationship between the nitrite reductase encoding genes (nirK and nirS homologs) and the in vitro ability to reduce nitrate was investigated by screening soil bacteria collected from the Plant Germplasm-Royal Initiation Project at Kanchanaburi Province, Thailand, for their denitrifying ability and then assaying for the presence of nirK and nirS homologs by PCR with degenerate primers.
Materials and methods
Denitrifying bacteria isolation and nitrite-nitrate reducing ability test
Soil samples (500 g) were collected at a depth of 15 cm below the undisturbed surface. Soil bacteria were screened and then the isolation of denitrifying bacteria was performed under anaerobic conditions by plating soil bacteria after dilution onto nutrient agar (Schalau) at 30°C. All bacterial isolates were transferred separately into individual Durham tubes containing nitrate broth and incubated at 30°C for 14 days in an anaerobic chamber (Bactron/Sheldon, US). Samples from each culture were collected at day 0 and 7, respectively. The nitrate reduction ability was analysed by using the alpha-naphthylamine method (Beishir 1996). The likely species identification of all denitrifying bacteria was performed using the API test kit according to the manufacturer’s conditions (BioMerieux). Then denitrifying bacteria were cultivated in nutrient broth at 30°C for 18–24 h. One ml of cell suspension was transferred to 150 ml of nitrate broth and incubated in an anaerobic chamber at 30°C with shaking at 200 rev/min for 7 days. Nitrate and nitrite levels were measured at day 0 and 7 using a nitrate–nitrogen and nitrite–nitrogen test kit (HACH, USA), respectively, as per the manufacturer’s instructions.
DNA extraction, PCR amplification, cloning, sequencing and phylogenetic analysis
Genomic DNA was extracted from the cultured bacteria as described by Ausubel et al. (2002). Fragments of the nirK and nirS genes were amplified using the primer pairs nirK1F-nirK5R for nirK and nirS1F-nirS6R for nirS (Braker et al. 1998) in a final volume of 50 μl containing 5 μl of 10× PCR buffer 200 μM of deoxyribonucleoside triphosphate 1.0 U of Taq polymerase 1 μM of both primers, 400 ng of bovine serum albumin μl−1 and 10–100 ng of extracted gDNA. PCR conditions consisted of 1 min at 95°C, followed by a 1 min primer-annealing step and 1 min at 72°C. After 35 cycles, a final 7 min incubation at 72°C was performed. Expected amplicons were cloned using the PCR-Script™ Amp Cloning Kit (Stratagene, USA), and transformant selection was performed according to the manufacturer’s instructions. Selected transformants were then screened for the correct plasmid inserts by nested PCR, using two sets of primers (Braker et al. 1998). Plasmids were extracted from transformants that contained the desired inserts by the Fast Plasmid™ Mini kit (Eppendorf, USA). DNA sequencing was performed commercially by pyrosequencing at Macrogen, Korea.
For each of the two gene fragments, the sequences were “blasted” and aligned by the National Center for Biotechnology Information BLASTN program and the ClustalX programmes, respectively. Phylogenetic analysis was performed with a neighbor-joining algorithm and distance calculation according to Jukes and Cantor using PAUP4.0*b. Neisseria gonorrhoeae aniA gene (accession no. M97926) and the nirN gene from Pseudomonas aeruginosa (accession no. D84475) were used as out groups for nirK and nirS, respectively. The tree topology was evaluated by bootstrap analysis using 1,000 replicates. ClustalX programs, respectively. Phylogenetic analysis was performed with a neighbor-joining algorithm and distance calculation according to Jukes and Cantor using PAUP4.0*b.
Results and discussion
Denitrification bacteria and the nitrate–nitrite reduction ability
From the 211 soil bacteria isolates, 13 were found to be capable of denitrification and were then evaluated for the ability to reduce nitrate, and to accumulate or reduce nitrite according to the method described by Wistreich and Lechtman (1988). These 13 bacterial isolates with denitrifying activity were classified as six species by the API test results (Table 1). API kit was selected primarily to identify genus in our samples since it can be used for general purposes for genus identification in vary groups of environmental bacteria since the originally widely used 16S rDNA primers may not be suitable for specific or some small groups of bacteria (see comment in Wang and Qian 2009). Three of the isolates have previously been identified as denitrifiers; Agrobacterium radiobacter, Pseudomonas aeruginosa and Pseudomonas stutzeri (Braker et al. 1998; Hallin and Lindgren 1999; Wang and Skipper 2004). The other three isolates, Burkholderia cepacia, Corynebacterium propinquum and Micrococcus lylae are thus identified as potentially new species of denitrifiers in this study, although alternative means of species identification for conformation, such as phylogenetic analysis of the 16S rRNA gene, should be performed. Agrobacterium radiobacter was the dominant species of the denitrifying bacteria from the collected soil, in as much as 6/13 isolates were from this species (Table 1). For the ability to reduce both nitrate and nitrite, two isolates were found to reduce nitrate rapidly (Pseudomonas aeruginosa, B11-3 and Corynebacterium propinquum, A31-18) and one isolate could do it but slowly (Burkholderia cepacia, C22-14). These three isolates did not show any accumulation of nitrite, while the others ten isolates were found to lead to nitrite accumulation and also to reduce nitrate slowly.
Table 1.
Species identification, PCR amplification of nitrite reductase genes, and the percent nitrate reduction by day 7 of culture of the 13 selected isolates of denitrifying bacteria
| Isolate | Organism | Nitrite reductase gene | % Nitrate reduction by day 7 | Amount of nitrate reduction (mg/l) | Amount of nitrite accumulated (mg/l) |
|---|---|---|---|---|---|
| C22-18 | Agrobacterium radiobacter | nirK | 22.41 | 65 | 156.0 |
| C23-5 | Agrobacterium radiobacter | nirK, nirS | 46.55 | 135 | 81.1 |
| C23-15 | Agrobacterium radiobacter | nirK | 29.31 | 85 | 141.0 |
| C32-7 | Agrobacterium radiobacter | nirK | 19.64 | 55 | 173.8 |
| C32-13 | Agrobacterium radiobacter | nirK | 2.5 | 7 | 158.0 |
| C33-1 | Agrobacterium radiobacter | nirK | 21.43 | 60 | 173.3 |
| C22-14 | Burkholderia capacia | – | 58.93 | 165 | 75.0 |
| A31-18 | Corynebacterium propinquum | – | 97.32 | 171.5 | 1.18 |
| C32-5 | Micrococcus lylae | nirS | 34.48 | 100 | 137.5 |
| C32-6 | Micrococcus lylae | nirS | 56.90 | 165 | 147.2 |
| B11-3 | Pseudomonas aeruginosa | nirS | 99.14 | 287.5 | 0.2 |
| C22-5 | Pseudomonas aeruginosa | – | 62.5 | 175 | 61.2 |
| C22-24 | Pseudomonas stutzeri | – | 67.24 | 195 | 6.95 |
| Control | 0.00 | ND | ND |
DNA extraction, PCR amplification, cloning, sequencing and phylogenetic analysis
The thirteen isolates of denitrifying bacteria were then screened for the presence of the cd 1- and Cu-nir genes (nirS and nirK, respectively) by PCR amplification using degenerate primers as outlined under the methods section. Five isolates yielded only nirK amplicons (all Agrobacterium radiobacter) while three isolates produced only a nirS amplicon. One isolate (Agrobacterium radiobacter isolate C23-5) apparently had both nirK and nirS genes, and four isolates failed to produce an amplicon for either of the two genes (Table 1). For those isolates producing amplicons of the expected size for nirK1F-nirK5R (514 bp) and nirS1F-nirS6R (890 bp), the amplicons were cloned and sequenced.
The partial consensus nirK and nirS nucleotide sequences from all isolates were aligned with the existent nirK and nirS sequences of denitrifying bacteria in the GenBank database. For nirK, this revealed 92–98% nucleotide sequence identity to the nirK of Pseudomonas aeruginosa (accession no. AY345247), whilst the nirS sequences revealed 92–99% sequence identity, confirming their likely correct identity as nirK and nirS gene homologues. However, to confirm gene function and structure, complete sequence analysis and protein expression will be performed next. Phylogenetic analysis separated the five isolates which contained nirK into one group, as expected given that they are the same bacterial species (A. radiobacter) from the same region (Fig. 1). However, this clade also included P. aeruginosa, suggesting either a highly conserved gene fragment (homoplasy from insufficient informative characters) or recent lateral gene transfer. Interestingly, the nirK sequence of isolate C23-5 which showed a lower sequence identity (92%) to the others in this group, although still in the same closely related group as the other A. radiobacter and P. aeruginosa, was placed distal to these in its own subclade (Fig. 2). Indeed, this close phylogenetic distance for the nirK homologs between A. radiobacter and P. aeruginosa is not supported by the nirS sequences (Fig. 2), although it is not possible to infer lateral gene transfer over artifacts from homoplasy. The nirS sequences separated into two main groups. The first consisted of isolates B11-3 (Pseudomonas aeruginosa) and C32-5 (Micrococcus lylae), along with existent sequences in the database from uncultured bacteria, B. cepacia and P. aeruginosa. The second well separated group consisted of the published sequences plus a different Micrococcus lylae isolate (C32-6) and a sole Agrobacterium radiobacter isolate (C23-5). The two isolates of Micrococcus lylae segregated into the two different well separated groups, again suggesting potential lateral gene transfer but this needs further work for confirmation including molecular species confirmation.
Fig. 1.
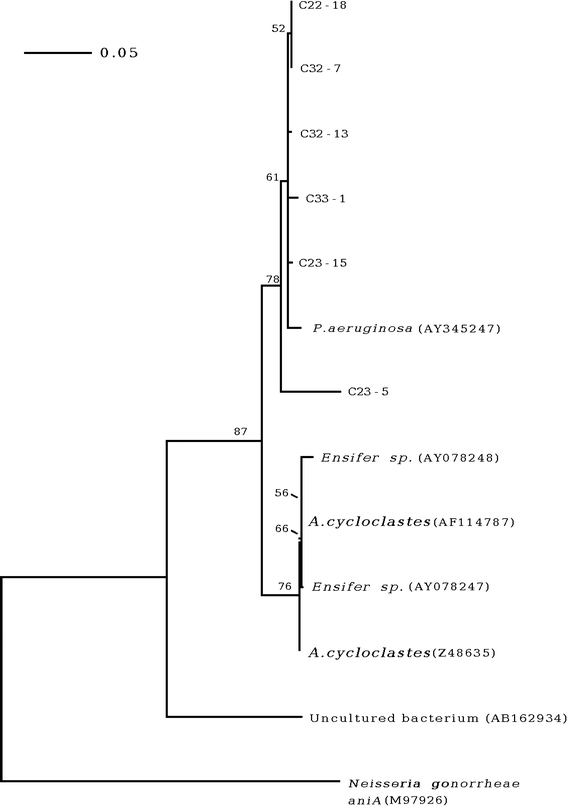
A rooted neighbor-joining phylogram of the partial nirK sequences. The scale bar indicates the expected number of changes per sequence position. For each node, bootstrap values greater than 50% are shown. The GenBank accession number is indicated in brackets after the name of the organism. The sequence of aniA from Neisseria gonorrhoeae (accession no. M97926) was used as the out group to root the phylogram
Fig. 2.
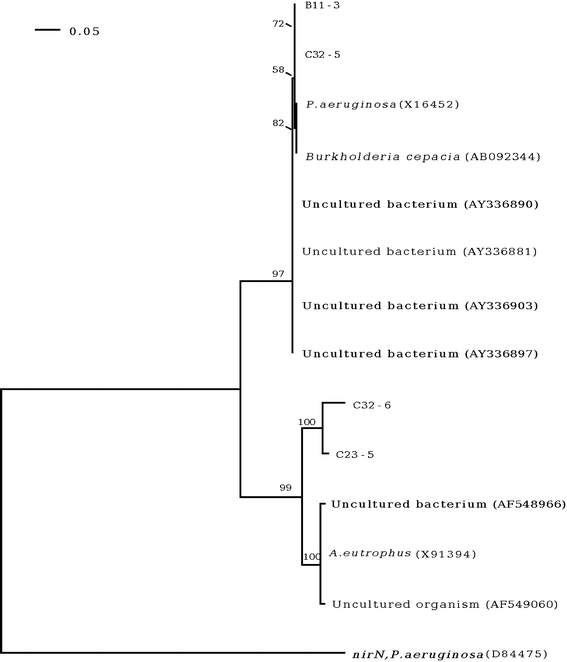
A rooted neighbor-joining phylogram of the partial nirS sequences. The scale bar indicates the expected number of changes per sequence position. For each node, bootstrap values greater than 50% are shown. The GenBank accession number is indicated in brackets after the name of the organism. The sequence of nirN from Pseudomonas aeruginosa was used as the out-group to root the phylogram
Acknowledgments
This research was supported by Rachadapisek Sompoj Fund of Chulalongkorn University, Bangkok, Thailand. We appreciate for Dr. Jittra Piapukiew about molecular technique advice. We would like to thank Dr. Robert Butcher from Publication Counselling Unit, Chulalongkorn University for kindly comments on manuscript English proof.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. 5. New York: Wiley; 2002. pp. 2–11. [Google Scholar]
- Beishir L. Microbiology in practice: a self-instructional laboratory course. 6. New York: Harper Collins College Publishers; 1996. pp. 318–319. [Google Scholar]
- Braker G, Fesefeldt A, Witzel KP. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braker G, Zhou J, Wu L, Devol AH, Tiedje JM. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/AEM.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin S, Lindgren PE. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S, Baudoin E, Lopez-Gutiérrez JC, Martin-Laurent F, Brauman A, Phillipot L. Quantification of denitrifying bacteria in soil by nirK gene targeted real-time PCR. J Microbiol Methods. 2004;59:327–335. doi: 10.1016/j.mimet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Heylen K, Gevers D, Vanparys B, Wittebole L, Geets J, Boon N, De Vos P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ Microbiol. 2006;8(11):2012–2021. doi: 10.1111/j.1462-2920.2006.01081.x. [DOI] [PubMed] [Google Scholar]
- Oakley BB, Francis CA, Robert KJ, Fuchman CA, Srinvasan S, Staley JT. Analysis of nitrite reductase (nirK and nirS) genes and cultivation reveal depauperate community of denitrifying bacteria in the Black Sea suboxic zone. Environ Microbiol. 2007;9(1):118–130. doi: 10.1111/j.1462-2920.2006.01121.x. [DOI] [PubMed] [Google Scholar]
- Priemé A, Braker G, Tiedje JM. Diversity of nitrite reductase (nirK and nirS) gene fragment in forested upland and wetland soil. Appl Environ Microbiol. 2002;68:1893–1900. doi: 10.1128/AEM.68.4.1893-1900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JJ, Heichen RS, Bottomley PJ, Cromack K, Jr, Myrold DD. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl Environ Microbiol. 2003;69:5974–5982. doi: 10.1128/AEM.69.10.5974-5982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Boehm AB, Francis CA. Denitrifier community composition along a nitrate and salinity gradient in a coastral aquifer. Appl Environ Microbiol. 2006;72(3):2102–2109. doi: 10.1128/AEM.72.3.2102-2109.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qian P-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. 2009;4(10):7401–7409. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Skipper HD. Identification of denitrifying rhizobacteria from bentgrass and bermudagrass golf greens. J Appl Microbiol. 2004;97:827–837. doi: 10.1111/j.1365-2672.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- Wistreich GA, Lechtman MD. Laboratory exercises in microbiology. 6. New York: Macmillan; 1988. pp. 220–221. [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


