Abstract
Mitochondrial DNA (mtDNA) is inherited as a protein–DNA complex (the nucleoid). We show that activation of the general amino acid response pathway in ρ+ and ρ– petite cells results in an increased number of nucleoids without an increase in mtDNA copy number. In ρ– cells, activation of the general amino acid response pathway results in increased intramolecular recombination between tandemly repeated sequences of ρ– mtDNA to produce small, circular oligomers that are packaged into individual nucleoids, resulting in an ∼10–fold increase in nucleoid number. The parsing of mtDNA into nucleoids due to general amino acid control requires Ilv5p, a mitochondrial protein that also functions in branched chain amino acid biosynthesis, and one or more factors required for mtDNA recombination. Two additional proteins known to function in mtDNA recombination, Abf2p and Mgt1p, are also required for parsing mtDNA into a larger number of nucleoids, although expression of these proteins is not under general amino acid control. Increased nucleoid number leads to increased mtDNA transmission, suggesting a mechanism to enhance mtDNA inheritance under amino acid starvation conditions.
Keywords: general amino acid control/mtDNA/nucleoid/recombination/yeast
Introduction
Mitochondria are essential organelles whose function in respiratory metabolism requires expression of the mitochondrial genome. Mitochondrial DNA (mtDNA) must therefore be faithfully inherited in order to insure propagation of respiratory-competent mitochondria. MtDNA is organized as a DNA–protein complex that can be visualized in cells with DNA-specific dyes as brightly staining, punctate structures termed nucleoids (Williamson and Fennell, 1979; Stevens, 1981). Early studies of inheritance of mtDNA in zygotic pedigrees indicated that the number of segregating units is smaller than the number of mtDNA molecules (Dujon, 1981); those findings plus some more recent evidence (Lockshon et al., 1995; Nunnari et al., 1997; Okamoto et al., 1998) make it likely that the nucleoid is the basic unit of mtDNA segregation. Very little is known about the protein composition of nucleoids, how they are organized, what controls their assembly and how they are transmitted to progeny cells.
Haploid cells of the yeast Saccharomyces cerevisiae with wild-type mitochondrial genomes (ρ+) contain some 10–20 nucleoids per cell, which is somewhat less than the number of genome equivalents of ρ+ mtDNA, estimated to be 25–50 (Williamson and Fennell, 1979). In respiratory-incompetent, ρ– petite cells, large segments of the ∼80 kb ρ+ mitochondrial genome are deleted, and the retained mtDNA sequences are amplified as tandem repeats. Although the amount of mtDNA in ρ– petites is about the same as in ρ+ cells (Dujon, 1981), ρ– petites generally have fewer nucleoids per cell, and these are larger and stain more brightly than those in ρ+ cells. These differences in nucleoid number and morphology probably reflect the larger sized molecules of ρ– mtDNA that are linked together through recombination between the repeat units (Lockshon et al., 1995).
In matings between cells with different mitochondrial genomes, progeny cells with pure mitochondrial genotypes (homoplasmic) appear rapidly among the diploid progeny, suggesting that only a fraction of the input mitochondrial genomes is transmitted to a bud (Birky et al., 1978). Recent studies on the sorting of mitochondrial constituents in zygotes suggest that nucleoid transmission is an active process whereby these structures are sorted preferentially to the diploid buds independently of the sorting of proteins of the mitochondrial matrix, inner and outer membranes (Nunnari et al., 1997; Okamoto et al., 1998). These observations suggest the existence of a nucleoid segregation apparatus. Finally, the inheritance of mtDNA depends on a daunting array of factors that function in mtDNA recombination, mitochondrial gene expression, biogenesis, morphology and metabolism, as well as on other factors whose precise functions are unknown (reviewed in Hermann and Shaw, 1998).
Some insights into the inheritance of mtDNA have been obtained from studies of an abundant mtDNA-binding protein, Abf2p. This protein is a member of the high mobility group (HMG) family of DNA-binding proteins and is essential for the transmission of ρ+ mtDNA in cells grown on fermentable carbon sources but is dispensable in cells grown on non-fermentable carbon sources (Diffley and Stillman, 1991; Megraw and Chae, 1993; Zelenaya-Troitskaya et al., 1998), i.e. conditions that select for respiratory-competent cells. These observations suggest that, unlike its mammalian counterpart, mtTFA (Parisi and Clayton, 1991), Abf2p plays little or no role in mitochondrial gene expression. Indeed, vertebrate orthologs contain a C–terminal extension, absent from Abf2p, that has been shown to be important for mtDNA transcription (Parisi et al., 1993; Dairaghi et al., 1995). Like other HMG proteins (Fisher et al., 1992; Landsman and Bustin, 1993), Abf2p can bend and wrap DNA (Diffley and Stillman, 1992) and is thus likely to function in DNA packaging. Additional evidence that Abf2p functions in mtDNA organization comes from observations that mtDNA nucleoids are more diffuse in ρ+ abf2Δ cells, and that nucleoids isolated from those cells have an altered distribution of proteins compared with nucleoids isolated from ρ+ ABF2 cells (Newman et al., 1996). Genetic and biochemical data also indicate that Abf2p is required for efficient mtDNA recombination by promoting or stabilizing Holliday junction recombination intermediates (MacAlpine et al., 1998; Zelenaya-Troitskaya et al., 1998). Together with studies showing a direct relationship between the density of mtDNA recombination junctions and the efficiency of mtDNA transmission (Lockshon et al., 1995), these data suggest that mtDNA recombination is an important factor in mtDNA inheritance.
Suppressors have been isolated that improve the mtDNA stability of ρ+ abf2Δ cells (Kao et al., 1996; Cho et al., 1998). One unusual high copy suppressor that we identified (Troitskaya et al., 1995) was ILV5, a gene encoding the mitochondrial matrix enzyme acetohydroxy acid reductoisomerase (Petersen et al., 1983). This enzyme catalyzes steps in the biosynthesis of isoleucine, leucine and valine. Expression of the ILV5 gene is regulated by the general amino acid control pathway and is a target for the transcriptional activator, Gcn4p (Hinnebusch, 1988). Thus, when cells are starved of isoleucine, leucine and valine, derepression of GCN4 occurs; this in turn increases the expression of ILV5, as well as other targets of the general amino acid control pathway, including genes encoding other enzymes functioning in branched chain amino acid biosynthesis (Holmberg and Petersen, 1988). We found that mtDNA is significantly more stable in abf2Δ ρ+ cells when they were grown on dextrose medium lacking isoleucine, leucine and valine, or in medium containing those amino acids if Gcn4p was expressed constitutively. Importantly, we found that mtDNA is also unstable in ilv5Δ ρ+ cells, leading to the production of ρ– petites (Troitskaya et al., 1995). These effects were strictly dependent on the presence of the ILV5 gene and not on the integrity of the branched chain amino acid biosynthetic pathway. These findings suggested that Ilv5p is a bifunctional protein with roles in branched chain amino biosynthesis and mtDNA stability.
Here we study the relationship between the number of mtDNA nucleoids and the number of individual mtDNA molecules. To our surprise, this parsing of mtDNA into nucleoids is regulated by the general amino acid control pathway, and can be separated into one or more activities affecting recombination of mtDNA and to an activity of Ilv5p that controls the organization of mtDNA molecules in nucleoids. We show that an increased number of nucleoids resulting from an activation of the general amino acid control pathway dramatically increases the transmission of mtDNA, suggesting that the general amino acid control pathway operates on mtDNA organization to increase mtDNA transmission under starvation conditions.
Results
The number of mtDNA nucleoids is regulated by the general amino acid control pathway
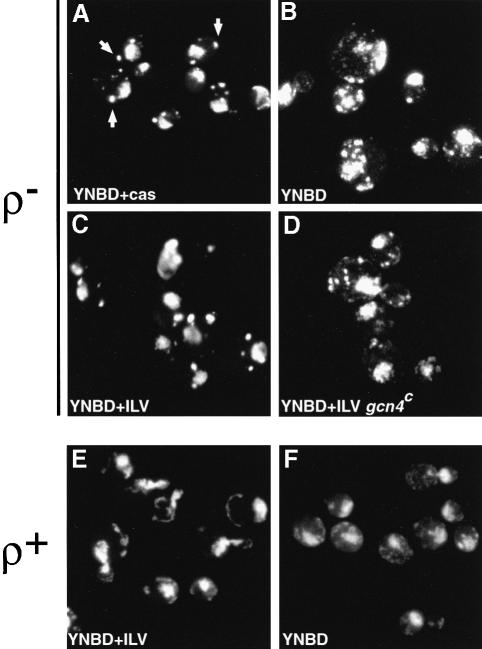
In preliminary experiments, we noticed that when ρ– petite strains were cultured in minimal dextrose medium (YNBD), there was a change in the distribution of mtDNA nucleoids when stained with the DNA-specific dye, 4′,6-diamidino-2-phenylindole (DAPI) compared with that observed when those strains were grown in dextrose medium containing casamino acids (YNBD+cas): it appeared that the number of nucleoids had increased. For example, instead of the few (∼5), brightly staining nucleoids typically observed for most ρ– petites when cultured in rich medium (Figure 1A), there were numerous (>50) small nucleoid (DAPI-stained) structures when that strain was cultured in YNBD medium (Figure 1B). (By direct microscopic observation, many small pinpoints of DAPI fluorescence were evident in those ρ– cells cultured in YNBD medium, much like a ‘starry night’. We note, however, that printed images do not capture fully this impression of a starry night.) The ρ– petite strain (HS40) used in these experiments is a hypersuppressive petite whose mitochondrial genome consists of a 760 kb tandem repeat of ori5, one of up to eight putative ori/rep sequences in ρ+ mtDNA. This effect is not peculiar to hypersuppressive petites, because we found an identical medium-dependent pattern of nucleoid redistribution in a neutral ρ– petite strain containing a 2 kb repeat of the VAR1 gene (not shown), which has no sequences in common with the mtDNA of HS40. As shown below, we also observed a similar nucleoid reorganization in ρ+ cells.
Fig. 1. The morphology and distribution of mtDNA nucleoids are regulated by general amino acid control. Cells were grown as indicated, fixed with ethanol and stained with DAPI. (A) Cells of strain ρ– HS40 grown in YNBD+cas (rich) medium contain a few large, bright DAPI-stained mtDNA nucleoids as indicated by the arrowheads. (B) Cells of strain ρ– HS40 grown on YNBD (minimal) medium have a greatly increased number of smaller nucleoids. (C) Cells of strain ρ– HS40 grown in YNBD medium supplemented with isoleucine, leucine and valine (YNBD+ILV) have a few large nucleoids. (D) Cells of strain ρ– HS40 harboring the plasmid, p238, from which Gcn4p is constitutively expressed, have large numbers of small nucleoids when grown in YNBD medium supplemented with isoleucine, leucine and valine. (E) Cells of strain ρ+ 14 WW cultured in YNBD+ILV medium have more and smaller nucleoids than do cells of the petite mutant grown in the same medium. (F) Cells of strain ρ+ 14 WW cultured in YNBD medium have an increased number of nucleoids.
How might these differences in culture conditions account for the change in the number of mtDNA nucleoids? When cultured on minimal medium, wild-type yeast cells undergo partial starvation of the branched chain amino acids isoleucine, leucine and valine, resulting in the induction of Gcn4p, the transcription factor that regulates the general amino acid control pathway (Hinnebusch, 1988). The induction of Gcn4p can be inhibited specifically by the presence of isoleucine, leucine and valine in the growth medium (multivalent repression). To determine whether the number of mtDNA nucleoids is influenced directly by general amino acid control of branched chain amino acid biosynthesis, we first determined the effect of supplementing the minimal growth medium with iso– leucine, leucine and valine on the nucleoid distribution in ρ– HS40 cells. As shown in Figure 1C, ρ– HS40 cells grown in YNBD medium supplemented with those amino acids have just a few large nucleoids, similar to the number observed when those cells were grown in the more complex medium, YNBD+cas (Figure 1A).
To confirm that the number of mtDNA nucleoids is affected specifically by Gcn4p regulation, ρ– HS40 cells were transformed with a centromeric expression plasmid, p238, containing the gcn4c mutant allele. The gcn4c allele has mutations in regulatory AUG codons upstream of the Gcn4p-coding sequence, so that Gcn4p is constitutively expressed and is insensitive to multivalent repression by isoleucine, leucine and valine (Mueller and Hinnebusch, 1986). Microscopic examination of the nucleoid morphology in those ρ– HS40 cells transformed with p238 and grown in the presence of isoleucine, leucine and valine showed an increase in the number of nucleoid structures (Figure 1D), similar to the pattern observed in non-transformed cells grown on minimal YNBD medium (Figure 1B). These data suggest that one or more factors under general amino acid control are responsible for these differences in nucleoid numbers.
Importantly, we have extended this phenomenon to ρ+ cells. There are more mtDNA nucleoids in ρ+ cells cultured in YNBD medium (Figure 1F) than in ρ+ cells grown in medium supplemented with isoleucine, leucine and valine (Figure 1E). In the supplemented medium, the ρ+ nucleoids are more numerous than are those in ρ– petite cells, and the bright spots appear to be more strung together and less punctate (compare Figure 1E with C). Because we were interested in examining possible molecular rearrangements that might be associated with the observed changes in nucleoid redistribution, most of the remaining experiments of this study focused on ρ– petite strains where the magnitude of the changes is greater.
General amino acid control influences mtDNA recombination
One obvious possibility to account for this increase in the number of mtDNA nucleoids in cells grown in minimal medium would be an increase in the total amount of mtDNA. However, direct measures of mtDNA content in cells grown in YNBD medium with or without casamino acids did not detect any significant differences (data not shown). Thus, a plausible hypothesis is that, in the case of ρ– petites, the number of individual mtDNA molecules has increased due to recombination events that reduce the mean number of genome repeats in each mtDNA molecule. Those molecules might then appear as distinct nucleoids, thus increasing their number without a net increase in the amount of mtDNA. For ρ+ mitochondrial genomes, resolution of molecules linked together by recombination junctions could also effectively increase the number of individual mtDNA molecules, giving rise to an increase in nucleoid number (see the following section).
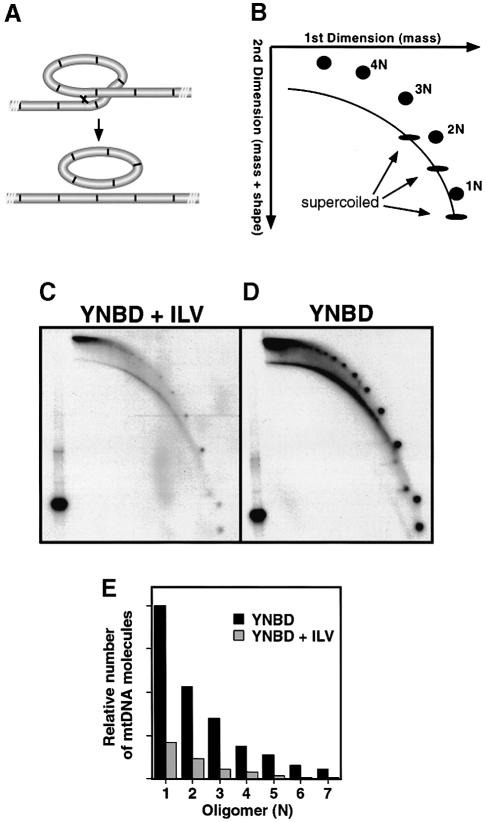
Homologous recombination between the tandem repeats of ρ– mtDNA would produce smaller, circular oligomers of the basic repeating unit (Figure 2A). To detect possible changes in the oligomeric state of ρ– mtDNA, we first used two-dimensional gel electrophoresis to separate the various molecular species of mtDNA. As illustrated in the schematic of Figure 2B, the first dimension of this gel system resolves DNA primarily according to mass, while the second dimension resolves DNA according to both mass and shape, and would resolve circular oligomers away from linear molecules (Brewer and Fangman, 1991). Following hybridization with ρ– HS40-specific probes, discrete circular supercoiled and relaxed oligomers and linear DNA molecules are discerned. A comparison of the oligomeric state of ρ– HS40 mtDNA isolated from cells grown on YNBD medium containing isoleucine, leucine and valine (Figure 2C) versus cells grown on unsupplemented YNBD medium (Figure 2D) shows that there was a much larger number of small, oligomeric, circular species in the mtDNA from cells grown on minimal YNBD medium than from cells grown on YNBD medium supplemented with isoleucine, leucine and valine. We estimate that the number of individual molecules of mtDNA increased ∼5-fold in the cells grown on minimal YNBD medium. Although 20 μg samples of cellular DNA are analyzed in Figure 2C and D, a large proportion of the mtDNA is present as high molecular weight oligomers (>15 repeats) that either do not enter the first dimension gel, or were lost during the excision of the first dimension gel lane. As a loading control, 1 μg of DNA was cleaved with EcoRV, linearizing all of the repeating units, and run in a single lane in the second dimension in each panel; each panel contains a comparable signal in that lane so that large differences in signal among the experimental lanes reflect different distributions of molecule sizes. The distribution of the number of molecules in the different oligomeric species was quantified as a function of cell growth conditions (Figure 2E). In both samples, the monomer is the most abundant small circular species. The numbers of higher oligomers (2N, 3N, 4N, etc.) fall off rapidly and to the same extent in both samples. However, there are significantly more molecules of each oligomeric species of mtDNA from the cells cultured in YNBD medium than from cells grown in medium supplemented with isoleucine, leucine and valine.
Fig. 2. The mtDNA of ρ– HS40 cells is resolved down to oligomeric sized repeats by activation of the general amino acid control pathway. (A) Diagram of tandemly repeated ρ– mtDNA undergoing intramolecular recombination to produce circular oligomers. (B) Diagram of the resolution of the various oligomeric states of ρ– mtDNA by two-dimensional gel electrophoresis. Oligomeric circles, linear DNA and supercoiled molecules are detected as discrete spots by hybridization with a ρ– HS40-specific probe. (C) and (D) Two-dimensional gel electrophoresis of undigested DNA extracted from cells grown on YNBD medium containing isoleucine, leucine and valine (C) or grown on unsupplemented YNBD medium (D). A 20 μg sample of DNA was resolved by two-dimensional gel electrophoresis and hybridized with a probe specific for ρ– HS40 mtDNA. As a loading control, 1 μg of total DNA from ρ– HS40 cells linearized by digestion with EcoRV was run only in the second dimension. The mtDNA repeat of HS40 contains a single EcoRV site so that all of the mtDNA in the loading control is observed as a 0.76 kb band, corresponding to the length of the unit repeat. (E) Relative amount of mtDNA oligomers in 20 μg samples of mtDNA from cells grown in YNBD medium with or without isoleucine, leucine and valine.
Nucleoid reorganization depends on Abf2p and Mgt1p
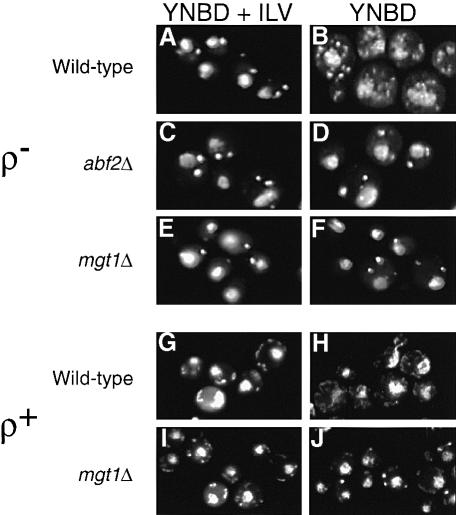
Efficient mtDNA recombination has been shown to require the HMG protein, Abf2p (MacAlpine et al., 1998; Zelenaya-Troitskaya et al., 1998), and Mgt1p (Zweifel and Fangman, 1991) (also known as Cce1p; Kleff et al., 1992), a cruciform-specific mitochondrial endonuclease that resolves Holliday junction recombination intermediates. To test whether there is a relationship between the number of nucleoids in cells and the number of individual molecules of mtDNA that might be controlled by the extent of intramolecular recombination between repeat units of ρ– mtDNA, we analyzed nucleoid number and the distribution of oligomers of mtDNA in ρ– HS40 cells with deletions of either the ABF2 (abf2Δ) or the MGT1 (mgt1Δ) gene. Neither of these genes is required for the stability of ρ– mtDNAs (Zweifel, 1991; Zelenaya-Troitskaya et al., 1998). Figure 3A and B shows the typical increase in the number of mtDNA nucleoids in wild-type ρ– HS40 cells grown in YNBD medium compared with the much smaller number of nucleoids in cells grown in YNBD medium supplemented with isoleucine, leucine and valine. In striking contrast, there was no difference in the number of nucleoids in abf2Δ or mgt1Δ cells grown in YNBD medium supplemented with isoleucine, leucine and valine versus YNBD medium alone: in both cases, only a few, brightly staining nucleoids were evident (Figure 3C–F). Similarly, for ρ+ mgt1Δ cells, the observed increase in mtDNA nucleoids was blocked when those cells were cultured in minimal medium lacking isoleucine, leucine and valine (Figure 3G and H). These observations suggest that mtDNA recombination plays an essential role in the dispersal of nucleoids in cells grown in medium lacking branched chain amino acids.
Fig. 3. Changes in nucleoid number require ABF2 and MGT1. The number of mtDNA nucleoids was examined by DAPI staining of ρ– HS40 wild-type (A and B), abf2Δ (C and D), mgt1Δ (E and F), ρ+ 14 WW wild-type (G and H) and mgt1Δ (I and J) cells grown in either YNBD medium supplemented with isoleucine, leucine and valine (YNBD+ILV) or in YNBD medium alone, as indicated.
To test whether the block in the increased number of mtDNA nucleoids was due to a block in the recombinational reduction of the mtDNA, DNA samples were prepared from ρ– HS40 cells of nuclear genotype ABF2 MGT1, abf2Δ or mgt1Δ, grown in minimal medium with or without supplementation with isoleucine, leucine and valine. These DNAs were analyzed on a standard one-dimensional agarose gel and hybridized with a probe specific for the ρ– HS40 mtDNA repeat. This one- dimensional system resolves small, circular oligomers and permits us to make direct comparisons of the oligomeric state of mtDNA in multiple samples. As noted above for the two-dimensional gel system, many DNA molecules are long oligomers of the basic repeating unit and do not enter this one-dimensional gel system, but the relevant smaller molecules are readily observed. Consistent with the two-dimensional analysis of the preceding section, there was a dramatic increase in the amount of small, circular oligomers of mtDNA when wild-type ρ– HS40 cells were cultured in YNBD medium lacking amino acids (Figure 4, lane 4 versus lane 1); however, the level of small oligomers of the ρ– HS40 mtDNA was markedly reduced in abf2Δ or mgt1Δ derivatives of the ρ– HS40 petite cultured in the same medium (lanes 5 and 6), and oligomers were essentially undetectable in abf2Δ or mgt1Δ cells cultured in YNBD medium supplemented with isoleucine, leucine and valine (lanes 2 and 3). A fraction (1/20) of the total uncut DNA was digested with EcoRV, which reduces the HS40 mtDNA down to its 760 bp repeating unit, and used as a loading control as shown at the bottom of Figure 4. Expression of the gcn4c allele in cells grown in medium containing isoleucine, leucine and valine produces the same DNA pattern as was obtained with wild-type cells cultured in unsupplemented YNBD medium (lane 7). Recombinational reduction produces, in addition to the well resolved, circular oligomers, a broad distribution of molecules that enter the gel, but are largely unresolved.
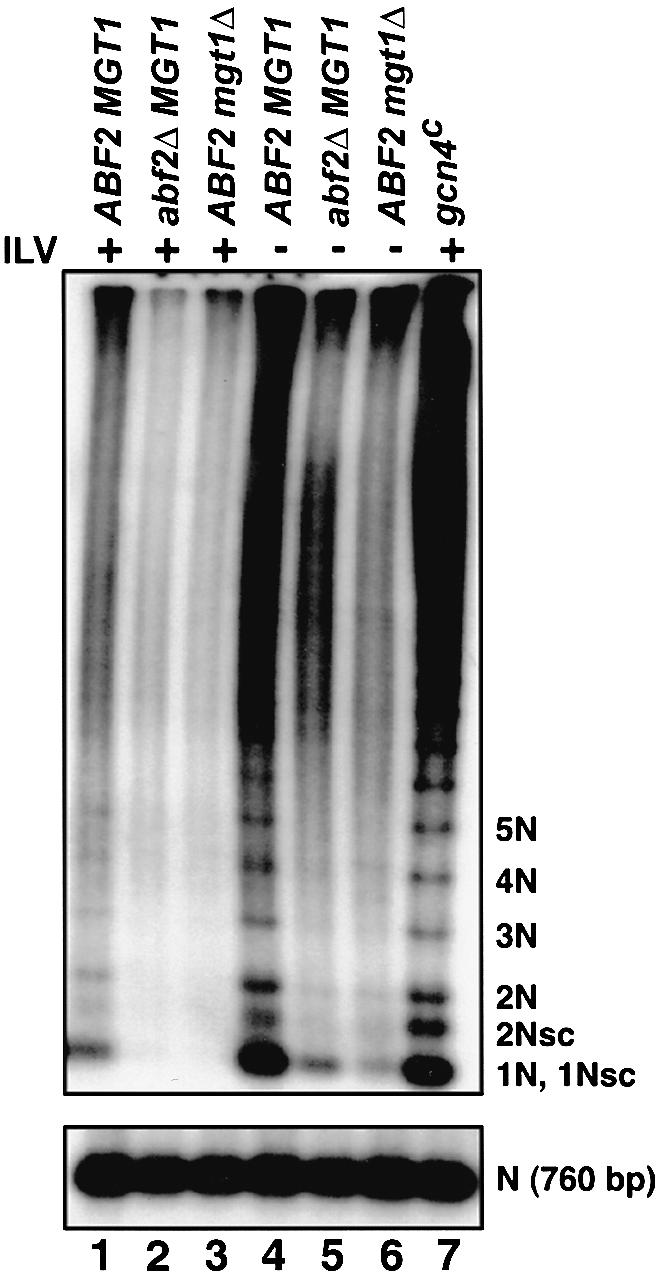
Fig. 4. The abf2Δ and mgt1Δ mutant alleles inhibit the production of small circular oligomers of ρ– HS40 mtDNA induced by activation of the general amino acid control pathway. One-dimensional gel electrophoresis of ∼20 μg samples of total DNA was used to resolve the different oligomeric forms of mtDNA. YNBD medium was supplemented with isoleucine, leucine and valine (ILV) as indicated in the figure. Supercoiled species are indicated as sc. As a loading control, 1 μg of total DNA was linearized by digestion with EcoRV and detected with an HS40 probe (lower panel).
Collectively, these results confirm the following: first, that the increase in the number of mtDNA nucleoids in cells grown on minimal medium correlates with an increase in the amount of smaller oligomers of the ρ– genome. Secondly, they show that recombinational reduction producing those smaller oligomers is under general amino acid control. Thirdly, they underscore the importance of both Abf2p and Mgt1p in mtDNA recombination. Finally, neither ABF2 nor MGT1 has consensus Gcn4p-binding sites in their 5′–flanking regions, and their expression is unaffected by the general amino acid control pathway (data not shown). Hence, the effects on recombination were not due to changes in the amount of Abf2p or Mgt1p.
Ilv5p is required for the GCN4-dependent increase in the number of mtDNA nucleoids
The modulation of mtDNA recombination and nucleoid number by the general amino acid control pathway is reminiscent of our previous observations that increased expression of ILV5, a GCN4- regulated gene encoding a mitochondrial matrix enzyme catalyzing a step in the biosynthesis of branched chain amino acids, can suppress the mtDNA instability phenotype of abf2Δ cells grown on non-selective (dextrose) medium (Troitskaya et al., 1995). This suppression could be achieved by increasing the dosage of the ILV5 gene, or by increasing its expression via GCN4-dependent activation of the general amino acid control pathway. Furthermore, deletion of ILV5 markedly increased the frequency of spontaneous petites and greatly exacerbated the mtDNA instability phenotype of abf2Δ cells. These effects were not due to a block in the branched chain amino acid biosynthetic pathway per se, because they were not observed in cells deleted for ILV2, which encodes a mitochondrial enzyme catalyzing the step in branched chain amino acid biosynthesis immediately preceding that of Ilv5p.
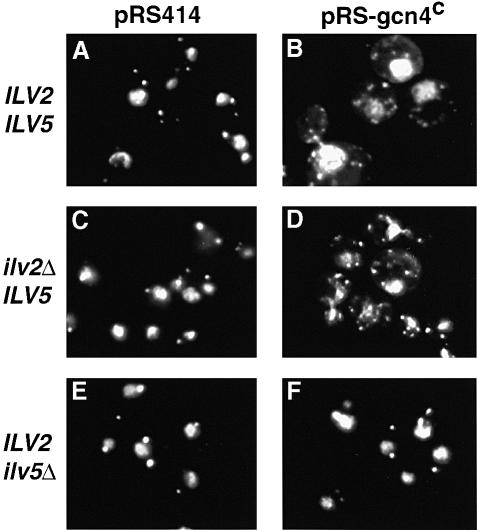
To determine whether ILV5 also functions in the control of nucleoid distribution and mtDNA recombination, ρ– HS40 cells with wild-type, ilv2Δ and ilv5Δ nuclear genotypes, each transformed with either a control plasmid, pRS414, or a plasmid containing the gcn4c allele, pRS-gcn4c, to activate the general amino acid response, were grown in YNBD medium supplemented with isoleucine, leucine and valine. In both the wild-type (Figure 5A and B) and ilv2Δ strains (Figure 5C and D), the constitutive expression of Gcn4p resulted in an increased number of nucleoids. The ilv5Δ strain, however, failed to undergo this nucleoid redistribution (Figure 5E and F). These results indicate that Ilv5p has a specific role in the control of mtDNA nucleoid number unrelated to the functionality of the branched chain amino acid biosynthetic pathway.
Fig. 5. Ilv5p is required for the general amino acid control-induced nucleoid redistribution. Wild-type, ilv2Δ and ilv5Δ ρ– HS40 petite cells transformed with either pRS414 or pRS-gcn4c, as indicated in the figure, were grown in YNBD+ILV medium and stained with DAPI.
Reductional recombination of mtDNA is independent of nucleoid organization
Is the failure to observe an increase in the number of mtDNA nucleoids in ilv5Δ ρ– HS40 cells expressing the gcn4c allele due to a block in the production of small, circular oligomers of the ρ– HS40 mtDNA? To determine whether reductional recombination was ILV5-dependent, the amount of small molecules was measured by one-dimensional gel electrophoresis in mtDNA from wild-type, ilv2Δ and ilv5Δ cells (Figure 6). To our surprise, the gcn4c-dependent resolution of mtDNA to small, circular oligomers, as documented above, occurred in all three strains despite the conclusion from our cytological observations that ILV5 was required for the dispersal of mtDNA nucleoids upon activation of the general amino acid control pathway. In the ilv2Δ background, recombinational reduction to smaller, circular oligomers also occurred but, as shown in the preceding section, this was accompanied by an increase in the number of nucleoids. These findings suggest that the reductional recombination of ρ– mtDNA and nucleoid reorganization and distribution are controlled by at least two GCN4-dependent factors, one of which is Ilv5p.
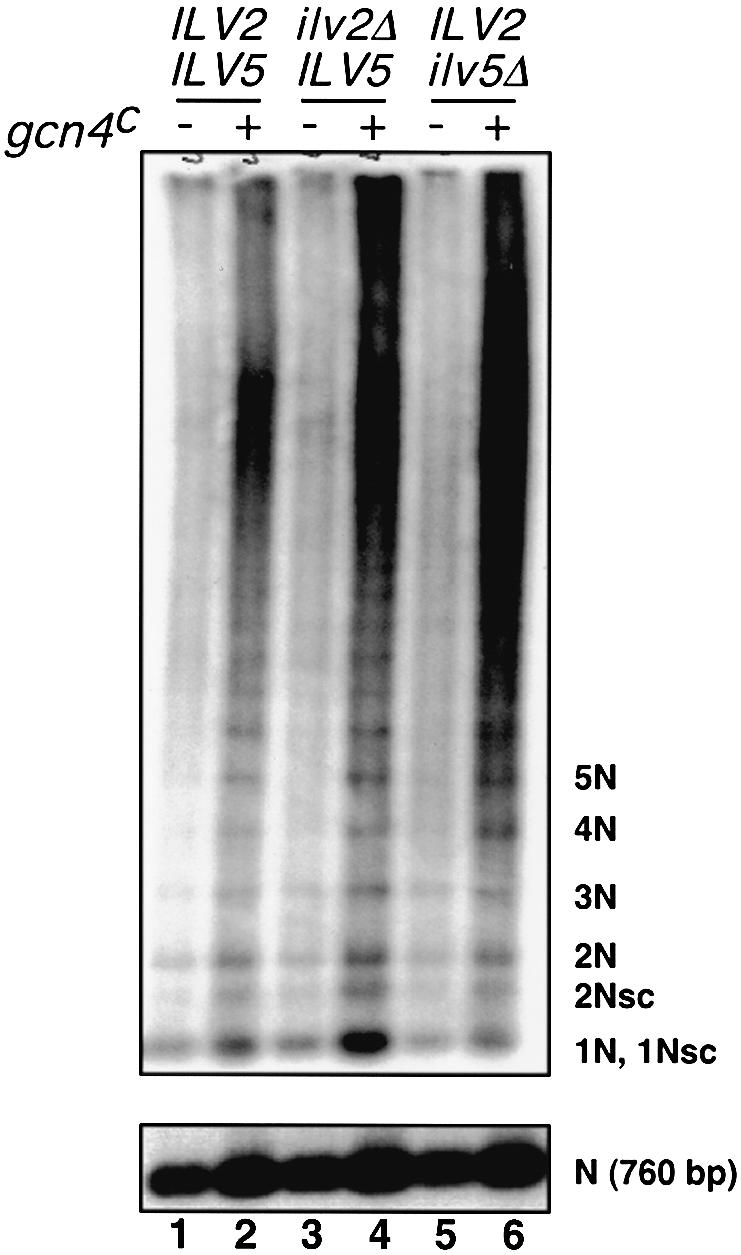
Fig. 6. The production of small, circular oligomers of ρ– HS40 mtDNA by constitutive expression of Gcn4p is not inhibited in ilv2Δ or ilv5Δ cells. One-dimensional gel electrophoresis of ∼20 μg samples of total DNA was used to resolve the different oligomeric forms of mtDNA found in wild-type, ilv2Δ and ilv5Δ cells grown in YNBD+ILV medium. The cells harbored either an empty vector (pRS416) or a vector expressing the constitutively expressed gcn4c allele (pRS416-gcn4c). As a loading control, 1/20 of the DNA sample used in the upper panel was linearized by digestion with EcoRV and detected with an HS40 probe (lower panel).
Constitutive Gcn4p expression results in increased transmission of the mtDNA of a neutral ρ– petite
As we have shown, a direct consequence of activation of the general amino acid control pathway was an increase in the number of nucleoids and individual mtDNA molecules. By analogy with multicopy plasmid systems (Nordstrom and Austin, 1989), this increase in the number of potential segregating units of mtDNA could result in an increase in transmission of ρ– DNAs in crosses between ρ– and ρ+ cells, and could also affect the suppressiveness of a petite genome. To test these notions, the neutral (non-suppressive) VAR1 petite harboring the wild-type (p164) or constitutively expressed gcn4c allele (p238) was mated to a ρ+ tester strain and the mating mixture was plated directly onto YNBD medium that selects for the diploid progeny. After 3 days of growth, the fraction of ρ– zygotic clones was scored by tetrazolium overlay (Ogur et al., 1957). Using this assay, 3–7% of the diploid progeny in both crosses were ρ–, which is about equal to the number of spontaneous ρ– colonies observed in the ρ+ tester strain. Thus, we conclude that activation of the general amino acid control pathway does not increase the suppressiveness of a neutral petite. However, we observed that when these crosses were carried out under conditions in which the general amino acid control pathway was activated, the diploid colonies appeared scalloped (data not shown), suggesting that some fraction of the cells in those colonies were ρ–. The implication of this observation is that activation of the general amino acid control pathway extends the heteroplasmic state between the ρ+ and ρ– genomes. (As a control, gcn4c expression in haploid ρ+ cells had no effect on the generation of spontaneous petites.)
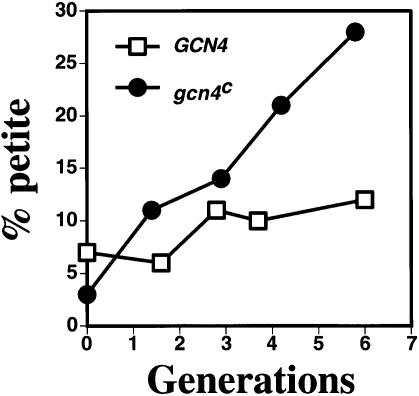
To test this possibility, a time course experiment was done in which the VAR1 petite strain, containing either the GCN4 or the gcn4c allele, was mated to the ρ+ tester for 3 h and the diploid progeny were allowed to outgrow for several generations. Aliquots of the mating mixture were plated at various time points to select for the diploid progeny, and the fraction of respiratory- incompetent colonies was analyzed (Figure 7). At generation zero, the percentage of ρ– petites in both crosses was about the same as that observed in cultures of the ρ+ tester strain. However, with each successive generation, an increasing number of ρ– colonies was observed in the gcn4c cross, which increased 9-fold by six generations. By contrast, the number of ρ– colonies in the GCN4 cross reached a plateau corresponding to an ∼1.7-fold increase in the percentage of petites after three generations. From these results, we conclude that the increased number of nucleoids resulting from activation of the general amino acid control pathway results in an increase in the transmission of mtDNA.
Fig. 7. The transmission of a neutral ρ– genome in a cross to a ρ+ tester strain is increased by constitutive GCN4 expression. A VAR1 petite strain, containing either the GCN4 (□) or the gcn4c (•) allele (p164 and p238, respectively), was mated to the ρ+ tester for 3 h and the diploid progeny were grown in liquid YNBD+ILV medium (containing the necessary nutritional supplements) before plating. Aliquots of the mating mixture were plated at various time points to select for the diploid progeny, and the fraction of respiratory-incompetent colonies was analyzed by TTC overlay (Ogur et al., 1957).
Discussion
MtDNA is packaged in cells as nucleoids, i.e. protein–DNA complexes, whose organization, assembly and transmission are not well understood. Earlier research showed a relationship between nucleoid organization and mtDNA recombination based on the observation that blocking the resolution of recombination junctions in ρ– mtDNA by deleting the MGT1 gene decreased the number of nucleoid structures and impaired mtDNA transmission (Lockshon et al., 1995). In the present study, we have found that not only are the number of individual mtDNA molecules and the number of mtDNA nucleoids tightly co-regulated, but, surprisingly, the regulation is mediated by factors under general amino acid control.
Many amino acid biosynthetic enzymes are induced when cells are starved for amino acids. De-repression of genes under general amino acid control results from an increase in the amount of the positive regulator, Gcn4p (Hinnebusch, 1992), which binds to consensus sites in the upstream regions of responsive genes to activate their transcription. Thus, activation of the general amino acid control pathway can occur either by starving cells for amino acids, or by artificially maintaining high levels of expression of Gcn4p in medium supplemented with amino acids. The latter can be accomplished conveniently by constitutively expressing Gcn4p from a mutant gcn4c allele in which translational control is lost by mutation of AUG regulatory codons upstream of the gcn4c reading frame (Mueller and Hinnebusch, 1986). We have used both methods to show that at least two factors subject to general amino acid control, and specifically to multivalent repression by isoleucine, leucine and valine, function in the parsing of mtDNA molecules into nucleoids. One of these factors is the mitochondrial enzyme, Ilv5p, whose only known function, prior to our previous findings that it could suppress the mtDNA instability phenotype of Δabf2 cells and was required for the stability of ρ+ mtDNA (Troitskaya et al., 1995), was the biosynthesis of branched chain amino acids. The current data suggest that Ilv5p functions additionally to maintain a stoichiometry between mtDNA and nucleoids. Activation of the general amino acid control pathway also induces one or more factors that increase mtDNA recombination. Neither Abf2p nor Mgt1p, two mitochondrial proteins known to function in mtDNA recombination (Lockshon et al., 1995; MacAlpine et al., 1998), is likely to be one of those factors, because their expression is not under general amino acid control.
Analysis of the sequence database reveals some 60 genes with potential Gcn4p regulatory binding sites. One potential candidate identified in the search was the open reading frame YOL095C, which encodes a putative mtDNA helicase (Schuldiner et al., 1998). Although YOL095C has five Gcn4p-binding sites in its 5′-flanking region, deletion of the gene had no effect on Gcn4p activation of nucleoid redistribution of ρ– mtDNA nucleoids (unpublished observations). Given the unexpected finding that Ilv5p is a bifunctional protein, other proteins whose known functions are seemingly unrelated to mtDNA events cannot be ruled out as candidates.
The packing of mtDNA into nucleoid structures is likely to involve the participation of a number of DNA-binding proteins, some of which may have unique specificity for DNA sequences or interactions with other DNA-binding proteins, while others may function more generally in nucleoid organization. In Escherichia coli, for example, at least four proteins play a role in nucleoid stucture by bending DNA; two of them, HNS and HU, bind relatively non-specifically to E.coli DNA, whereas FIS and IHF are site-specific (Schmid, 1990). Because the E.coli DNA-packing protein, HU, though not a member of the HMG family of DNA-binding proteins, can suppress the mtDNA instability phenotype of ρ+ abf2Δ cells when expressed in those cells and targeted to mitochondria (Megraw and Chae, 1993; Megraw et al., 1994), Abf2p is likely to be in the latter category. Moreover, from abundance measurements, Abf2p may bind every 15–30 bp of mtDNA (Diffley and Stillman, 1991). A common property of HU as well as of many HMG proteins, including Abf2p, is that they can bend DNA and induce supercoiling in the presence of a topoisomerase activity (Landsman and Bustin, 1993). We have, in fact, detected supercoiled topoisomers among the population of small, circular oligomers of ρ– mtDNA generated in cells in which the general amino acid control pathway was activated. Although the fraction of ρ– mtDNA that exists as small, circular oligomers of the repeat unit is greatly diminished in abf2Δ cells, supercoiled forms can nevertheless be detected in that (small) population (data not shown). It is tempting to speculate that Ilv5p may have a similar and overlapping function with Abf2p in mtDNA packaging and perhaps work in concert with Abf2p in nucleoid assembly and organization. Consistent with this view are the findings that overexpression of Ilv5p can suppress the instability of ρ+ mtDNA in abf2Δ cells (Troitskaya et al., 1995) and that the mtDNA instability phenotype of abf2Δ cells is greatly exacerbated in the abf2Δ ilv5Δ double mutant (Troitskaya et al., 1995).
It is noteworthy that the vast majority of petites that are produced when abf2Δ cells are grown on non-selective (glucose) medium are ρ° petites (Diffley and Stillman, 1991; Troitskaya et al., 1995). Although phenotypically identical, ρ° and ρ– petites probably arise by fundamentally different mechanisms, the former through a failure to transmit mtDNA to the progeny, and the latter through homologous and non-homologous recombination events giving rise to deleted molecules that, once formed, are generally stable (Dujon, 1981). We have noted previously that in ilv5Δ strains, the vast majority of petites that are produced are ρ– rather than ρ° cells (Troitskaya et al., 1995). Possibly, Ilv5p organizes mtDNA in such a way as to suppress intramolecular recombination events that lead to the production of ρ– petites. Taken together, the data would suggest that Ilv5p and Abf2p have overlapping but non-identical functions in the organization of mtDNA in nucleoids.
If Ilv5p functions directly in nucleoid organization or assembly, it would be expected to interact with mtDNA. Indeed, we have found that Ilv5p can be cross-linked to ρ+ mtDNA in organello with formaldehyde and is relatively abundant among the population of proteins detected by this method as interacting with mtDNA (B.Kaufman, unpublished observations). The finding that there are fewer mtDNA nucleoids in ilv5Δ cells, even though activation of the general amino acid control pathway in those cells results in a large increase in the number of individual mtDNA molecules, suggests that Ilv5p may provide a mechanism for counting mtDNA molecules in the assembly of nucleoid structures. One possibility for such a parsing mechanism is that Ilv5p ‘caps’ mtDNA nucleoids in a processive assembly process, thus suppressing potential internucleoid interactions that could occur from multivalent interactions among some nucleoid proteins.
The increase in the number of mtDNA nucleoids and individual mtDNA molecules (small circular oligomers) in ρ– petite cells can be correlated directly with an increase in recombination across the tandemly repeated sequences of the ρ– mitochondrial genome induced by activation of the general amino acid response pathway. Blocking this intramolecular recombination, for example by deleting the MGT1 gene encoding the mitochondrial cruciform-cutting endonuclease (Kleff et al., 1992), blocks the increase in nucleoid number. How then can we account for a similar increase in the number of ρ+ mtDNA nucleoids when ρ+ mtDNA lacks extensive repeated sequences? Moreover, if such intramolecular recombination events were to occur in ρ+ mtDNA, there should have been an increase in the production of ρ– petites when ρ+ cells were cultured in medium lacking amino acids—an outcome that was not observed. One plausible explanation for the increase in nucleoids in ρ+ cells cultured in minimal medium is that ρ+ mtDNAs linked together by recombination intermediates are resolved to individual molecules in cells grown under amino acid starvation conditions. Although the steady-state level of ρ+ mtDNAs with recombination junctions in wild-type cells is much lower than is observed for ρ– mitochondrial genomes and thus difficult to detect by two-dimensional gel electrophoresis (Lockshon et al., 1995; MacAlpine et al., 1998), a number of observations suggest that a significant fraction of ρ+ mtDNA is indeed linked together through recombination junctions in cells grown in rich medium. First, the number of nucleoids in ρ+ cells is less than the number of mtDNA genome equivalents (Williamson and Fennell, 1979). Secondly, pulse-field gel electrophoretic analysis of ρ+ mtDNA shows that a large fraction of ρ+ mitochondrial DNA migrates as greater than unit length molecules (Bendich, 1996). Finally, as we show here, the increase in ρ+ mtDNA nucleoids in cells grown in minimal medium is dependent on Mgt1p, whose only known activity is to cleave Holliday junction recombination intermediates, suggesting that resolution of recombination intermediates is a factor influencing nucleoid number and reorganization in ρ+ cells.
Our findings raise an obvious question: why involve the general amino acid control pathway in mtDNA organization? A possible answer to this question comes from the finding that the increase in the number of mtDNA nucleoids caused by activation of the general amino acid control pathway resulted in a significant extension of the heteroplasmic state of newly issued diploids from a cross between a neutral ρ– petite and ρ+ cells. Since ρ+ mtDNA nucleoid number is also modulated by general amino acid control, and given the accumulated evidence that nucleoids are the segregating unit of mtDNA, we propose that under starvation conditions, cells may enhance their survivability not only by activating amino acid biosynthetic pathways but also by increasing the likelihood that progeny cells inherit a full complement of mtDNA as a result of changes in nucleoid organization.
Materials and methods
Strains, growth conditions and cell matings
The strains used in this study were derivatives of 14 WW (MATα ade2 trp1 ura3-52 leu2 cit1::leu2). The petite genomes HS40 and VAR1 were placed in this background using cytoduction as described previously (Zelenaya-Troitskaya et al., 1998). The ABF2, MGT1, ILV5 and ILV2 genes were also disrupted as previously described (Troitskaya et al., 1995; MacAlpine et al., 1998; Zelenaya-Troitskaya et al., 1998) creating 14 Δilv5, 14 abf2Δ, 14 mgt1Δ and 14 ilv2Δ. For the mitochondrial genome transmission experiments, strain DBY747 (MAT a his3, leu2, trp1, ura3) was used as the ρ+ tester. All cells were grown at 30°C in either YNBD+cas (0.67% yeast nitrogen base, 3% dextrose and 1% casamino acids), YNBD+ILV (0.67% yeast nitrogen base, 3% dextrose and 340, 550 and 430 μg/ml of isoleucine, leucine and valine, respectively) or YNBD (0.67% yeast nitrogen base and 3% dextrose). Additional amino acid supplements were added as required.
Plasmids and transformations
Plasmids p164 and p238 contain the wild-type and constitutively expressed gcn4c allele, respectively. The gcn4c allele was removed from the plasmid p238 (a gift from E.Hannig) and cloned into PstI–SalI sites of the CEN vector pRS414, yielding pRS-gcn4c. All yeast transformations were done using the one-step transformation procedure (Chen et al., 1992).
Mitochondrial genome transmission
The transmission of petite genomes was assayed by pre-growing both 14 WW VAR1 ρ– and DBY747 ρ+ in YNBD+ILV media. Strain 14 WW VAR1 ρ– harbored either plasmid p238 or p164. Matings were carried out for 3 h on YNBD+ILV using 2 × 107cells from each strain. Cells were then transferred to fresh liquid media (YNBD+ILV) and allowed to grow in the absence of selection. Aliquots from various time points were plated onto selective media for diploids. The fraction of petites in the diploid population was determined by the 2,3,5-triphenyl tetrazolium chloride (TTC) overlay procedure (Ogur et al., 1957).
DNA isolation and gel electrophoresis
Total DNA was prepared as described previously with the exception that no enrichment for single-stranded DNA intermediates was used (MacAlpine et al., 1998). To separate the oligomeric forms of mtDNA, 20 μg of total DNA was fractionated by electrophoresis in the first dimension in 0.5% agarose gels (Seakem LE, FMC) at 1 V/cm. The second dimension was carried out in 1% agarose gels by electrophoresis at 2.5 V/cm at 4°C. DNA was transferred to nylon membranes and hybridization was carried out as described previously (MacAlpine et al., 1998). The probe was a cloned 760 bp fragment containing the entire HS40 genome (pBluescript HS40), released by digestion with HindIII and radiolabeled by random priming with [α–32P]dATP.
Microscopy
Cells were washed twice with double-distilled H2O and then fixed with 1 ml of 95% ethanol. DAPI was added to a final concentration of 1 μg/ml, and the cells were immediately washed again with double-distilled H2O several times. Imaging was done using a Leica microscope (model DMRXE; Deerfield, IL) equipped with an HBO 100 W/2 mercury arc lamp and ×100 Pan-Aprochromat objective using the >425 nm long pass emission filter for DAPI. Images were captured with either Kodak Tri-X 400 high speed 35 mm film (Eastman Kodak, Rochester, NY) or a color-chilled three charge-coupled device camera system (model C5810; Hamamatsu Photonics, Bridgewater, NJ) and then processed using Adobe Photoshop 5.0 (Adobe Systems, Inc., San Jose, CA).
Acknowledgments
Acknowledgements
We thank E.Hannig (UT Dallas) for the gift of the gcn4c plasmid, p238. This work was supported by grant GM33510 from the National Institutes of Health. D.M. was supported by training grant T32-GM08203 from the National Institutes of Health.
References
- Bendich A.J. (1996) Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol., 255, 564–588. [DOI] [PubMed] [Google Scholar]
- Birky C.W., Strausberg, R.L., Perlman, P.S. and Forster, J.L. (1978) Vegetative segregation in yeast: estimating parameters using a random model. Mol. Gen. Genet., 158, 251–261. [Google Scholar]
- Brewer B.J. and Fangman, W.L. (1991) Mapping replication origins in yeast chromosomes. BioEssays, 13, 17–22. [DOI] [PubMed] [Google Scholar]
- Chen D.-C., Yang, B.-C. and Kuo, T.-T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet., 21, 83–84. [DOI] [PubMed] [Google Scholar]
- Cho J.H., Ha, S.J., Kao, L.R., Megraw, T.L. and Chae, C.-B. (1998) A novel DNA-binding protein bound to the mitochondrial inner membrane restores the null mutation of mitochondrial histone Abf2p in Saccharomyces cerevisiae.Mol. Cell. Biol., 18, 5712–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairaghi D., Shadel, G. and Clayton, D. (1995) Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol., 249, 11–28. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. and Stillman, B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F.X. and Stillman, B. (1992) DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem., 267, 3368–3374. [PubMed] [Google Scholar]
- Dujon B. (1981) Mitochondrial genetics and functions. In Strathern,J.N., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 505–635. [Google Scholar]
- Fisher R.P., Lisowsky, T., Parisi, M.A. and Clayton, D.A. (1992) DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem., 267, 3358–3367. [PubMed] [Google Scholar]
- Hermann G.J. and Shaw, J.M. (1998) Mitochondrial dynamics in yeast. Annu. Rev. Cell. Dev. Biol., 14, 265–303. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1988) Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae.Microbiol. Rev., 52, 248–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1992) General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In Jones,E.W., Pringle,J.R. and Broach,J.R. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 319–414. [Google Scholar]
- Holmberg S. and Petersen, J.G.L. (1988) Regulation of isoleucine–valine biosynthesis in Saccharomyces cerevisiae.Curr. Genet., 13, 207–217. [DOI] [PubMed] [Google Scholar]
- Kao L.R., Megraw, T.L. and Chae, C.B. (1996) SHM1: a multicopy suppressor of a temperature-sensitive null mutation in the HMG1-like abf2 gene. Yeast, 12, 1239–1250. [DOI] [PubMed] [Google Scholar]
- Kleff S., Kemper, B. and Sternglanz, R. (1992) Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J., 11, 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman D. and Bustin, M. (1993) A signature for the HMG-1 box DNA-binding proteins. BioEssays, 15, 1–8. [DOI] [PubMed] [Google Scholar]
- Lockshon D., Zweifel, S.G., Freeman-Cook, L.L., Lorimer, H.E., Brewer, B.J. and Fangman, W.L. (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell, 81, 947–955. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Perlman, P.S. and Butow, R.A. (1998) The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo.Proc. Natl Acad. Sci. USA, 95, 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L. and Chae, C.B. (1993) Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem., 268, 12758–12763. [PubMed] [Google Scholar]
- Megraw T.L., Kao, L.R. and Chae, C.B. (1994) The mitochondrial histone HM: an evolutionary link between bacterial HU and nuclear HMG1 proteins. Biochimie, 76, 909–916. [DOI] [PubMed] [Google Scholar]
- Mueller P.P. and Hinnebusch, A. (1986) Multiple upstream AUG codons mediate translational control of GCN4. Cell, 45, 201–207. [DOI] [PubMed] [Google Scholar]
- Newman S.M., Zelenaya-Troitskaya, O., Perlman, P.S. and Butow, R.A. (1996) Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG-box protein, Abf2p. Nucleic Acids Res., 24, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K. and Austin, S.J. (1989) Mechanisms that contribute to the stress segregation of plasmids. Annu. Rev. Genet., 23, 37–69. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W. and Walter, P. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell, 8, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogur M., St. John, R. and Nagai, S. (1957) Tetrazolium overlay technique for populations studies of respiration deficiency in yeast genetics. Science, 125, 928–929. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Perlman, P.S. and Butow, R.A. (1998) The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol., 142, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M.A. and Clayton, D.A. (1991) Similarity of human mitochondrial trancription factor 1 to high mobility group proteins. Science, 252, 965–969. [DOI] [PubMed] [Google Scholar]
- Parisi M.A., Xu, B.J. and Clayton, D.A. (1993) A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro.Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J.G.L., Kielland-Brand, M.C., Holmberg, S. and Nilsson-Tillgren, T. (1983) Approaches to genetic improvement of brewers yeast. Cloning of the isoleucine–valine genes. Carlsberg Res. Commun., 48, 21–34. [Google Scholar]
- Schmid M. (1990) More than just ‘histone-like’ proteins. Cell, 63, 451–453. [DOI] [PubMed] [Google Scholar]
- Schuldiner O., Yanover, C. and Benvenisty, N. (1998) Computer analysis of the entire budding yeast genome for putative targets of the GCN4 transcription factor. Curr. Genet., 33, 16–20. [DOI] [PubMed] [Google Scholar]
- Stevens B. (1981) Mitochondrial structure. In Strathern,J.N., Jones,E.W. and Broach,J.R. (eds), The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 471–504. [Google Scholar]
- Troitskaya O., Perlman, P.S. and Butow, R.A. (1995) ILV5 encodes a bifunctional mitochondrial protein involved in branched chain amino acid biosynthesis and maintenance of mitochondrial DNA. EMBO J., 14, 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.H. and Fennell, D.J. (1979) Visualization of yeast mitochondrial DNA with the fluorescent stain ‘DAPI’. Methods Enzymol., 56, 728–733. [DOI] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O., Newman, S.M., Okamoto, K., Perlman, P.S. and Butow, R.A. (1998) Functions of the HMG box protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae.Genetics, 148, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel S.G. and Fangman, W.L. (1991) A nuclear mutation reversing a biased transmission of yeast mitochondrial DNA. Genetics, 128, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]