Abstract
The Escherichia coli AlkA protein is a base excision repair glycosylase that removes a variety of alkylated bases from DNA. The 2.5 Å crystal structure of AlkA complexed to DNA shows a large distortion in the bound DNA. The enzyme flips a 1–azaribose abasic nucleotide out of DNA and induces a 66° bend in the DNA with a marked widening of the minor groove. The position of the 1–azaribose in the enzyme active site suggests an SN1-type mechanism for the glycosylase reaction, in which the essential catalytic Asp238 provides direct assistance for base removal. Catalytic selectivity might result from the enhanced stacking of positively charged, alkylated bases against the aromatic side chain of Trp272 in conjunction with the relative ease of cleaving the weakened glycosylic bond of these modified nucleotides. The structure of the AlkA–DNA complex offers the first glimpse of a helix–hairpin–helix (HhH) glycosylase complexed to DNA. Modeling studies suggest that other HhH glycosylases can bind to DNA in a similar manner.
Keywords: AlkA/glycosidase mechanism/helix–hairpin–helix/3-methyladenine DNA glycosylase/protein–DNA complex
Introduction
The chemical instability of DNA poses a challenge to the long-term maintenance and inheritance of genetic material. A variety of environmental toxins and cellular agents can react with DNA and chemically alkylate the bases. These added alkyl groups on the bases block replicative polymerases and/or interfere with the binding of regulatory proteins to DNA (Friedberg et al., 1995), causing widespread cellular responses (Jelinsky and Samson, 1999), including the activation of cell cycle checkpoints and/or programmed cell death (Engelward et al., 1998). Cells have devised a number of DNA repair strategies to restore the original DNA structure after a chemical insult, including direct reversal, nucleotide excision repair and base excision repair (BER). Most single base modifications in DNA are corrected by BER. In the first step of the BER pathway, a lesion-specific DNA N-glycosylase removes the damaged base by cleaving the glycosylic bond between the base and deoxyribose of the DNA backbone. The abasic DNA site produced by this reaction is excised and a DNA polymerase fills the resulting gap to restore the original sequence.
Base excision glycosylases locate damaged bases within a vast excess of unmodified DNA, then expose a target nucleotide for cleavage of the glycosylic bond by a reaction that does not require a high energy cofactor. We know little about how DNA glycosylases search for DNA damage or about the mechanism and energetics of distorting DNA structure to capture the modified nucleotide in the active site of the glycosylase. Crystal structures of uracil-specific glycosylases (Slupphaug et al., 1996; Barrett et al., 1998), a thymine dimer glycosylase (Vassylyev et al., 1995) and the human 3-methyladenine DNA glycosylase (Lau et al., 1998) complexed to DNA substrates or inhibitors have revealed different types of DNA distortions that insert an unpaired nucleotide of the DNA substrate into a pocket on the enzyme surface by a process termed base flipping or nucleotide flipping (Roberts and Cheng, 1998). Damaged bases might be more susceptible than normal bases to being flipped out of the DNA helix and into the glycosylase active site. For example, a chemical modification could weaken the [π]–electron stacking interactions between the modified base and its neighbors in the DNA double helix, making it easier to expose the modified base for enzymatic excision from DNA. Alternatively, a DNA glycosylase might flip nucleotides out of the DNA helix indiscriminately, but only those damaged bases that fit precisely into the substrate-binding site of the enzyme would be subject to hydrolysis.
Escherichia coli expresses two BER glycosylases that excise alkylation-damaged bases from DNA (Thomas et al., 1982). The constitutively expressed E.coli Tag protein (3–methyladenine DNA glycosylase I) is relatively specific for the removal of 3–methyladenine (Thomas et al., 1982), whereas the inducible AlkA protein (3–methyladenine DNA glycosylase II) removes a structurally diverse group of alkylated bases from DNA (Lindahl et al., 1988). The crystal structure of AlkA revealed a broad cleft located between two protein subdomains that contains a catalytically essential aspartic acid residue (Labahn et al., 1996; Yamagata et al., 1996). We previously proposed that this wide active site cleft could accommodate the differently shaped nucleotide substrates of the AlkA protein in a flipped out conformation (Labahn et al., 1996). The crystal structure of AlkA revealed an unexpected structural homology with another DNA glycosylase, endonuclease III (Endo III) (Kuo et al., 1994; Thayer et al., 1995), providing the initial evidence for a superfamily of BER glycosylases with very little sequence similarity yet remarkably similar folds. The helix–hairpin–helix (HhH) superfamily of DNA glycosylases (Nash et al., 1996) is named for a short sequence motif that forms a compact α–helical structure located on the rim of the active site pockets of Endo III (Thayer et al., 1995) and AlkA (Labahn et al., 1996; Yamagata et al., 1996). The HhH motif is also found in a large number of other proteins that bind to DNA without specificity for the base sequence (Doherty et al., 1996). It has been proposed that residues within the HhH motif of the BER glycosylases might have a role in base flipping by BER glycosylases, but until now a structure of an HhH protein–DNA complex was lacking.
Here we report the 2.5 Å resolution crystal structure of the E.coli AlkA protein complexed to DNA containing a modified abasic nucleotide, 1–azaribose, a possible transition state mimic of the glycosylase reaction. In the protein–DNA complex, the 1–azaribose is flipped out of the DNA helix and inserted into the active site cleft of the enzyme. A leucine intercalates into the minor groove of the DNA, where it wedges the neighboring base pairs apart and severely distorts the DNA structure around the flipped out abasic nucleotide. The position of the abasic nucleotide within AlkA's active site suggests an alternative mechanism for the glycosylase reaction, in which the catalytically essential residue Asp238 provides direct nucleophilic assistance for an SN1-type reaction that excises the damaged base. Structural modeling based on the AlkA–DNA crystal structure and sequence comparisons with other members of the HhH DNA glycosylase superfamily strongly suggest that these proteins bind to DNA substrates in a similar manner.
Results and discussion
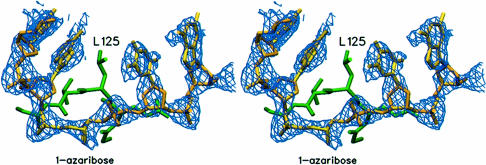
The AlkA protein was crystallized in complex with an inhibitory DNA containing 1–azaribose, an abasic deoxyribonucleotide analog with a nitrogen substituted at the C1′ position and a carbon in place of O4′ (Makino and Ichikawa, 1998). The 1-azaribose inhibitor was designed as a possible mimic of the transition state for glycosylic bond cleavage, and it binds to AlkA with high affinity (KDNA = ∼100 pM). The crystal structure of AlkA complexed to a DNA duplex containing 1-azaribose was determined with phases obtained from molecular replacement using a model of the unliganded AlkA protein (Labahn et al., 1996). Two protein–DNA complexes occupy the asymmetric unit of the crystals, and phases calculated from two AlkA protomers correctly positioned in the crystal showed clear density for the DNA bound to each protein molecule (Figure 1). The DNA model was fitted to the electron density and refined to a crystallographic R–factor of 0.25 (Rfree = 0.29), using X–ray data extending to a resolution limit of 2.5 Å (Table I). The two protein–DNA complexes in the asymmetric unit are essentially identical, with an r.m.s. deviation of 0.25 Å for the protein monomers (calculated for all atoms) and 0.39 Å for the DNA monomers (all atoms).
Fig. 1. Unbiased Fo – Fc difference electron density for the DNA. This electron density map was calculated with phases from the protein model after two molecules of the AlkA dimer were positioned in the asymmetric unit by molecular replacement and rigid body refinement. The resulting electron density for the bound DNA in the region of the active site clearly shows the flipped out 1–azaribose. The protein is shown in green and the fitted DNA is shown in yellow. Figures 1, 2, 4 and 5 were created with the program SETOR (Evans, 1990).
Table I. Data collection and refinement statistics.
| Unique reflections | 27 877 |
| Completenessa | 97.2 (100) |
| Resolution range (Å) | 30.0–2.5 |
| Rmerge (%)b | 8.5 (21.6) |
| Rworkc | 25% |
| Rfreec | 29% |
| R.m.s. deviation from ideal geometry | |
| bond lengths (Å) | 0.0126 |
| bond angles (°) | 1.45 |
aValues in parentheses are for the outermost shell of data (2.59–2.5 Å).
bRmerge=Σ|Iobs – <I>|/ΣIobs
cRwork= Σ||Fo| – |Fc||/Σ|Fo|, Rfree is the same parameter calculated with the reserved reflections.
DNA bending and nucleotide flipping
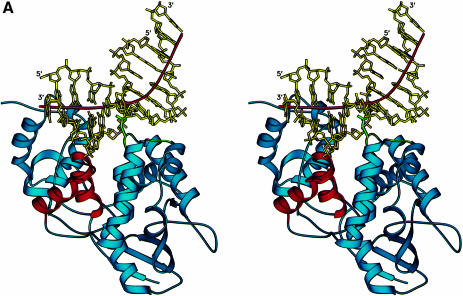
The structure of the AlkA protein–DNA complex reveals a unique mode of binding to DNA that exposes a nucleotide substrate to the enzyme active site. The bound DNA is highly distorted (Figure 2), with a bend centered at the site of the flipped out abasic nucleotide and an immensely widened minor groove. The DNA is held in complex with the AlkA protein by a combination of polar and non-polar interactions, although there are remarkably few positively charged residues on the protein's DNA-binding surface that could interact with the DNA backbone. One striking feature of the structure is that the protein is able to exert such a distortion on DNA with relatively few contacts to the phosphodiester backbone. Except for the single interaction of Lys170 with a DNA phosphate, all polar interactions with the protein involve the DNA strand containing the 1-azaribose (Figure 2B). This is similar to the human uracil-DNA glycosylase (hUDG), which predominantly contacts with the uracil-containing strand of DNA (Parikh et al., 1998). In contrast to AlkA, UDG is active against both single- and double-stranded DNA. AlkA relies on van der Waals interactions with the minor groove to bind to DNA substrates, and the protein makes comparatively few hydrogen bonds to the DNA. These minor groove interactions are the probable explanation for AlkA's strong preference for double-stranded DNA substrates.
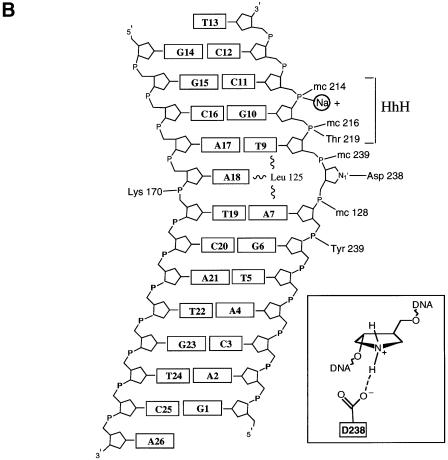
Fig. 2. (A) AlkA-induced distortion of DNA. A 66° bend in the DNA results from the insertion of Leu125 and loops αD–αE and αG–αH (shown in green) into the minor groove. The DNA is anchored to the protein by the interactions with the HhH motif (shown in red). The local helical axis of the DNA is shown by a red line. (B) Schematic diagram of the AlkA–DNA contacts. The 1–azaribose abasic nucleotide is in an extrahelical conformation with N1′ of the sugar positioned 3.2 Å from the carboxylate oxygen of Asp238 (inset). Except for Lys170, all hydrogen-bonding contacts are made with the 1–azaribose-containing strand of the DNA. The HhH motif acts to anchor the DNA to the protein by providing several hydrogen bonds as well as a metal-mediated interaction. Having relatively few polar interactions with the DNA, AlkA also relies on van der Waals interactions for binding energy (see Figure 3). Residues with main chain atoms contacting the DNA are indicated by the prefix ‘mc’.
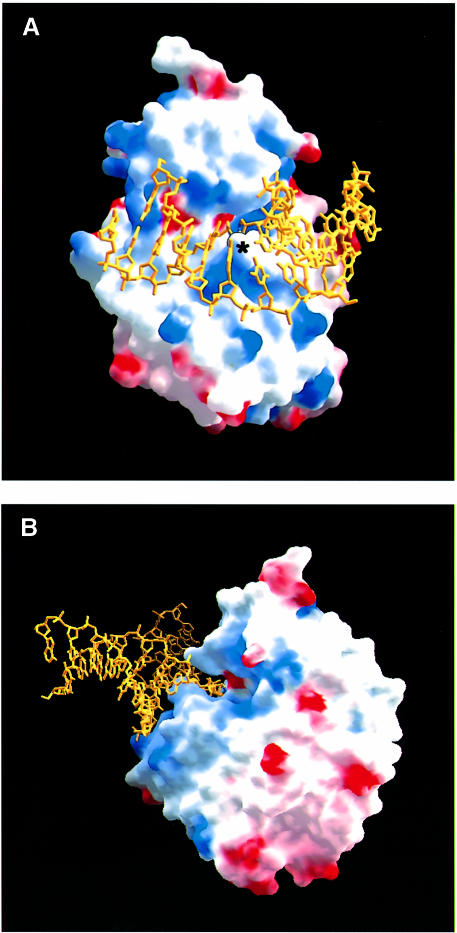
The predominantly non-polar DNA-binding surface of AlkA consists of an active site cleft that is flanked by a ‘wedge’ comprising the loops between helices G and H (αG–αH) and between helices D and E (αD–αE) (Figure 3). The tip of the wedge is capped by Leu125 in the αD–αE loop, which juts into the minor groove and displaces the base targeted for excision. The structurally homologous loop between helices α2 and α3 of the HhH glycosylase MutY was previously suggested to be a minor groove reading motif (Guan et al., 1998). At the base of the wedge is Pro175 of the αG–αH loop. Pro175 is forced into the minor groove, where it makes van der Waals contacts with the deoxyribose sugar of position A18 located opposite the 1-azaribose abasic nucleotide (Figure 2), resulting in an ∼2–fold expansion of the minor groove width (∼15.5 Å wide). The intercalating leucine (Leu125) creates a remarkable bend of ∼66° in the bound DNA (calculated with CURVES; Lavery and Sklenar, 1989) that causes the DNA to bend away from the protein (Figures 2 and 3). The DNA bend disrupts base pair stacking interactions on either side of the flipped out nucleotide. The adenine base at position A18 opposite the 1–azaribose is pushed 1.4 Å into the major groove, while the base pairs flanking the abasic site (A7–T19 and T9–A17; Figure 2B) are unstacked. The absence of specific protein interactions with base A18 is consistent with AlkA's lack of specificity for the base opposite a substrate base (Saparbaev et al., 1995). The large DNA distortion is probably a consequence of the insertion of Leu125 into the DNA helix and the anchoring of DNA with the HhH motif. On the 3′ side of the flipped out nucleotide, the DNA is held in place by interactions with the HhH motif (Figure 2). The bend in the DNA relieves the torsional strain caused by the dramatic widening of the minor groove. A similar protein-induced bending of DNA has been observed in complexes with high mobility group (HMG) proteins. The relatively compact DNA-binding domain of the HMG proteins binds to the minor groove of sharply bent DNA, inserting a hydrophobic residue between two base pairs (Love et al., 1995; Ohndorf et al., 1999). Extensive bending of DNA is also seen in the DNA complex of the base excision glycosylases UDG and T4 endonuclease V. However, endonuclease V flips out adenine on the strand opposite cis, syn–thymine dimers, and the protein makes numerous contacts to both strands of DNA (Vassylyev et al., 1995). Other DNA glycosylase enzymes such as the human 3–methyladenine DNA glycosylase (AAG) and the mismatch-specific uracil DNA glycosylase (MUG) distort DNA comparatively little, bending their DNA targets by <20° (Slupphaug et al., 1996; Barrett et al., 1998; Lau et al., 1998).
Fig. 3. (A) The binding surface of AlkA colored according to its electrostatic potential (blue, positively charged; red, negatively charged) shows a relatively uncharged surface contacting the DNA. The intercalating Leu125 is indicated by an asterisk, and the catalytic Asp238 is indicated by the small red patch above the asterisk and adjacent to Leu125. (B) A rotated view of the AlkA–DNA complex showing that the abasic 1–azaribose has been rotated out of the DNA and into the aromatic active site cleft of the enzyme. Leu125 has been inserted into the gap created by the flipped out nucleotide. The minor groove has been widened substantially by the protein, contributing to the significant distortion of the DNA. Figures 1 and 6 were generated with the program GRASP (Nicholls et al., 1993).
The HhH motif and nucleotide flipping
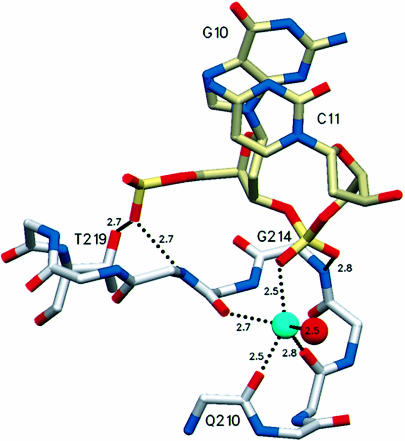
Of the limited number of polar interactions between AlkA and the DNA, most are contributed by the HhH motif (Figure 2). Residues 202–227, which form the HhH motif of AlkA, tightly anchor the DNA to the protein through hydrogen bonds and a metal-mediated interaction. The main chain amide nitrogens of residues 214, 216 and 219 donate hydrogen bonds to the phosphodiester backbone of DNA at positions C10 and C11 (Figure 4). HhH binding is strengthened further by a metal that contacts the phosphate of nucleotide C11. The metal ion is coordinated by the main chain carbonyl oxygens of residues 210, 212 and 215. Two of the three remaining metal coordination sites are occupied by a phosphate oxygen of nucleotide C11 and by a water molecule. The ligated metal was modeled as a sodium atom because that is the dominant cation in the crystallization conditions, and the 2.5 Å average distance between the metal and its oxygen ligands is well suited for sodium and it rules out a water at this position. Furthermore, the HhH motifs of DNA polymerase β bind sodium and potassium in preference to divalent metals (Pelletier and Sawaya, 1996). The DNA phosphate oxygen ligand is apparently required for the stable binding of a metal to the HhH motif because the metal is absent from crystal structures of AlkA (Labahn et al., 1996; Yamagata et al., 1996), Endo III (Thayer et al., 1995) and MutY (Guan et al., 1998), all determined at high resolution in the absence of DNA. The crystal structure of the AlkA–DNA complex shows that the HhH motif is not directly responsible for flipping nucleotide substrates out of DNA, but instead it serves as a stable platform for the protein-induced distortion of DNA structure caused by the intercalating leucine side chain (Figure 3).
Fig. 4. The HhH motif serves to anchor the DNA to the protein through hydrogen bonds with Thr219 and the main chain amides of residues 216 and 214. The HhH motif is positioned adjacent to the active site (cf. Figure 2B) but it does not participate directly in base flipping or in catalysis. The hairpin turn of the HhH ligates a metal that contacts the DNA phosphate backbone and serves to organize the hairpin for additional hydrogen-bonding interactions with the DNA. The sodium ion modeled here (light blue) is coordinated by the main chain carbonyl oxygens of residues 215, 212 and 210, the phosphate oxygen of the DNA and a water molecule. One remaining potential site of metal coordination is unoccupied.
The 1-azaribose is flipped out of the DNA and into the enzyme active site by rotation about the P–O5′ bond (α angle) and about the O3′–P bond (ζ angle) of the phosphodiester backbone. The nitrogen of the 1–azaribose ring is situated 3.2 Å from the carboxylate of Asp238, a catalytically essential residue (Labahn et al., 1996; Yamagata et al., 1996). The direct interaction of Asp238 with the 1–azaribose moiety probably contributes substantially to the high affinity and specificity of binding to DNA containing this modified abasic nucleotide, which was designed to mimic the carbocation character of the transition state of N-glycohydrolase reactions (Schramm et al., 1994; Makino and Ichikawa, 1998). Leu125 of the αD–αE loop is pushed into the minor groove where it occupies the position vacated by the flipped out 1-azaribose nucleotide (Figure 3). The geometry of this protein-induced distortion of DNA is quite different from that caused by hUDG (Slupphaug et al., 1996; Parikh et al., 1998). UDG ‘pinches’ the DNA backbone at the site of the flipped out uridine, compressing the interphosphate distance and creating a zig-zag conformation in the phosphodiester backbone. In contrast, the DNA backbone is fully extended around the flipped out 1-azaribose sugar in the AlkA–DNA complex. Thus, UDG and AlkA induce very different distortions in their DNA substrates. Biochemical and structural evidence suggest that certain alkylated bases cause little perturbation of DNA structure (Ezaz-Nikpay and Verdine, 1994), supporting the notion that AlkA must actively rotate nucleotides into the extrahelical conformation observed in the crystal structure. The extreme distortion of the bound DNA presumably lowers the energetic cost of unstacking a substrate nucleotide and disrupting its interactions with a base-pairing partner in order to flip the nucleotide into the enzyme active site.
A model for base recognition and catalysis
We have modeled a nucleoside substrate in the AlkA-binding pocket by superimposing a 3–methyladenine nucleoside on the 1–azaribose in the crystal structure (Figure 5). The modeled 3–methyladenine base stacks against Trp272, and C1′ of its ribose ring is situated near Asp238. The stacking of the electron-deficient, positively charged 3-methyladenine base against the aromatic side chain of Trp272 constitutes a π–cation interaction (Gallivan and Dougherty, 1999) that would stabilize the extrahelical conformation of the alkylated nucleotide, as previously proposed (Labahn et al., 1996). The binding site identified by modeling has an open architecture that is consistent with AlkA's ability to excise from DNA many different and structurally diverse alkylated bases, and the modeled substrate base makes no detectable hydrogen bonds with residues of the AlkA active site. Arg22 is the only polar amino acid in the vicinity of the proposed binding site for the base, but it cannot be oriented correctly for hydrogen-bonding interactions with 3-methyl– adenine. The proposed binding site for 3–methyladenine in the AlkA structure broadly overlaps the binding site for an adenine base soaked into crystals of another HhH superfamily glycosylase, MutY (Guan et al., 1998). The adenine base stacks against Met185 of MutY, the residue that aligns with Trp272 of the AlkA structure (Figure 7).
Fig. 5. A 3–methyladenine substrate modeled in AlkA's active site. 3–methyladenine (pink) was superimposed on the 1–azaribose moiety in the crystal structure of the AlkA–DNA complex. In the resulting model, the 3–methyladenine base stacks face-to-face against Trp272 and makes edge-on contacts with Tyr222. The open architecture of AlkA's substrate-binding pocket would accommodate many types of modified bases. Trp218 is located behind the ribose of the flipped out nucleotide, leaving no room for a water nucleophile (cf. Figure 6).
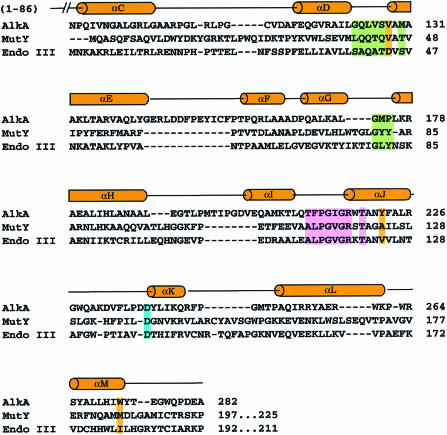
Fig. 7. Structure-based sequence alignment of HhH base excision glycosylases. The sequences of AlkA, MutY and Endo III are aligned based on the best superposition of Cα atoms in the crystal structures, as determined by the program DALI (Holm and Sander, 1993). The secondary structure of AlkA is shown above the sequence. Members of the HhH superfamily not only share a similar fold, but also have a common surface chemistry in the region contacting DNA in the AlkA–DNA complex (cf. Figure 3). The areas shaded green are the residues in AlkA that are within van der Waals contact distance of the minor groove of the DNA. The residues of the HhH motif that make contact with the DNA are shaded pink and the catalytic aspartate is shaded blue. Residues lining the putative base-binding pocket are shaded orange (cf. Figure 5). It is likely that members of the HhH glycosylase superfamily all bind to DNA and expose a substrate base in a similar fashion.
Whereas most BER glycosylases are specific for a particular type of damaged base, AlkA and the mammalian 3–methyladenine DNA glycosylases are unusual in that they recognize and excise a structurally diverse group of alkylated bases from DNA (Wyatt et al., 1999). Despite this broad specificity for a variety of alkylated bases, closely related base modifications such as 8–oxoguanine are not substrates. The basis for the broad substrate range of the alkylation-specific BER enzymes is not understood. The modified bases that are processed efficiently by AlkA have a delocalized positive charge (Lindahl, 1982) and we have suggested previously that the enzyme binds selectively to electron-deficient, alkylated bases through strong [π]–cation interactions with electron-rich aromatic side chains in the enzyme active site (Labahn et al., 1996). The limited extent of AlkA's DNA interaction surface, comprising ∼600 Å2 of buried protein surface calculated with a 1.4 Å probe, and the lack of base-specific contacts to DNA both favor the binding and distortion of double-stranded DNA without regard to its sequence. AlkA excises unmodified purine bases from DNA in vitro, albeit at rates that are orders of magnitude slower than the excision of alkylated bases (Berdal et al., 1998). This finding confirms that unmodified purine bases can be inserted into AlkA's active site in a catalytically competent orientation. Base stacking interactions with Trp272 and edge-on contacts with Tyr222 might stabilize these normal bases in the active site long enough for catalysis to occur. The lack of any specific hydrogen-bonding interactions between the modeled base and the enzyme active site (Figure 5) suggests that AlkA's exceedingly low activity towards non-alkylated substrates, and its correspondingly high selectivity for certain alkylated nucleosides, might result from factors other than its selectivity for binding to substrates. AlkA's preferred substrates are electron-deficient purines with a weakened glycosylic bond that is readily cleaved under acidic conditions (Lawley and Brookes, 1963). As suggested by Seeberg and co-workers (Berdal et al., 1998), AlkA's enzymatic selectivity might be explained by the chemical lability of the glycosylic bond in these destabilized substrates, rather than the selective recognition and binding of these modified bases to the enzyme. These authors suggest that AlkA can bind to substrate and non-substrate bases alike, but only nucleotides with a weakened glycosylic bond are cleaved efficiently. Consistent with this hypothesis, alkylated bases such as 3,N4–ethenocytosine and 1,N6–ethenoadenine, which are not electron deficient and have stable glycosylic bonds, are not excised readily from DNA by AlkA (Saparbaev et al., 1995; Hang et al., 1997; Saparbaev and Laval, 1998).
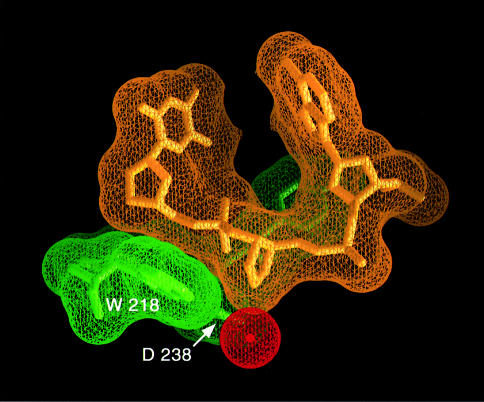
It was assumed previously that the conserved, catalytically essential aspartic acid of HhH superfamily glycosylases (Asp238 of AlkA) might activate a water nucleophile for the attack on the back of C1′, causing release of a damaged base from DNA by a bimolecular nucleophilic displacement (SN2) mechanism (Sun et al., 1995; Labahn et al., 1996; Scharer et al., 1998). The SN2 mechanism requires a water that is positioned for the attack on the back of the deoxyribose C1′ carbon, between the substrate nucleoside and the general base, the conserved aspartic acid. The crystal structure of the human 3–methyladenine DNA glycosylase complexed to a pyrrolidine-containing DNA revealed a bound water in the active site, located between a conserved glutamic acid and the back of a flipped out nucleoside. This strongly implies that an SN2-type mechanism is used for glycosylic bond cleavage (Lau et al., 1998). However, the crystal structure of AlkA complexed to the 1–azaribose DNA is not consistent with an SN2 direct displacement mechanism. N1′ of the 1–azaribose ring, corresponding to the C1′ position of a modeled substrate (Figure 5), is in direct contact with the carboxylate of Asp238 (3.2 Å O–N distance), leaving no room for a water nucleophile (Figure 6). Although it is possible that a substrate nucleoside could bind in an orientation different from that of the 1–azaribose abasic inhibitor, the extensive number of contacts to the flipped out abasic nucleotide and the surrounding positions of the DNA make this an unlikely possibility. Alternatively, the AlkA-catalyzed reaction might proceed by a different mechanism. The observed direct interaction of Asp238 with the anomeric position of the sugar could offset the charge of a carbocation intermediate in an SN1-type cleavage reaction. Double displacement mechanisms have been characterized for a number of glycosidases (Ford et al., 1974; Sinnott, 1990). A general acid protonates the leaving group of the substrate sugar and thereby facilitates glycosylic bond cleavage by generating a resonance-stabilized oxonium ion. An ionized carboxylate of an aspartic acid can stabilize the positive oxonium ion either by forming a covalent glycosyl–enzyme complex or by charge–charge interactions (Sinnott, 1990). In the case of lysozyme, no covalent intermediate between the stabilizing carboxy oxygen and the oxonium ion is suggested because of the ∼3 Å distance between the two atoms. Following bond cleavage, a nearby water is activated by the deprotonated general acid (now functioning as a general base), and the resulting hydroxide ion combines with the charged sugar to complete the reaction.
Fig. 6. Mechanistic implications of the AlkA active site. The closely interacting van der Waals surfaces of the protein (green) and the DNA (yellow) leave no room between the deoxyribose of a flipped out nucleotide and Trp218 to position a water molecule (red) for an attack on the back of the glycosylic bond. This is strong evidence against a direct displacement (SN2) mechanism of glycosylic bond cleavage (see text for details).
A similar mechanism might be applicable to AlkA. However, AlkA lacks an obvious residue that could fulfill the role of a general acid to protonate the leaving group and later activate a water molecule. Neutral substrates require this catalytic assistance to eject the leaving group. However, AlkA's positively charged, alkylated substrates are pre-protonated and might not require this type of assistance. The properly positioned carboxylate group of AlkA's Asp238 could be all that is required to stabilize a carbocation intermediate, by either ionic or covalent interactions, thus promoting the release of an alkylated base. The lack of a general acid in AlkA's active site provides a means of catalytic selectivity. Positively charged bases, which do not require protonation and have a weakened glycosylic bond, can be removed effectively, and in turn it may function as a general base to activate a nearby water molecule. Unmodified bases require protonation by a general acid prior to hydrolysis and therefore are poor substrates. Other HhH glycosylases that catalyze the removal of neutral bases from DNA are likely to require assistance from a general acid located near the leaving group. It has been proposed that Glu37 of MutY serves this role (Guan et al., 1998).
DNA binding by other HhH superfamily glycosylases
Crystal structures of AlkA (Labahn et al., 1996; Yamagata et al., 1996), Endo III (Kuo et al., 1994; Thayer et al., 1995) and MutY (Guan et al., 1998) have shown remarkably similar protein folds despite their dissimilar amino acid sequences, aside from the HhH motif and catalytically important aspartic acid. Other repair enzymes, such as 8–oxoguanine DNA glycosylase, have the HhH sequence motif and a nearby conserved aspartic acid (Nash et al., 1996) and they are therefore likely to have a similar protein fold. The evolutionary conservation of the HhH glycosylase fold implies that it is an effective scaffold for DNA binding and base flipping. AlkA has an N–terminal domain that is not present in the other recognized HhH DNA glycosylases (Labahn et al., 1996; Yamagata et al., 1996). However, the N-terminal domain is located away from the DNA-binding surface of AlkA and its structure is unchanged when AlkA binds to DNA, so it is unlikely to influence DNA binding activity. We have modeled the MutY–DNA and Endo III–DNA complexes by superimposing the unliganded proteins onto the structure of AlkA complexed to DNA. This modeling exercise shows that MutY and Endo III could bind to DNA targets with a flipped out nucleotide in the same manner as AlkA. All three enzymes have a predominantly non-polar surface within the proposed region of DNA contact, and the DNA from the AlkA structure fits the other enzymes without any major steric clashes. The position of the minor groove wedge is evident in all three structures. Gln42 of MutY and Gln41 of Endo III align with Leu125 of AlkA, they are in the proper position to bend the DNA and assist in nucleotide flipping, and non-conservative substitutions at these positions decrease base excision activity (Guan et al., 1998). Catalytic aspartic acids of AlkA (Asp238), MutY (Asp138) and Endo III (Asp138) are positioned similarly and in close proximity to the flipped out abasic nucleotide. Lys191 was previously shown to contribute to Endo III's DNA-binding activity (Thayer et al., 1995). The sharp bend in the DNA from the AlkA structure, when superimposed on Endo III, brings the phosphodiester backbone into close proximity with Lys191. A lesser bend in the DNA would not permit this interaction.
Conclusions
Until now, the active site for members of the HhH superfamily had been inferred from mutagenesis of AlkA and Endo III to be located within the cleft at the subdomain interface. Additionally, the structure of adenine bound to MutY suggested a binding pocket for an extrahelical base. The structure of the AlkA–DNA complex not only confirms this location of the active site, but it reveals a new mode of DNA binding and base flipping that involves a sharp bend in the DNA with the unstacking of the base pairs flanking the flipped out nucleotide. AlkA recognizes DNA through non-specific van der Waals contacts and a limited number of hydrogen bonds. The conserved HhH motif participates in a metal ion-mediated interaction with the phosphodiester backbone that positions the target nucleotide to be flipped into the enzyme active site by the minor groove intercalator, Leu125. These interactions result in a strong distortion of the DNA in the form of a 66° bend and widening of the minor groove to >15 Å. Comparisons of the AlkA–DNA structure with other members of the HhH glycosylase superfamily show that these enzymes not only have a homologous fold but share a common surface chemistry in the DNA-contacting regions. It is likely that all members of the HhH superfamily bind to and distort DNA substrates in a similar fashion.
Materials and methods
Crystallization and X-ray data collection
Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer and purified by anion-exchange HPLC (Poros HQ10 medium; PerSeptive Biosystems). 1–Azaribose-containing DNA was prepared from 1–azaribose (Makino and Ichikawa, 1998) by the procedure of Deng et al. (1997). The sequence of the 1–azaribose-containing strand is 5′–GACATGAZTGCC–3′, where Z represents the 1–azaribose. The complementary strand is 5′–GGCAATCATGTCA–3′. The strands were annealed (10 mM MES pH 6.5, 20 mM NaCl) and mixed with purified AlkA protein (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 2 mM dithiothreitol, 0.1 mM EDTA) in a 1:1 molar ratio to yield a protein–DNA complex of 8 mg/ml protein.
Crystals of the complex were obtained by the vapor diffusion method in hanging drops. Equal volumes of the protein–DNA solution were mixed with well solution containing 15% PEG 4K, 100 mM HEPES pH 7.5, 100 mM NaCl, 50 mM MgCl2 and 8% ethylene glycol. Crystals do not grow under these conditions in the absence of DNA or protein. Prior to data collection, the crystals were equilibrated for 2–4 h in 18% PEG 4K, 100 mM HEPES pH 7.5, 100 mM NaCl, 50 mM MgCl2 and 12% ethylene glycol and then for 1 h in the same solution with 20% ethylene glycol. Crystals were then mounted in a nylon loop and flash frozen in liquid nitrogen. The crystals belong to space group P3121 with unit cell dimensions a = b = 82.4 Å and c = 199.7 Å. Two AlkA–DNA complexes occupy the asymmetric unit and they are related by 2–fold rotation about a non-crystallographic symmetry axis oriented normal to the c–axis of the unit cell. X–ray data were collected using a laboratory Cu X–ray source as well as beamline X–25 of the National Synchrotron Light Source (Upton, NY).
Phasing and refinement
X-ray data were processed with DENZO/SCALEPACK (Otwinowski and Minor, 1997). Phases for the data were obtained by molecular replacement using the unliganded AlkA protein (Labahn et al., 1996) as a search model. Molecular replacement was performed using the Crystallography and NMR System (CNS version 0.5; Brünger et al., 1998), and the best solution after rigid body refinement had an R–factor of 44%. The initial Fo – Fc density maps calculated with this model showed clear density for the DNA. The DNA model was fitted to the density and the structure was refined using CNS. The preferred torsion angles of the 1–azaribose abasic nucleotide were determined empirically by fitting the abasic nucleotide into the unbiased electron density phased with the protein model and using the resulting torsion angles as targets during refinement. The success of the model refinement at each stage was gauged by the change in the free R–factor (Brünger, 1992) and confirmed by simulated annealing–omit procedures (Brünger et al., 1998). The DNA model was built into the electron density using the graphics program O (Jones and Kjeldgaard, 1992). Coordinates of the AlkA–DNA complex have been deposited in the Protein Data Bank (pdbid# 1DIZ).
Acknowledgments
Acknowledgements
We thank Hyock Joo Kwon, Michael Sawaya and Dane Walther for their assistance with X–ray diffraction experiments, and the staff at beamline X–25 of the National Synchrotron Light Source (NSLS; Upton, NY) for their advice on data collection. We are grateful to Greg Verdine and members of his research group for their continued advice and assistance with studies of DNA base excision repair. This work was funded by grants from the National Institute of General Medical Sciences (T.E. and Y.I.), the Giovanni Armenise Foundation for Advanced Scientific Research and the Harvard Center for Structural Biology. T.H. is supported by the Environmental Health Sciences Training Program at the Harvard School for Public Health (NIEHS training grant).
References
- Barrett T.E., Savva, R., Panayotou, G., Barlow, T., Brown, T., Jiricny, J. and Pearl, L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- Berdal K.G., Johansen, R.F. and Seeberg, E. (1998) Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J., 17, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998)Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Deng L., Scharer, O.D. and Verdine, G.L. (1997) Unusually strong binding of a designed transition-state analog to a base-excision DNA repair protein. J. Am. Chem. Soc., 119, 7865–7866. [Google Scholar]
- Doherty A.J., Serpell, L.C. and Ponting, C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward B.P., Allan, J.M., Dreslin, A.J., Kelly, J.D., Wu, M.M., Gold, B. and Samson, L.D. (1998) A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. J. Biol. Chem., 273, 5412–5418. [DOI] [PubMed] [Google Scholar]
- Evans S.V. (1990) SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Ezaz-Nikpay K. and Verdine, G.L. (1994) The effects of N7-methylguanine on duplex DNA structure. Chem. Biol., 1, 235–240. [DOI] [PubMed] [Google Scholar]
- Ford L.O., Johnson, L.N., Machin, P.A., Phillips, D.C. and Tijan, R. (1974) Crystal structure of a lysozyme–tetrasaccharide lactone complex. J. Mol. Biol., 88, 349–371. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Gallivan J.P. and Dougherty, D.A. (1999) Cation–[π] interactions in structural biology. Proc. Natl Acad. Sci. USA, 96, 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Manuel, R.C., Arvai, A.S., Parikh, S.S., Mol, C.D., Miller, J.H., Lloyd, S. and Tainer, J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Hang B., Singer, B., Margison, G.P. and Elder, R.H. (1997) Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc. Natl Acad. Sci. USA, 94, 12869–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander, C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jelinsky S.A. and Samson, L.D. (1999) Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl Acad. Sci. USA, 96, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A. and Kjeldgaard,M. (1992) O—the manual. Uppsala, Sweden: http://kaktus.kemi.aau.dk.
- Kuo C.F., McRee, D.E., Fisher, C.L., O'Handley, S.F., Cunningham, R.P. and Tainer, J.A. (1994) Atomic structure of the DNA repair [4Fe–4S] enzyme endonuclease III. Science, 258, 434–440. [DOI] [PubMed] [Google Scholar]
- Labahn J., Scharer, O.D., Long, A., Ezaz-Nikpay, K., Verdine, G.L. and Ellenberger, T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- Lau A.Y., Scharer, O.D., Samson, L., Verdine, G.L. and Ellenberger, T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell, 95, 249–258. [DOI] [PubMed] [Google Scholar]
- Lavery R. and Sklenar, H. (1989) Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- Lawley P.D. and Brookes, P. (1963) Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J., 89, 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. (1982) DNA repair enzymes. Annu. Rev. Biochem., 51, 61–87. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick, B., Sekiguchi, M. and Nakabeppu, Y. (1988) Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem., 57, 133–157. [DOI] [PubMed] [Google Scholar]
- Love J.J., Li, X., Case, D.A., Giese, K., Grosschedl, R. and Wright, P.E. (1995) Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]
- Makino K. and Ichikawa, Y. (1998) Synthesis of a 2-deoxy-ribose type 1-N-iminosugar. Tetrahedron Lett., 39, 8245–8248. [Google Scholar]
- Nash H.M., Bruner, S.D., Scharer, O.D., Kawate, T., Addona, T.A., Spooner, E., Lane, W.S. and Verdine, G.L. (1996) Cloning of a yeast 8–oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj, R. and Honig, B. (1993) GRASP—graphical representation and analysis of surface properties. Biophys. J., 64, A116. [Google Scholar]
- Ohndorf U.M., Rould, M.A., He, Q., Pabo, C.O. and Lippard, S.J. (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature, 399, 708–712. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parikh S.S., Mol, C.D., Slupphaug, G., Bharati, S., Krokan, H.E. and Tainer, J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier H. and Sawaya, M.R. (1996) Characterization of the metal ion binding helix–hairpin–helix motifs in human DNA polymerase β by X-ray structural analysis. Biochemistry, 35, 12778–12787. [DOI] [PubMed] [Google Scholar]
- Roberts R.J. and Cheng, X. (1998) Base flipping. Annu. Rev. Biochem., 67, 181–198. [DOI] [PubMed] [Google Scholar]
- Saparbaev M. and Laval, J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M., Kleibl, K. and Laval, J. (1995) Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res., 23, 3750–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer O.D., Nash, H.M., Jiricny, J., Laval, J. and Verdine, G.L. (1998) Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Biol. Chem., 273, 8592–8597. [DOI] [PubMed] [Google Scholar]
- Schramm V.L., Horenstein, B.A. and Kline, P.C. (1994) Transition state analysis and inhibitor design for enzymatic reactions. J. Biol. Chem., 269, 18259–18262. [PubMed] [Google Scholar]
- Sinnott M.L. (1990) Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev., 90, 1171–1202. [Google Scholar]
- Slupphaug G., Mol, C.D., Kavli, B., Arvai, A.S., Krokan, H.E. and Tainer, J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- Sun B., Latham, K.A., Dodson, M.L. and Lloyd, R.S. (1995) Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme–DNA imino intermediates. J. Biol. Chem., 270, 19501–19508. [DOI] [PubMed] [Google Scholar]
- Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Yang, C.H. and Goldthwait, D.A. (1982) Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry, 21, 1162–1169. [DOI] [PubMed] [Google Scholar]
- Vassylyev D.G., Kashiwagi, T., Mikami, Y., Ariyoshi, M., Iwai, S., Ohtsuka, E. and Morikawa, K. (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell, 83, 773–782. [DOI] [PubMed] [Google Scholar]
- Wyatt M.D., Allan, J.M., Lau, A.Y., Ellenberger, T.E. and Samson, L.D. (1999) 3-Methyladenine DNA glycosylases: structure, function and biological importance. BioEssays, 21, 668–676. [DOI] [PubMed] [Google Scholar]
- Yamagata Y., et al. (1996)Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli.Cell, 86, 311–319. [DOI] [PubMed] [Google Scholar]