Abstract
Structural characterization of a sulfate complex with an azamacrocycle suggests that one sulfate is encapsulated in the macrocyclic cavity with eight hydrogen bonds; a significant selectivity of the host was observed for sulfate over halides, nitrate and perchlorate as evaluated by 1H NMR studies in water.
Sulfate is present in the biological system and plays a crucial role in many biochemical processes,1 such as in biosynthesis2 and sulfate binding proteins.3 Sulfate is also a known inorganic pollutant in the environment.4 For example, contamination of the Hanford nuclear waste site by this anion has been a matter of increased concern, hampering the vitrification process.5 However, as compared to other oxoanions, such as phosphate and nitrate, this anion has been less studied with synthetic hosts.6 Therefore, the design of suitable hosts for selective recognition of sulfate remains a challenge in the area of anion binding chemistry.7
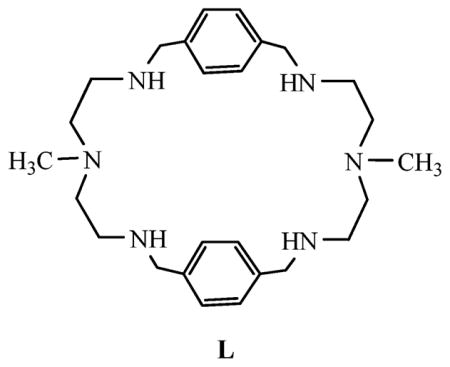
Encapsulated sulfate was crystallographically characterized in self-assembled metal–organic cage hosts, polyamides,8 ureas9 or indoles.10 In these cases, an encapsulation occurred within the crystal lattices formed by a host matrix or between hosts. Based on the theoretical calculations, Hay and coworkers suggested that sulfate can form up to twelve hydrogen bonds11 that were also structurally shown in a metal–organic framework functionalized with tris-urea binding sites.12 In nature, a seven-coordinate sulfate structure was observed in a sulfate binding protein, where three of four oxygens were linked with two hydrogen bonds.13 As observed in many cases, the oxygen atoms in oxoanions are generally held by two hydrogen bonds due to the directionality of the oxygen lone pairs.14 This makes the sulfate an ideal anion to accept eight hydrogen bonds via four oxygen atoms. However, to the best of our knowledge, with the exception of a sandwich complex of sulfate with monocyclic tetraamide15 and an encapsulated sulfate complex with tricyclic hexaamide,8a there is no structural example in which sulfate is octacoordinated with a synthetic host. Herein, we report the structural characterization of an octacoordinated sulfate encapsulated in the intramolecular cavity of an azamacrocycle L, and the 1H NMR binding studies showing a significant selectivity for sulfate over other anions in water.
Although, azamacrocycles containing multiple binding sites have been shown to be effective systems for binding of a variety of anions, their uses as sulfate binding hosts are quite limited.16 In an effort to achieve selective anion receptors, we decided to examine the anion binding ability of L that was synthesized from the condensation of N-methyl-2,2′-diaminodiethylamine and terephthalaldehyde in methanol followed by the reduction with NaBH4.17 X-Ray quality crystals of the sulfate complex were grown from a slow evaporation of the solution of L in water in presence of H2SO4 at pH 2.0.
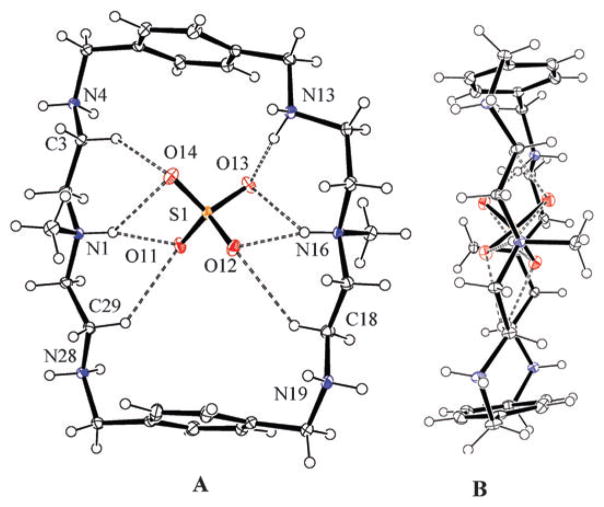
Structural elucidation by X-ray diffraction analysis‡ reveals that the complex crystallizes in its triclinic space group to yield a molecular formula, [H6L(SO4)]·2SO4·4H2O in which one divalent sulfate is encapsulated (Fig. 1). The macrocycle is essentially planar ellipsoid with two methyl groups lying in a trans configuration. All the six nitrogen atoms are protonated and the charges are balanced by three sulfate anions.
Fig. 1.
ORTEP views [H6L(SO4)]4+ motif showing hydrogen bonded encapsulated sulfate: (A) overhead view and (B) side view. External water and sulfate are not shown for clarity. Thermal ellipsoids are drawn at the 40% probability level.
In the macrocyclic unit, both protons on the tertiary amines are pointed towards the cavity and are available to interact with two oxygens of the encapsulated sulfate via single NH· · ·O bond (see Fig. 1A and Table 1). Clearly, the participation of tertiary protons in bridging the internal anion makes the N· · ·N distance shorter (6.985 Å ) than the distance between the two aromatic rings (9.678 Å ). As shown in Fig. 1A, each oxygen atom of the internal sulfate accepts two hydrogen bonds, one from tertiary amine (N13 or N16) and another from secondary amine (N13) or methylene group (C8, C18 or C29). Therefore, a total of eight coordination numbers of sulfate are obtained from the intermolecular hydrogen bonding interactions, five from NH· · ·O and three from CH· · ·O bonds. The existence of CH· · ·O bonds is well documented in synthetic molecules18 as well as in many natural systems.19 The other secondary ammonium protons on N4, N19 and N28 are pointing outward forming hydrogen bonds with external sulfates and water molecules (see supporting materials†). Indeed, an equal distribution of hydrogen bonds to all four oxygens assists the internal sulfate anion to sit at the center of the macrocyclic cavity (Fig. 1B). Structural characterization has been reported for sulfate and nitrate complexes of an m-xylyl based macrocycle,16a for a perchlorate complex of a thiophene based macrocycle20 or for a bromide complex of L.16 In these structures, the macrocycle, however, binds an anion from both sides of the ring. In the present structure, the sulfate anion is completely encapsulated in the cavity via both NH and CH interactions.
Table 1.
Selected hydrogen bonding parameters (A, °) for SO42− binding in L
| D—H· · ·O | H· · ·O | D· · ·O | ∠DHO |
|---|---|---|---|
| N1–H11· · ·O11 | 1.80(2) | 2.6882(17) | 162(2) |
| N1–H11· · ·O14 | 2.54(2) | 3.2328(18) | 132.7(17) |
| N13–H131· · ·O13 | 1.73(2) | 2.7022(17) | 176(2) |
| N16–H161· · ·O12 | 1.83(2) | 2.6676(17) | 162(2) |
| N16–H161· · ·O13 | 2.50(2) | 3.1591(18) | 133.4(17) |
| C3–H3B· · ·O14 | 2.465 | 3.205 | 135.78 |
| C18–H18A· · ·O12 | 2.493 | 3.155 | 128.78 |
| C29–H29A· · ·O11 | 2.489 | 3.207 | 131.86 |
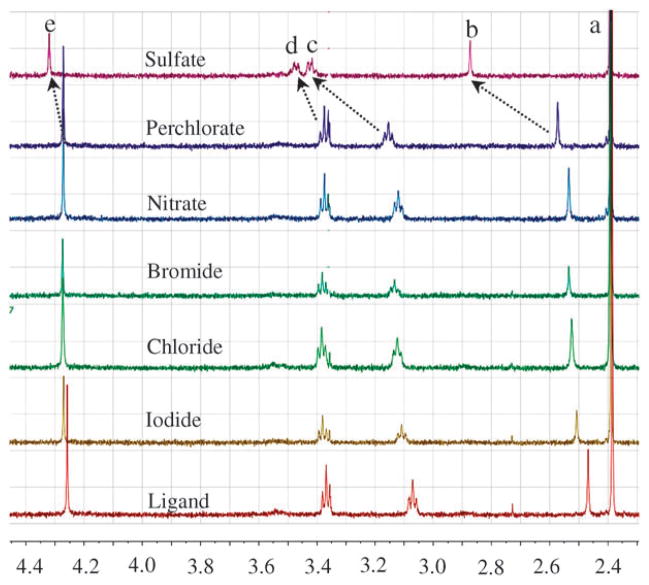
The binding properties of the receptor for anions with different shapes and sizes (Cl−, Br−, I−, ClO4−, NO3− and SO42−) were investigated by 1H NMR studies using [H6L](Ts)6 (prepared from the addition of six equivalents of tosylic acid to L) inD2O at pH 2.1. As shown in Fig. 2, the addition of one equivalent of sodium sulfate resulted in a significant downfield shift of the proton signals of L in the 1H NMR spectrum, indicating a strong interaction between the cationic macrocycle and the anionic guest. The highest changes were obtained for the protons Hb and Hc that are connected with the tertiary nitrogens. This observation suggested the possible interaction of central charged nitrogen atoms21 in binding of the anion that was further confirmed by the structural analysis of the sulfate complex. In addition, aromatic protons of the p-xylyl subunits were also shifted and could be due to the spatial proximity of the corresponding protons to the negatively charged anion in the complex. However, the addition of other anions resulted in a small shift, indicating a weaker interaction of the anion with L.
Fig. 2.
Partial 1H NMR spectra of [H6L](Ts)6 (2 mM) in presence of one equivalent of different anions in D2O at pH 2.1. a = TsCH3, b = NCH3, c = NCH2, d = NCH2CH2 and e = ArCH2.
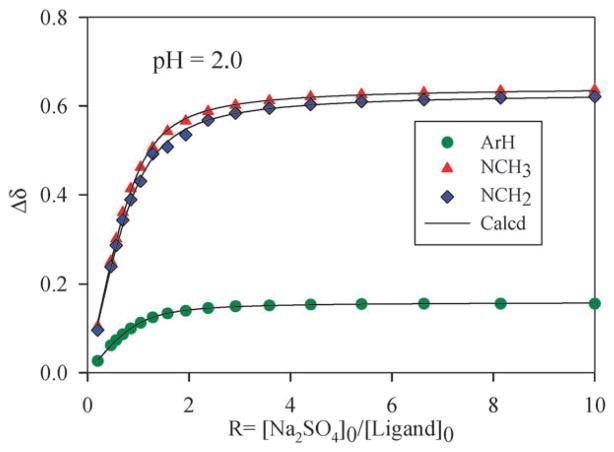
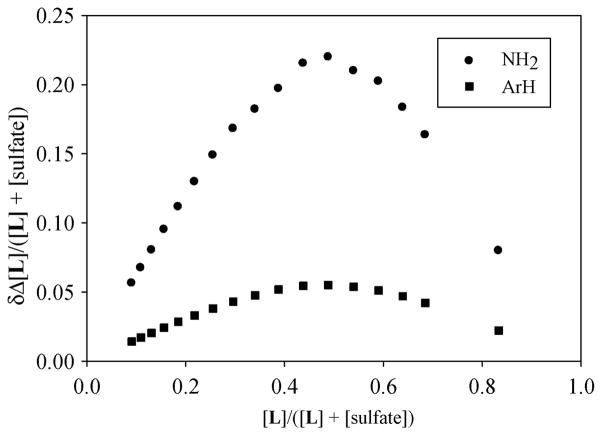
In order to determine the binding constants, the ligand (2 mM) was titrated with an increasing amount of the respective anion (20 mM) in D2O at pH 2.1. The variation in shift changes for several protons against the anion concentration gave the best fit for a 1 : 1 binding model22 (Fig. 3). As summarized in Table 2, the ligand showed a significant selectivity for sulfate over other anions examined. The observed high binding for sulfate was due to the presence of two charges as opposed to one charge in other anions. Therefore, the binding was primarily dominated by electrostatic interactions as anticipated in competitive polar solvents like water. The 1 : 1 stoichiometry of the sulfate complex in solution was further verified by the Job plot displaying a sharp maximum at an equimolar ratio of the anion and L (Fig. 4).
Fig. 3.
1H NMR titration curves for sulfate binding with [H6L](Ts)6 (2 mM) in D2O at pH = 2.1. Net changes in the chemical shifts of different chemical shifts are shown against the increasing amount of Na2SO4 (20 mM).
Table 2.
Association constants (K) of the anion complexes determined by 1H NMR titration at pH 2.1
| Anion | K, M−1 | Anion | K, M−1 |
|---|---|---|---|
| Sulfate | 3600 | Chloride | 15 |
| Nitrate | 120 | Bromide | 90 |
| Perchlorate | >20 | Iodide | 25 |
Fig. 4.
Job plot of L with Na2SO4 (20 in D2O at pH 2.1) showing a maximum at 0.5 mole fraction confirming a 1 : 1 complex formation.
In summary, we report a simple azamacrocycle that shows a strong selectively for sulfate over halides and other oxoanions in water. The solid state structure described herein provides insight into the binding interactions in the complex, which represents a rare example of the encapsulated sulfate within the intramolecular cavity that is highly complementary to tetrahedral divalent sulfate, and capable of donating a multiple hydrogen bonds to form an octacoordinate sulfate complex.
Research sponsored by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy via contract with Oak Ridge National Laboratory (MAH). The project described was supported by Grant Number G12RR013459 from the National Center for Research Resources. This material is based upon work supported by the National Science Foundation under CHE-0821357.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: One crystallographic data in CIF format, the synthetic procedure in pdf format. CCDC 777812. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c0cc01699c
Crystal data for for [H6L(SO4)]·2SO4·4H2O: C26H56N6O16S3, M = 804.95, triclinic, a = 12.283(2) Å, b = 12.289(2) Å, c = 13.315(3) Å, α = 105.301(3)°, β = 99.994(3)°, γ = 103.542(3)°, V=1824.4(6)Å3, T=100(2) K, space group P, Z=2, μ(Mo-Kα) = 0.281 mm−1, 16 902 reflections measured, 8778 independent reflections (Rint = 0.0405). The final R1 values were 0.0425 (I > 2σ(I)). The final wR(F2) values were 0.1100 (I > 2σ(I)). The final R1 values were 0.0462 (all data). The final wR(F2) values were 0.1130 (all data). CCDC 777812.
Notes and references
- 1.(a) Atwood JL, Holman KT, Steed JW. Chem Commun. 1996:1401. [Google Scholar]; (b) Bianchi A, García-España E, Bowman-James K, editors. Supramolecular Chemistry of Anions. Wiley-VCH; New York: 1997. [Google Scholar]; (c) Gale PA. Coord Chem Rev. 2003;240:191. [Google Scholar]; (d) Cormode DP, Murray SS, Cowley AR, Beer PD. Dalton Trans. 2006:5135. doi: 10.1039/b609817g. [DOI] [PubMed] [Google Scholar]
- 2.Young RW. J Cell Biol. 1973;57:175. doi: 10.1083/jcb.57.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raboudi N, Julian J, Rohde LH, Carson DD. J Biol Chem. 1992;267:11930. [PubMed] [Google Scholar]
- 4.(a) Barth MC, Church AT. J Geophys Res. 1999;104:30231. [Google Scholar]; (b) Heizer WD, Sandler RS, Seal E, Murrai SC, Busby MG, Schliebe BG, Pusek SN. Dig Dis Sci. 1997;42:1055. doi: 10.1023/a:1018801522760. [DOI] [PubMed] [Google Scholar]; (c) Fowler CJ, Haverlock TJ, Moyer BA, Shriver JA, Gross DE, Marquez M, Sessler JL, Hossain MA, Bowman-James K. J Am Chem Soc. 2008;130:14386. doi: 10.1021/ja806511b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sessler JL, Katayev E, Pantos GD, Ustynyuk YA. Chem Commun. 2004:1276. doi: 10.1039/b403665d. [DOI] [PubMed] [Google Scholar]
- 6.Mullen KM, Beer PD. Chem Soc Rev. 2009;38:1701. doi: 10.1039/b806041j. [DOI] [PubMed] [Google Scholar]
- 7.(a) Bondy CR, Gale PA, Loeb SJ. J Am Chem Soc. 2004;126:5030. doi: 10.1021/ja039712q. [DOI] [PubMed] [Google Scholar]; (b) Wu B, Liang J, Yang J, Jia C, Yang XJ, Zhang H, Tang N, Janiak C. Chem Commun. 2008:1762. doi: 10.1039/b719019k. [DOI] [PubMed] [Google Scholar]; (c) Fisher MG, Gale PA, Light ME, Loeb SJ. Chem Commun. 2008:5695. doi: 10.1039/b816002c. [DOI] [PubMed] [Google Scholar]
- 8.(a) Hossain MA, Llinares JM, Powell D, Bowman-James K. Inorg Chem. 2001;40:2936. doi: 10.1021/ic015508x. [DOI] [PubMed] [Google Scholar]; (b) Kang SO, Day VW, Bowman-James K. Org Lett. 2009;11:3654. doi: 10.1021/ol9014249. [DOI] [PubMed] [Google Scholar]
- 9.(a) Jose DA, Kumar DK, Ganguly B, Das A. Inorg Chem. 2007;46:5817. doi: 10.1021/ic062466+. [DOI] [PubMed] [Google Scholar]; (b) Zhuge F, Wu B, Liang J, Yang J, Liu Y, Jia C, Janiak C, Tang N, Yang X. Inorg Chem. 2009;48:10249. doi: 10.1021/ic9012685. [DOI] [PubMed] [Google Scholar]; (c) Custelcean R, Barsano J, Bonnesen PV, Kertesz V, Hay BP. Angew Chem, Int Ed. 2009;48:4025. doi: 10.1002/anie.200900108. [DOI] [PubMed] [Google Scholar]
- 10.(a) Kim J, Juwarker H, Liu X, Lah MS, Jeong KS. Chem Commun. 2010;46:764. doi: 10.1039/b919519j. [DOI] [PubMed] [Google Scholar]; (b) Gale PA, Hiscock JR, Jie CZ, Hursthouse MB, Light ME. Chem Sci. 2010;1:215. [Google Scholar]
- 11.Hay BP, Firman TK, Moyer BA. J Am Chem Soc. 2005;127:1810. doi: 10.1021/ja043995k. [DOI] [PubMed] [Google Scholar]
- 12.Custelcean R, Moyer BA, Hay BP. Chem Commun. 2005:5971. doi: 10.1039/b511809c. [DOI] [PubMed] [Google Scholar]
- 13.Pflugrath JW, Quiocho FA. Nature. 1985;314:257. doi: 10.1038/314257a0. [DOI] [PubMed] [Google Scholar]
- 14.Bowman-James K. Acc Chem Res. 2005;38:671. doi: 10.1021/ar040071t. [DOI] [PubMed] [Google Scholar]
- 15.Kang SO, Hossain MA, Powell D, Bowman-James K. Chem Commun. 2005:328. doi: 10.1039/b411904e. [DOI] [PubMed] [Google Scholar]
- 16.(a) Clifford T, Dnaby A, Llinares JM, Mason S, Alcock NW, Powell D, Aguilar JA, García-España E, Bowman-James K. Inorg Chem. 2001;40:4710. doi: 10.1021/ic010135l. [DOI] [PubMed] [Google Scholar]; (b) Saeed MA, Powell DR, Fronczek FR, Hossain MA. Tetrahedron Lett. 2010;51:4233–4236. doi: 10.1016/j.tetlet.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson D, Dey KR, Fronczek FR, Hossain MA. Tetrahedron Lett. 2009;50:6537. doi: 10.1016/j.tetlet.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vega IED, Gale PA, Light ME, Loeb SJ. Chem Commun. 2005:4913. doi: 10.1039/b510506d. [DOI] [PubMed] [Google Scholar]
- 19.Scheiner S, Kar T, Gu Y. J Biol Chem. 2001;276:9832. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 20.Saeed MA, Thompson JJ, Fronczek FR, Hossain MA. CrystEngComm. 2010;12:674. doi: 10.1039/b918152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Saeed MA, Fronczek FR, Hossain MA. Chem Commun. 2009:6409. doi: 10.1039/b916099j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Saeed MA, Fronczek FR, Huang MJ, Hossain MA. Chem Commun. 2010;46:404. doi: 10.1039/b923013k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider HJ, Kramer R, Simova S, Schneider U. J Am Chem Soc. 1988;110:6442. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.