Abstract
Mutations in CAPN3/Capn3, which codes for skeletal muscle-specific calpain-3/p94 protease, are responsible for limb-girdle muscular dystrophy type 2A (LGMD2A). Using “knock-in” (referred to as Capn3CS/CS) mice, in which the endogenous calpain-3 is replaced with a mutant calpain-3:C129S, that is proteolytically inactive but structurally intact calpain-3, our previous studies demonstrated that loss of calpain-3 protease activity caused muscular dystrophy. However, compared to Capn3 null (Capn3−/−) mice, Capn3CS/CS mice showed less severe dystrophic symptoms. This suggests that calpain-3 also has a non-proteolytic function. This study aimed to elucidate non-protease calpain-3 functions, by comparison of Capn3CS/CS mice with Capn3−/− mice. We found that calpain-3 is a component of the sarcoplasmic reticulum (SR), and that calpain-3 interacts with, but does not proteolyze, typical SR components such as ryanodine receptor and calsequestrin. Furthermore, Capn3CS/CS mice showed that the non-enzymatic role of calpain-3 is required for proper Ca2+ efflux from the SR to cytosol during contraction of muscles. These results indicate that calpain-3 functions as a non-enzymatic element for the Ca2+ efflux machinery in the SR rather than as a protease. Thus, defects of the non-enzymatic function of calpain-3 must also be involved in pathogenesis of LGMD2A.
Keywords: limb-girdle muscular dystrophy type 2A, calpainopathy, ryanodine receptor, protease, proteolysis
Introduction
Calpains are Ca2+-requiring intracellular cysteine proteases that proteolyze substrates at specific sites to modulate substrate functions.1,2 Calpain-3 (also called p94) is predominantly expressed in skeletal muscle 3. In humans, CAPN3 has been found to be responsible for limb-girdle muscular dystrophy type 2A (LGMD2A, also called calpainopathy).4 Consistent with this, Capn3 knock-out (Capn3−/−) mice show a muscular dystrophy (MD) phenotype.5,6 We have shown that transgenic over-expression of mutant C129S calpain-3 which is structurally intact results also in a myopathy phenotype,7 and that calpain-3 knock-in mice (Capn3CS/CS) also showed MD.8 These results indicate that calpain-3 protease activity is essential for skeletal muscle functions.
In sarcomeres, calpain-3 directly binds to connectin/titin at the M-lines and the N2A regions9 where multiple proteins form complexes to respond to myofibrillar signal transduction systems.10,11 Interestingly, calpain-3 changes its localization from the M-lines to the N2A regions as sarcomeres extend.8,12 Although calpain-3 protease activity was dispensable for binding to connectin/titin,13 FRAP studies revealed that mobility of calpain-3 between the M-lines and the cytosol became compromised when its protease activity was lost.8 Capn3CS/CS mice showed an impaired adaptation to physical stress, accompanied by deficiency in exercise-dependent up-regulation of muscle ankyrin-repeat protein-2 and heat-shock proteins, which are normally observed in WT mice.8 Altogether, calpain-3 changes its localization in myocytes depending on sarcomere conditions to mediate signal transduction in response to external stress, and its protease activity maintains myocyte homeostasis, disruption of which leads to muscular dystrophy.8
The dystrophic symptoms of Capn3CS/CS mice are less severe than those reported for Capn3−/− mice; the phenotype observed in Capn3−/−, but not Capn3CS/CS, mice include increased serum creatine kinase (CK) and abnormal sarcomere organization,5,6,8 indicating that the severe phenotype of Capn3−/− mice was induced by the loss of calpain-3 protein in addition to the basic MD phenotype caused by the loss of its proteolytic activity. Accordingly work with Capn3−/− mice, suggests a structural role of calpain-3 in the integrity of the triad by binding to aldolase A.14 The Capn3CS/CS mouse strain is well suited to address novel functions of calpain-3, because it lacks only the proteolytic function of the calpain-3 protein, whereas the non-proteolytic functions are intact. By comparing present results with Capn3CS/CS with previous Capn3−/− data we found that calpain-3 plays a non-proteolytic role involved in Ca2+ efflux from the sarcoplasmic reticulum (SR). Both non-proteolytic and proteolytic functions of calpain-3 must play roles in the pathogenesis of LGMD2A.
Results
Identification of calpain-3 IS2 region interacting molecules
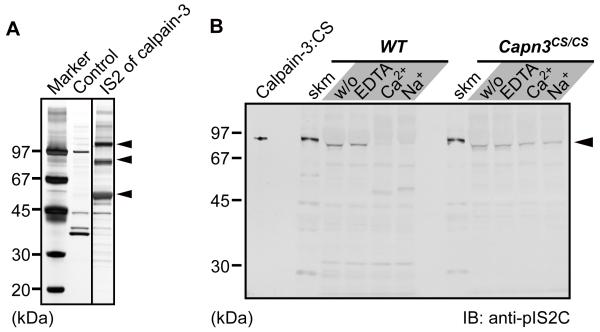
The phenotype of pathologically elevated serum CK levels and disorganized sarcomere structures was observed only in Capn3−/− mice, and not in Capn3CS/CS mice.5,6,8 To examine how inactive calpain-3 protein protects mice from severe symptoms observed in Capn3−/− mice, we searched calpain-3 interacting molecules in skeletal muscles by pull-down assay using an affinity column conjugated with a peptide corresponding to the IS2 region, one of the three calpain-3-characterizing regions.3 Three major bands interacted with the IS2 peptide were visualized with silver staining (Fig. 1A). Subsequent mass-spectrometry analysis identified these molecules (110 kDa, 90 kDa, and 55 kDa) as sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 1a (SERCA1a), phosphofructose kinase muscle type (PFKM), and calsequestrin 1 (CSQ1), respectively. These molecules are known components of the SR. Indeed, full-length calpain-3 was detected in the microsomal fraction isolated from skeletal muscle, which contains SR and T-tubule fractions (Fig. 1B, closed arrowhead). As calpain-3 is proteolytically activated in the presence of either Ca2+ or Na+,15 we examined the autolysis of calpain-3 in the SR. When this microsomal fraction from WT mice was treated with either Ca2+ or Na+, the calpain-3 signal disappeared, in contrast to that from Capn3CS/CS mice (Fig. 1B), showing that the detected band by anti-pIS2C antibody corresponded to full-length calpain-3 protein.
Fig. 1.
Identification of calpain-3 IS2 region interacting molecules.
(A) Identification of calpain-3-specific insertion 2 (IS2)-interacting proteins. Skeletal muscle lysates were pulled down with an IS2 peptide column, and subsequently the trapped proteins were electrophoresed and detected by silver staining. Three major bands at 110, 90, and 55 kDa (arrowheads) were identified as SERCA1a, phosphofructose kinase muscle type, and CSQ1, respectively, by MALDI-TOF/TOF analysis.
(B) Immunoblot detection of calpain-3 in a microsomal fraction. Microsomal fractions isolated from WT and Capn3CS/CS skeletal muscles were incubated with either 10 mM EDTA, 5 mM CaCl2, or 150 mM NaCl, at 30°C for 0 min (w/o) or 30 min. calpain-3:CS indicates the lysate from COS7 cells exogenously expressing human calpain-3:C129S. Slightly faster migration of the calpain-3 bands in the microsomal fractions than in total muscle fractions (skm) is because a large amount of SERCA1a migrating just above calpain-3 on the gel pushed calpain-3 downwards.
Calpain-3 is a component of the SR
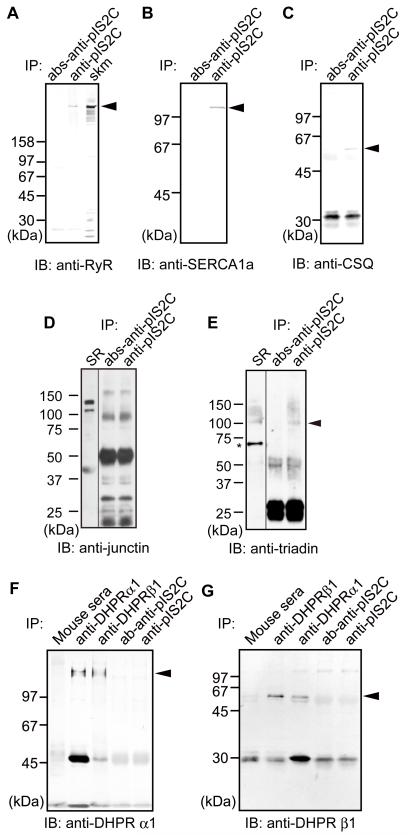
To examine whether calpain-3 is in the SR or the T-tubule, we immunoprecipitated (IPed) the lysed microsomal fraction with an anti-IS2C antibody. Immunoprecipitates (IPs) were then analyzed with antibodies directed to major components of the SR (ryanodine receptor (RyR), SERCA1a, CSQ1, junctin and triadin) and T-tubule (dihydropyridine receptors (DHPR) α1 and β1). As a result, RyR, SERCA1a, CSQ1, and triadin co-IPed with calpain-3, whereas junctin, DHPRα1 and β1 did not (DHPRα1 and β1 were co-IPed with each other, indicating functionality of the IP assay used) (Fig. 2). These results suggest that calpain-3 associates with the molecules present in the SR rather than in the T-tubule, although we have not identified molecule(s) that directly interact with calpain-3 in the SR.
Fig. 2.
Calpain-3 associates with protein complexes in the SR.
IP of microsomal fractions with anti-pIS2C antibody analyzed by western blot using antibodies against RyR (A), SERCA1a (B), CSQ1(C), junctin (D), triadin (E), DHPRα1 (F) and DHPRβ1 (G). Peptide-absorbed anti-calpain-3 (pIS2C) antibody (abs-anti-pIS2C) and normal mouse sera were used as negative controls. Junctin is one of the isoforms of aspartyl/asparaginyl α-hydroxylase (ASPH). Apparent molecular weight of junctin is about 40,000, although its calculated molecular weight is 26,000.37 Closed arrowheads, indicated proteins; IB, immunoblot; skm, total muscle fractions as a positive control for RyR; SR, microsome fraction as a positive control for junctin and triadin; *non-specific band detected by anti-triadin antibody used.
Calpain-3 did not proteolyze molecules in the SR components
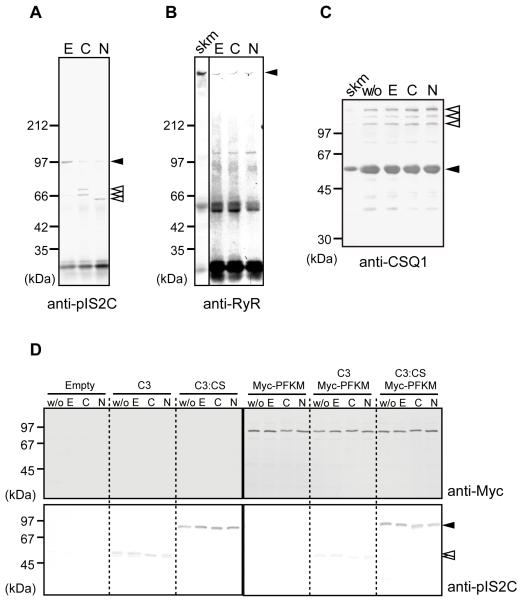
To determine if the SR components are proteolyzed by calpain-3, the IP complex obtained with anti-pIS2C antibody was incubated with either Ca2+ or Na+. Full-length calpain-3 (Fig. 3A, closed arrowhead) in this complex was autolyzed after the addition of Na+ as well as Ca2+ (Fig. 3A, open arrowheads). In contrast, RyR did not show any breakdown product detectable with anti-RyR antibody (Fig. 3B). Nor did CSQ1 (Fig. 3C) in the microsomal fraction or PFKM (Fig. 3D) exogenously expressed in the COS cells, indicating that these major SR proteins are not substrates for calpain-3.
Fig. 3.
Calpain-3 did not proteolyze molecules in the SR.
(A-B) IPs of microsomal fractions from WT mice by anti-pIS2C antibody were incubated with 10 mM EDTA (E), 5 mM CaCl2 (C), or 150 mM NaCl (N), and detected by anti-IS2C (A) or anti-RyR (B). In the presence of Ca2+ or Na+, calpain-3 was autolyzed. Open arrowheads, autolytic fragments of calpain-3; closed arrowheads, full-length calpain-3 (A), and full-length RyR (B). No RyR fragments were observed (B).
(C) The microsomal fraction from WT mice was treated with none (w/o), 10 mM EDTA (E), 5 mM CaCl2 (C), or 150 mM NaCl (N), and was subjected to western blot analysis using anti-CSQ1 antibody. No breakdown product for CSQ1 was observed. Closed arrowhead, CSQ1; open arrowheads, CSQ-like proteins;38 skm, total skeletal muscle lysate.
(D) Lysates of COS7 cells transfected with empty pcDNA3.1 vector (Empty), or expression vectors for calpain-3 (C3), calpain-3:C129S (C3:CS), and/or Myc-PFKM, were incubated with 10 mM EDTA (E), 5 mM CaCl2 (C), or 150 mM NaCl (N). “w/o” indicates no incubation control. No proteolysis of Myc-PFKM was observed in any lane when detected with anti-Myc antibody. Calpain-3 was detected by anti-pIS2C antibody (closed and open arrowheads indicate full-length and autolytic fragments of calpain-3, respectively). Note that wild type calpain-3 autolyzed without incubation, but that further band shift was observed when incubated with CaCl2 or NaCl.
Calpain-3 protease activity is dispensable for its localization in the SR and for triad function
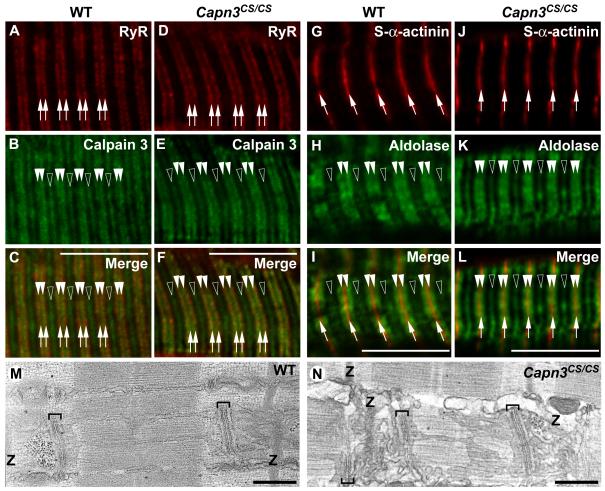
To investigate whether calpain-3 proteolytic activity is required for targeting to the SR or not, skeletal muscle longitudinal cryosections from WT and Capn3CS/CS mice were analyzed with antibodies against calpain-3 and RyR. In both WT calpain-3 and Capn3CS/CS mice, RyR showed doublets structures along the Z-bands where calpain-3 was partially co-localized with RyR (Fig. 4A-F), indicating that protease activity is not required for sorting of calpain-3 to the SR.
Fig. 4.
Localizations of RyR and aldolase A were not affected by a loss of calpain-3 protease activity.
(A-L) Longitudinal cryostat sections of EDL from WT (A-C, G-I) and Capn3CS/CS (D-F, J-L) mice were reacted with antibodies against RyR (A, D), calpain-3 (B, E), s-α-actinin (G, J), and aldolase A (H, K). RyR (arrows in A, C, D, F) partially co-localized with calpain-3 in the N2A regions of both WT and Capn3CS/CS (closed arrowheads in B, C, E, F). In addition to the N2A regions, calpain-3 was detected in the M-lines as we previously reported8 (open arrowheads in B and E). Localizations of aldolase A in both WT and Capn3CS/CS showed the identical patterns, i.e. doublets along the Z-bands (closed arrowheads in G, I, J, L), and near the M-lines (open arrowheads in H, I, K, L). Bars, 10 μm.
(M-N) Representative electron microscopic images of longitudinal sections of EDL from WT (M) and Capn3CS/CS (N) mice. The positions of the Z-bands in the sarcomeres are indicated as “Z”. Brackets indicate triads, in which a T-tubule is sandwiched between two junctional SR cisternae. The triad locates at the A-I junctions of the sarcomeres in both types of mice. The longitudinal SR runs in parallel with myofibrils between two triads. Bars, 0.5 μm.
As calpain-3 was suggested to be involved in recruitment of aldolase A to the triads,14 the structural integrity of aldolase A was examined with its specific antibody. Aldolase A was detected as doublets structures along the Z-bands, and in the M-line regions of both WT and Capn3CS/CS muscles (Fig. 4G-L). Localization of aldolase A in the triads of Capn3CS/CS muscles is consistent with the idea that calpain-3 is a structural component anchoring aldolase A to the triads,14 and that the proteolytic activity of calpain-3 is not necessary for this function.
We further observed the triad structure in skeletal muscles from WT and Capn3CS/CS mice with electron microscopy to reveal the impact of the loss of calpain-3 protease activity on the triad. Consistent with our light microscopic observations, the triad structures, in which two junctional SR cisternae face the T-tubule segment at the A-I junction in the sarcomeres, showed no marked difference between WT and Capn3CS/CS mice (brackets in Fig. 4M-N).
Proteolytic activity of calpain-3 is dispensable for Ca2+ efflux from SR
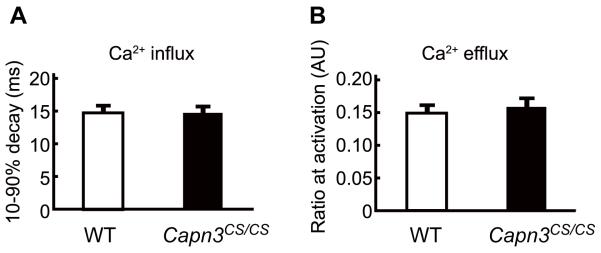
We investigated the non-proteolytic functions of calpain-3 in the SR. Ca2+ kinetics of WT and Capn3CS/CS fibers were examined by the same method as used in Kramerova et al.14 who reported that Capn3−/− mice have reduced Ca2+ release from the SR but normal uptake kinetics. Surprisingly, no significant differences were seen between WT and Capn3CS/CS mice in neither Ca2+ release nor re-uptake (Fig. 5). Taking all results into account, we conclude that calpain-3 protease activity is dispensable for Ca2+ kinetics, but not the calpain-3 molecule itself, revealing that calpain-3 is an essential component of the SR.
Fig. 5.
Proteolytic function of Calpain-3 is not required for SR calcium kinetics.
(A, B) Myofibers isolated from FDB muscle of WT and Capn3CS/CS mice were loaded with Fura2FF/AM, twitch activated and Ca2+ transients were measured. The time required for the decay of Fura2FF/AM fluorescence from 90 to 10% of its peak was used to determine the speed of Ca2+ influx from the cytosol to the SR (A). Ca2+ efflux from the SR to the cytosol was measured by the ratio of Ca2+ bound to Ca2+ unbound fluorescence upon myofiber stimulation (B). There was no significant difference in Ca2+ influx and speed of efflux between WT and Capn3CS/CS. The numbers of measured myofibers from WT and Capn3CS/CS were 24 and 22, respectively. Error bars, SE.
Discussion
In this study we demonstrated that calpain-3 is a component of the SR. Furthermore, the difference in SR functions between Capn3CS/CS and Capn3−/− mice elucidated a non-proteolytic function of calpain-3 in Ca2+ release from the SR, suggesting that calpain-3 is a member of the RyR associated protein complex in the SR.16 In addition to the previous study14 that showed that aldolase A was recruited to the SR component by calpain-3, our results suggest that calpain-3 plays a role in stabilizing RyR associated protein complexes in the SR. When Ca2+ is discharged to the cytosol, it is possible that calpain-3 functions as a fine tuning element in coordination with RyR and other molecules in the SR. Sorcin, five EF-hand motifs (called “penta EF-hand motif (PEF) domain”) containing Ca2+ binding molecule, has been reported to modulate the open probability of RyR through Ca2+/calmodulin-dependent protein kinase II.17,18 As calpain-3 also has PEF domain, calpain-3 might have a similar effect.
Besides calpain-3, μ- and m-calpains also exist in the SR.19 Our result that calpain-3 did not proteolyze RyR suggests that the role of calpain-3 in the SR is different from that of both μ- and m-calpains, which proteolyze RyR in vitro.19 In this study, CSQ1 was not cleaved by calpain-3 as well, although our previous proteomic study showed that CSQ1 is one of the calpain-3 candidate substrates.8 In myofibrils, calpain-3 protease activity is suppressed when calpain-3 binds to connectin/titin.20 In a similar fashion, the suppression of calpain-3 protease activity might occur in the intact SR structure so as not to be autolyzed quickly, although a molecule directly binding to calapin-3 has not yet been identified in the SR. Calpain-3 itself is activated in the SR complex in vitro, but it did not proteolyze major SR components, suggesting that calpain-3 down-regulates itself without degrading SR components when the proteolytic function of calpain-3 is no longer required, and/or that calpain-3 keeps its conformational integrity even if calpain-3 molecule is nicked by autolysis.21
CSQ1 was shown to co-immunoprecipitate with calpain-3, even though CSQ1 and calpain-3 are considered to localize differently in the SR lumen and the cytosol, respectively. As triadin is a transmembrane molecule of the SR and interacts with both RyR in the cytosol and CSQ1 in the SR lumen, triadin may associate calpain-3 with CSQ1.22 In other words, it is possible that calpain-3 may interact with RyR and/or triadin in the RyR-triadin-CSQ1 complex that spans the SR membrane. Alternatively, it is also possible that calpain-3 is located in the luminal SR, since other calpain members have been reported to exist in luminal compartments of organelles such as mitochondria.23,24,25,26 Further study is required to clarify the interaction of calpain-3 with CSQ1.
The triad structure is located in the A-I junctions, which is close to the N2A region, in the sarcomere of skeletal muscles.27 Electron microscopic observation demonstrated that the distance between the triads and the Z-bands depends on the sarcomere length, suggesting that an elastic myofibrillar component is responsible for maintaining the triads in the proper positions.28 However, it still remains unclear what links the triads to the myofibrils. We hypothesize that calpain-3 is involved in linking the triads to myofibrils. This hypothesis is based on the results that calpain-3 directly interacts with connectin/titin at the N2A regions,9 and that calpain-3 is a component of the SR and the triads (ref. 14 and this study). Consistent with this, the triad is located at the N2A region in skeletal muscle cells where calpain-3 is expressed, while it is located at the Z-bands in cardiac cells where calpain-3 is not expressed.3,29 Furthermore, Capn3−/− mice showed reduced amounts of RyR and aldolase A in the SR.14
In the SR fraction, calpain-3 and RyR account for approximately 1% and 5% of the total protein, respectively.30,31 As their molecular weights are 94,000 and 560,000, respectively, their molar ratio in the SR is about 1:1. However, calpain-3 interacts with RyR, SERCA1a, and aldolase A, and may exist in the SR lumen. Thus, not all RyRs are expected to be associated with calpain-3. Taken together, it is possible that calpain-3 is one of the molecules that tether the triads at their specific position to connectin/titin in skeletal muscles (Fig. 6).
Fig. 6.
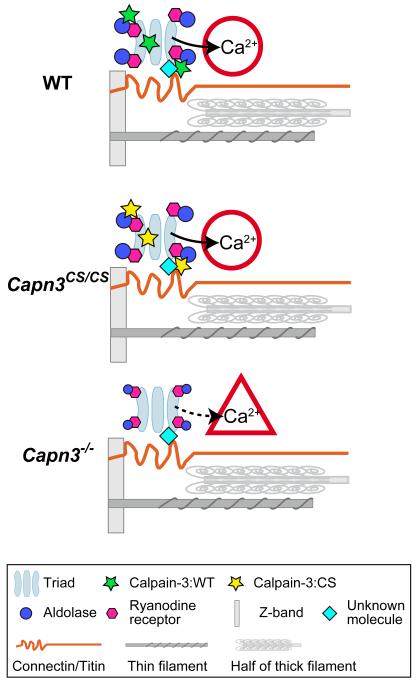
Model of calpain-3 structural functions in the SR.
The triads and major myofibrillar components are shown in a half sarcomere. (A) In WT mice, calpain-3 molecules (with proteolytic activity suppressed by an unknown mechanism) are structurally associated with SR components, and normal Ca2+ efflux from the SR takes place. (B) In Capn3CS/CS mice, normal Ca2+ efflux from the SR also takes place because calpain-3:C129S is proteolytically inactive calpain-3 but structurally intact. (C) In Capn3−/− mice, however, the calpain-3 molecules is lost and as a result Ca2+ efflux from the SR is reduced.14
Calpain-3 binds to connectin/titin at the M-lines and to s-α-actinin at the Z-bands, where muscle stress is transmitted to adjacent sarcomeres and extracellular regions. Thus, besides the SR, calpain-3 may function as a structural element in myofibrils to stabilize sarcomere components. Indeed, the absence of calpain-3 molecule in Capn3−/− mouse muscles caused disassembly of thick filaments.5 In contrast, our Capn3CS/CS mice showed properly aligned thick filaments,8 suggesting that, regardless of proteolytic activity, calpain-3 protein plays a role as a structural element of myofibrils as well as of the SR.
Recently, calpain-6, a unique non-proteolytic member of the mammalian calpain family with a naturally substituted catalytic residue, was reported to be involved in regulation of microtubules and microfilaments.33 As calpain-6 has no proteolytic activity, it is also likely to function as a structural element. Thus, a novel and universal function of calpains appears to be that of a scaffold by interacting with various molecules, as shown by their wide range of substrate specificity. Calpain-3, however, has very rapid and strong autolytic activity,32 and, therefore, there should be some mechanism for suppression of this activity in order for it to be functional as a structural protein. Elucidation of this mechanism will be an important future research topic.
In conclusion, using Capn3CS/CS mice, we demonstrate that calpain-3 has enzymatic and non-enzymatic roles in skeletal muscles, both of which are necessary for proper function of skeletal muscles. Comparison of Capn3CS/CS and Capn3−/− mice is unique and advantageous for elucidation of novel non-proteolytic functions of calpain-3. This comparison conclusively establishes non-proteolytic roles of calpain-3 that must be considered for pathogenic mechanisms of LGMD2A. For example, a wide range of MD severities were reported for LGMD2A, and, in general, patients with still detectable calpain-3 in western blot analysis have a less severe phenotype. This could reflect that in these patients structural functions of calpain-3 remained to some extent intact, resulting in less severe phenotypes. Thus, the non-proteolytic function of calpain-3 should be considered when designing therapeutics for the treatment of LGMD2A.
Materials and methods
Experimental animals
All procedures using experimental animals were approved by the Experimental Animal Care and Use Committee of Rinshoken, and the animals were treated according to the committee’s guidelines. C57BL/6 mice were purchased from Nihon CLEA Inc.
To generate Capn3CS/CS mice, a missense mutation that results in a Cys129Ser substitution and a neomycin-resistance gene (neoR) flanked by loxP sequences were introduced in exon 3 and intron 3, respectively, of the Capn3 gene. Capn3CS/CS mice were backcrossed with C57BL/6 J mice for more than 8 generations. Mouse genotypes were determined by PCR analyses.8 The SR fractions were prepared using skeletal muscles isolated from forelimb, hindlimb, and gluteal region of 7-20 week old mice.
Pull-down assay
Skeletal muscles were lysed with DG buffer (1% digitonin, 150 mM CsCl, 10 mM triethanolamine, 1mM EDTA, 10 μg/ml aprotinin, 2 mM PMSF) at 4°C for 1 hr. After centrifugation at 23,000×g for 15 min, the supernatant was incubated with human calpain-3-IS2 peptide (VKKKNKPIIFV-C)-bound Sepharose 4B at 4°C for 1 hr. Non-bound Sepharose 4B was used as control. Resins were extensively washed with the DG buffer, and eluted with 0.2 M glycine[pH2.5]. Bands were visualized with the Silver Staining Kit, Protein (GE Healthcare).
Identification of proteins by mass spectrometry
Gel slices containing silver-stained bands of interest were reduced, alkylated and digested by trypsin overnight at 37°C. Peptides were extracted from the gels with 50% acetonitrile in 0.1% trifluoroacetic acid (TFA) solution, purified with ZipTip (Millipore), mixed with matrix solution (5 mg/ml α-cyano-4-hydroxycinnamic acid, 50% acetonitrile, 0.1% TFA), and spotted onto a matrix-assisted laser desorption ionization (MALDI) target plate. Spotted samples were analyzed by MALDI-time-of-flight (TOF) mass-spectrometry (ABI 4800), and peptides were identified from the MS/MS spectra using a GPS Workstation (ABI).
Expression in COS7 cells
For expression of calpain-3 in COS7 cells, cDNA for full-length human calpain-3 or its C129S mutant in pSRD vector as previously described20 was used. The cDNA corresponding to phosphofructose kinase muscle type (PFKM) with an N-terminal Myc tag was cloned into pcDNA3.1 (Invitrogen) vector for expression of PFKM in COS7 cells. These expression vectors were transfected into COS7 cells with an electroporation method by Gene Pulser (Bio-Rad) as previously described.20
Immunoprecipitation and proteolysis assay of calpain-3
The microsomal fraction, which contains the SR and the t-tubule components, was prepared from skeletal muscles according to the previous report34 with slight modifications. In brief, a sucrose gradient centrifugation was omitted at the final step of the preparation. The microsome fraction was lysed with DG buffer, and then the supernatant fraction was obtained after centrifugation of lysed microsome fraction and passing through 0.22 μm filter membranes (Millipore). To minimize non-specific binding, the microsomal fraction was incubated with protein-G-Sepharose 4FF (GE) at 4°C for 1 hr. After centrifugation, the supernatant was incubated with either anti-calpain-3 (pIS2C) or peptide-absorbed anti-pIS2C antibody at 4°C for 1 hr. Subsequently, protein-G-Sepharose was added to each sample and reacted at 4°C for 1 hr. After washing with DG buffer, IPed molecules were eluted with antigen peptides for anti-pIS2C antibody, or directly boiled with SDS sample buffer.
For the calpain-3 proteolysis assay, skeletal muscle lysates, microsomal fractions, or COS7 cell lysates were suspended in TED buffer (10 mM Tris/Cl[pH7.5], 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.1 μM leupeptin, 40 μM bestatin, 1.5 μM aprotinin, and 0.7 μM calpastatin (Takara)), and incubated at 30°C for 30 min in the presence of 10 mM EDTA, 5 mM CaCl2, or 150 mM NaCl.
Western blot analysis
Western blot analysis was performed as previously described.8 Skeletal muscles were lysed with TED buffer. Primary antibodies are listed in Table 1. Peroxidase-conjugated goat-IgG or mouse-IgG were used as secondary antibodies. Subsequently, band signal was visualized with either POD immunostain set (Wako pure chemical industries, Ltd.), or ECL plus (GE Healthcare).
Table 1.
Antibodies used in this study
| Antigen name | Dilution (Blot) | Dilution (Histology) |
Company/Reference |
|---|---|---|---|
| pIS2C (calpain-3) | 1:3000 | 1:300 | 12 |
| RyR | 1:5000 | 1:500 | Affinity BioReagents |
| DHPR α1 | 1:500 | - | Affinity BioReagents |
| DHPR β1 | 1:500 | - | Developmental Studies Hybridoma Bank |
| CSQ | 1:2000 | - | Affinity BioReagents |
| sarcomeric-α-actinin | - | 1:1000 | Sigma |
| aldolase A | - | 1:1000 | Rockland |
| Myc | 1:1000 | - | clone#9E10; Developmental Studies Hybridoma Bank |
| triadin | 1:500 | - | Affinity BioReagents |
| ASPH (junctin) | 1:500 | - | Santa Cruz |
not used in this study
Histology
Immunochemical labeling were performed as previously reported.35 Primary antibodies were listed in Table 1. Alexa 488 or 555 conjugated goat-IgG or mouse-IgG were used as secondary antibodies. Fluorescence signal was observed on a laser scanning confocal microscope (Zeiss LSM510). The images were recorded and processed with LSM imaging software.
Electron Microscopy
EDL muscles were fixed with 2% glutaraldehyde, postfixed with 1% OsO4, and embedded in Epon. Ultrathin sections were stained with uranyl acetate and examined using a JEM 1200EX transmission electron microscope (JEOL).
Measurement of Ca2+ transients
Ca2+ efflux and influx were measured as described previously.36 Briefly, myofibers were isolated from flexor digitorum brevis muscles of WT and Capn3CS/CS mice by collagenase treatment (Worthington Biochem.). Isolated single myofibers were loaded with 4 μM Fura2FF/AM (Teflabs) for 45 minutes at 20°C. Under a microscope (Olympus IX70), dissociated intact myofibers were field simulated and then the fluorescence ratio at 360/380 nm excitation was determined to calculate the Ca2+ transient. To inhibit actomyosin interaction and reduce motion artifacts, 10 μM of N-benzyl-p-toluenesulphonamide (BTS) was added. These methods are exactly the same as described in ref. 14, and were performed by the same scientists (Drs. Ottenheijm and Granzier) at almost the same time, and using identical reagents and experimental conditions.
Acknowledgements
We are grateful to all the members of the Calpain Project (Rinshoken) for valuable support and discussions. This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology [MEXT.KAKENHI 18076007 to H.S.]; Japan Society for the Promotion of Science [JSPS.KAKENHI 20500369 to K.O., 20370055 to H.S.]; the Ministry of Health, Labor and Welfare [Research Grant (20B-13) for Nervous and Mental Disorders to H.S.]; the Takeda Science Foundation research grant to H.S.; the National Institutes of Health [NIH 062881 to H.G.].
Abbreviations
- CSQ
calsequestrin
- EDL
extensor digitorum longus
- IP
immunoprecipitated
- Capn3CS/CS
a genotype with both calpain-3 gene alleles with a mutation corresponding to C129S
- MD
muscular dystrophy
- PFKM
phosphofructose kinase muscle type
- RyR
ryanodine receptor
- s-α-actinin
sarcomeric-α-actinin
- SERCA
sarco/endoplasmic reticulum Ca2+ ATPase
- SR
sarcoplasmic reticulum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53:S12–18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- 3.Sorimachi H, Imajoh-Ohmi S, Emori Y, Kawasaki H, Ohno S, Minami Y, Suzuki K. Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J. Biol. Chem. 1989;264:20106–20111. [PubMed] [Google Scholar]
- 4.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, Hillaire D, Passos-Bueno M-R, Zats M, Tischfield JA, Fardeau M, Jackson CE, Cohen D, Beckmann JS. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 5.Kramerova I, Kudryashova E, Tidball JG, Spencer MJ. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 6.Richard I, Roudaut C, Marchand S, Baghdiguian S, Herasse M, Stockholm D, Ono Y, Suel L, Bourg N, Sorimachi H, Lefranc G, Fardeau M, Sebille A, Beckmann JS. Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated IkappaBalpha/nuclear factor kappaB pathway perturbation in mice. J. Cell Biol. 2000;151:1583–1590. doi: 10.1083/jcb.151.7.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tagawa K, Taya C, Hayashi Y, Nakagawa M, Ono Y, Fukuda R, Karasuyama H, Toyama-Sorimachi N, Katsui Y, Hata S, Ishiura S, Nonaka I, Seyama Y, Arahata K, Yonekawa H, Sorimachi H, Suzuki K. Myopathy phenotype of transgenic mice expressing active site-mutated inactive p94 skeletal muscle-specific calpain, the gene product responsible for limb girdle muscular dystrophy type 2A. Hum. Mol. Genet. 2000;9:1393–1402. doi: 10.1093/hmg/9.9.1393. [DOI] [PubMed] [Google Scholar]
- 8.Ojima K, Kawabata Y, Nakao H, Nakao K, Doi N, Kitamura F, Ono Y, Hata S, Suzuki H, Kawahara H, Bogomolovas J, Witt C, Ottenheijm C, Labeit S, Granzier H, Toyama-Sorimachi N, Sorimachi M, Suzuki K, Maeda T, Abe K, Aiba A, Sorimachi H. Dynamic distribution of muscle-specific calpain in mice has a key role in physical-stress adaptation and is impaired in muscular dystrophy. J Clin Invest. 2010;120:2672–83. doi: 10.1172/JCI40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorimachi H, Kinbara K, Kimura S, Takahashi M, Ishiura S, Sasagawa N, Sorimachi N, Shimada H, Tagawa K, Maruyama K, Suzuki K. Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J. Biol. Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 10.Granzier HL, Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004;94:284–95. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 11.Linke WA, Kruger M. The giant protein titin as an integrator of myocyte signaling pathways. Physiology (Bethesda) 2010;25:186–98. doi: 10.1152/physiol.00005.2010. [DOI] [PubMed] [Google Scholar]
- 12.Ojima K, Ono Y, Doi N, Yoshioka K, Kawabata Y, Labeit S, Sorimachi H. Myogenic stage, sarcomere length, and protease activity modulate localization of muscle-specific calpain. J. Biol. Chem. 2007;282:14493–1504. doi: 10.1074/jbc.M610806200. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi C, Ono Y, Doi N, Kitamura F, Tagami M, Mineki R, Arai T, Taguchi H, Yanagida M, Hirner S, Labeit D, Labeit S, Sorimachi H. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J. Biol. Chem. 2008;283:14801–14. doi: 10.1074/jbc.M708262200. [DOI] [PubMed] [Google Scholar]
- 14.Kramerova I, Kudryashova E, Wu B, Ottenheijm C, Granzier H, Spencer MJ. Novel role of calpain-3 in the triad-associated protein complex regulating calcium release in skeletal muscle. Hum. Mol. Genet. 2008;17:3271–80. doi: 10.1093/hmg/ddn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono Y, Ojima K, Torii F, Takaya E, Doi N, Nakagawa K, Hata S, Abe K, Sorimachi H. Skeletal muscle-specific calpain is an intracellular Na+-dependent protease. J Biol Chem. 2010;285:22986–98. doi: 10.1074/jbc.M110.126946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzini-Armstrong C, Protasi F. Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiological Reviews. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 17.Lokuta AJ, Meyers MB, Sander PR, Fishman GI, Valdivia HH. Modulation of cardiac ryanodine receptors by sorcin. J. Biol. Chem. 1997;272:25333–8. doi: 10.1074/jbc.272.40.25333. [DOI] [PubMed] [Google Scholar]
- 18.Anthony DF, Beattie J, Paul A, Currie S. Interaction of calcium/calmodulin-dependent protein kinase IIdeltaC with sorcin indirectly modulates ryanodine receptor function in cardiac myocytes. J Mol Cell Cardiol. 2007;43:492–503. doi: 10.1016/j.yjmcc.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist JS, Wang KK, Katz S, Belcastro AN. Calcium-activated neutral protease effects upon skeletal muscle sarcoplasmic reticulum protein structure and calcium release. J. Biol. Chem. 1992;267:20857–65. [PubMed] [Google Scholar]
- 20.Ono Y, Torii F, Ojima K, Doi N, Yoshioka K, Kawabata Y, Labeit D, Labeit S, Suzuki K, Abe K, Maeda T, Sorimachi H. Suppressed disassembly of autolyzing p94/CAPN3 by N2A connectin/titin in a genetic reporter system. J. Biol. Chem. 2006;281:18519–18531. doi: 10.1074/jbc.M601029200. [DOI] [PubMed] [Google Scholar]
- 21.Ono Y, Kakinuma K, Torii F, Irie A, Nakagawa K, Labeit S, Abe K, Suzuki K, Sorimachi H. Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J. Biol. Chem. 2004;279:2761–2771. doi: 10.1074/jbc.M308789200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Kelley J, Schmeisser G, Kobayashi YM, Jones LR. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J Biol Chem. 1997;272:23389–97. doi: 10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 23.Badugu R, Garcia M, Bondada V, Joshi A, Geddes JW. N terminus of calpain 1 is a mitochondrial targeting sequence. J. Biol. Chem. 2008;283:3409–17. doi: 10.1074/jbc.M706851200. [DOI] [PubMed] [Google Scholar]
- 24.Ozaki T, Yamashita T, Ishiguro SI. Mitochondrial m-calpain plays a role in the release of truncated apoptosis-inducing factor from the mitochondria. Biochim. Biophys. Acta. 2009;1793:1848–1859. doi: 10.1016/j.bbamcr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Arrington DD, Van Vleet TR, Schnellmann RG. Calpain 10: a mitochondrial calpain and its role in calcium-induced mitochondrial dysfunction. Am. J. Physiol. Cell Physiol. 2006;291:C1159–71. doi: 10.1152/ajpcell.00207.2006. [DOI] [PubMed] [Google Scholar]
- 26.Giguere CJ, Covington MD, Schnellmann RG. Mitochondrial calpain 10 activity and expression in the kidney of multiple species. Biochem. Biophys. Res. Commun. 2008;366:258–62. doi: 10.1016/j.bbrc.2007.11.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flucher BE, Takekura H, Franzini-Armstrong C. Development of the excitation-contraction coupling apparatus in skeletal muscle: association of sarcoplasmic reticulum and transverse tubules with myofibrils. Dev Biol. 1993;160:135–47. doi: 10.1006/dbio.1993.1292. [DOI] [PubMed] [Google Scholar]
- 28.Takekura H, Kasuga N, Yoshioka T. Influences of sarcomere length and selective elimination of myosin filaments on the localization and orientation of triads in rat muscle fibres. J Muscle Res Cell Motil. 1996;17:235–42. doi: 10.1007/BF00124245. [DOI] [PubMed] [Google Scholar]
- 29.Engel AG, Franzini-Armstrong C. Myology. 3rd edition. 2004. the. [Google Scholar]
- 30.Ilian MA, Morton JD, Bekhit AE, Roberts N, Palmer B, Sorimachi H, Bickerstaffe R. Effect of preslaughter feed withdrawal period on longissimus tenderness and the expression of calpains in the ovine. J. Agric. Food Chem. 2001;49:1990–8. doi: 10.1021/jf0010026. [DOI] [PubMed] [Google Scholar]
- 31.Imagawa T, Smith JS, Coronado R, Campbell KP. Purified ryanodine receptor from skeletal muscle sarcoplasmic reticulum is the Ca2+-permeable pore of the calcium release channel. J Biol Chem. 1987;262:16636–43. [PubMed] [Google Scholar]
- 32.Ono Y, Hayashi C, Doi N, Kitamura F, Shindo M, Kudo K, Tsubata T, Yanagida M, Sorimachi H. Comprehensive survey of p94/calpain 3 substrates by comparative proteomics--possible regulation of protein synthesis by p94. Biotechnol. J. 2007;2:565–76. doi: 10.1002/biot.200700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonami K, Kurihara Y, Aburatani H, Uchijima Y, Asano T, Kurihara H. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol. Cell. Biol. 2007;27:2548–2561. doi: 10.1128/MCB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito A, Seiler S, Chu A, Fleischer S. Preparation and morphology of sarcoplasmic reticulum terminal cisternae from rabbit skeletal muscle. J Cell Biol. 1984;99:875–85. doi: 10.1083/jcb.99.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ojima K, Uezumi A, Miyoshi H, Masuda S, Morita Y, Fukase A, Hattori A, Nakauchi H, Miyagoe-Suzuki Y, Takeda S. Mac-1(low) early myeloid cells in the bone marrow-derived SP fraction migrate into injured skeletal muscle and participate in muscle regeneration. Biochem. Biophys. Res. Commun. 2004;321:1050–1061. doi: 10.1016/j.bbrc.2004.07.069. [DOI] [PubMed] [Google Scholar]
- 36.Capote J, Bolanos P, Schuhmeier RP, Melzer W, Caputo C. Calcium transients in developing mouse skeletal muscle fibres. J. Physiol. 2005;564:451–64. doi: 10.1113/jphysiol.2004.081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover L, Quinn S, Ryan M, Pette D, Ohlendieck K. Supramolecular calsequestrin complex. Eur J Biochem. 2002;269:4607–16. doi: 10.1046/j.1432-1033.2002.03160.x. [DOI] [PubMed] [Google Scholar]
- 38.Knudson CM, Chaudhari N, Sharp AH, Powell JA, Beam KG, Campbell KP. Specific absence of the alpha 1 subunit of the dihydropyridine receptor in mice with muscular dysgenesis. J Biol Chem. 1989;264:1345–8. [PubMed] [Google Scholar]