SUMMARY
The virulence of Gram-positive bacteria is enhanced by toxins like the Streptococcus pyogenes β-NAD+ glycohydrolase known as SPN. SPN-producing strains of S. pyogenes additionally express the protein immunity factor for SPN (IFS), which forms an inhibitory complex with SPN. We have determined crystal structures of the SPN-IFS complex and IFS alone, revealing that SPN is structurally related to ADP-ribosyl transferases but lacks the canonical binding site for protein substrates. SPN is instead a highly efficient glycohydrolase with the potential to deplete cellular levels of β-NAD+. The protective effect of IFS involves an extensive interaction with the SPN active site that blocks access to β-NAD+. The conformation of IFS changes upon binding to SPN, with repacking of an extended C-terminal α-helix into a compact shape. IFS is an attractive target for the development of novel bacteriocidal compounds functioning by blocking the bacterium's self-immunity to the SPN toxin.
INTRODUCTION
The virulence of many bacteria is enhanced by powerful toxins that are injected into host cells where they co-opt cellular physiology by modulating signaling pathways, reorganizing the cytoskeleton, causing programmed cell death, or changing cellular metabolism {Bhavsar, 2007 #221}. The Gram-positive bacterium Streptococcus pyogenes causes a variety of human diseases ranging from superficial and self-limiting infections (pharyngitis, impetigo) to conditions that are highly destructive to tissue and life-threatening (necrotizing faciitis), including those caused by dysregulation of the immune system (rheumatic fever, acute glomerulonephritis) (Cunningham, 2000). Group A Streptococcus spp. encode a pore-forming protein Streptolysin O (SLO) that functions as a conduit to inject the toxin SPN (S. pyogenes β-NAD+ glycohydrolase) into the host cell. Once inside the host cell, SPN alters cellular functions and induces a cytotoxic response that ultimately causes cell death (Bricker et al., 2002; Madden et al., 2001).
The mechanism by which SPN alters host cell physiology is not understood, although the purified recombinant enzyme has robust β-NAD+-glycohydrolase (GHase) activity and appears to lack activity as an ADP-ribosyl transferase (RTase) or a ADP-ribosyl cyclase (ACase) (Ghosh et al., 2010), despite earlier reports of these activities in cell extracts from SPN-producing strains (Grushoff et al., 1975; Karasawa et al., 1995; Stevens et al., 2000). GHase activity converts β-NAD+ to nicotinamide and ADP-ribose (ADPR) and has the potential to deplete cellular stores of the essential cofactor and signaling molecule, β-NAD+ {Michos, 2006 #213}. Purified RTase enzymes typically show low levels of GHase activity in the absence of a protein substrate, whereas SPN has robust GHase activity (Ghosh et al., 2010) in agreement with other prokaryotic enzymes known as pure glycohydrolases {Everse, 1975 #215;Mather, 1969 #217}.
The SPN protein consists of two domains. The N-terminal domain (residues 41–190) is required for secretion of SPN through the SLO pore (Ghosh and Caparon, 2006). The C-terminal domain (residues 191–451) contains the active site and its GHase activity in vitro is comparable to full length SPN (ibid.). Although SPN is a pure GHase, the C-terminal domain contains amino acid sequence motifs that are hallmarks of multi-functional ACases (Ghosh and Caparon, 2006) and RTases (see below), raising questions about the origins of selective GHase activity. The N-terminal domain of SPN is not conserved in ACases and RTases and has a predicted jelly-roll fold, similar to those of glycan-binding proteins (Ghosh and Caparon, 2006).
The SPN GHase is highly toxic when expressed in Escherichia coli and presumably depletes cellular stores of β-NAD+. In Streptococcus pyogenes, SPN is expressed in a complex with the Immunity Factor for SPN (IFS) that functions as an antitoxin by blocking SPN enzymatic activity as a competitive inhibitor of β-NAD+ binding to SPN (Kimoto et al., 2006; Meehl et al., 2005). IFS lacks a signal sequence and is located exclusively in the cytoplasm of S. pyogenes, so the protective effect of IFS likely results from inhibiting GHase activity of pre-secretory SPN pools that fail to be exported from the bacterium (Kimoto et al., 2006; Meehl et al., 2005). The biological importance of SPN immunity is illustrated by the fact that the gene for IFS is absolutely essential for the viability of SPN-producing S. pyogenes strains (Meehl et al., 2005) and for overexpression of SPN in a heterologous host like E. coli (Kimoto et al., 2006; Meehl et al., 2005).
To understand the physical basis for SPN's enzymatic activities and its inhibition by IFS, we determined the crystal structure of the catalytically active SPNct domain complexed to IFS (Figure 1). Our analysis reveals that the protein fold of SPN is remarkably similar to canonical RTases like cholera toxin and diphtheria toxin, including several conserved sequence motifs in the active site (Figure 2). However, structural variations in SPN can explain its failure to ADP-glycosylate protein substrates (Figure 3) while retaining robust glycohydrolase activity. The structure of the SPN-IFS complex reveals how IFS inhibits SPN enzymatic activity by binding over the active site and completely blocking access to β-NAD+ (Figures 1 and 4). The structure of IFS determined in the absence of SPN reveals an alternative conformation of a C-terminal α-helical domain that is incompatible with binding to SPN (Figure 5). The conformational switch required for IFS to bind to SPN could be exploited by developing novel antimicrobials that block immunity of group A Streptococci to SPN and cause toxicity in SPN-producing strains.
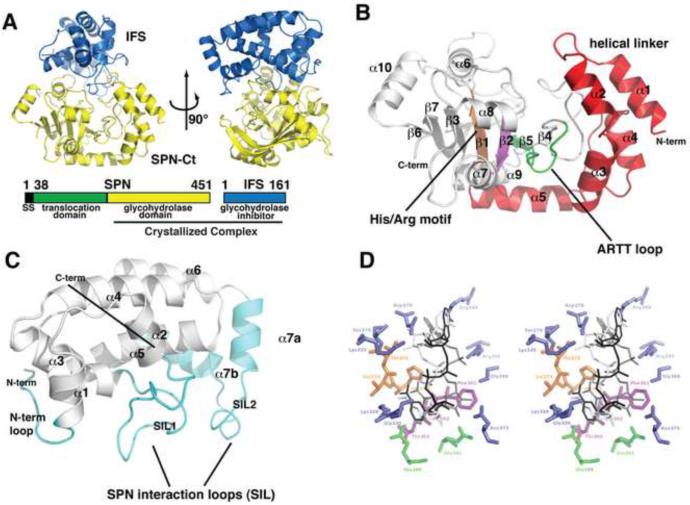
Figure 1.
Domain and three-dimensional structure of the SPNct-IFS complex. A. Orthogonal views of the SPNct-IFS complex depicted as ribbon structures. The orientations of SPNct and IFS in the left and right panel are approximately the same as those in (C) and (D), respectively. Full length SPN spans 451 amino acids and is made up of three regions: a secretion signal (black), translocation domain (green) and glycohydrolase domain (yellow). When SPN is secreted, its secretion signal is cleaved creating a mature fully active SPN. IFS remains in the bacterial cytoplasm when SPN is injected into the host cell. The glycohydrolase domain of SPN (SPNct) was crystallized in complex with full length IFS. B. The putative active site of SPN with NAD modeled in CTx (white), DTx (grey) and iota toxin (black) binding modes. Residues that are in hydrogen-bonding distance are highlight in blue while conserved residues found in the ARTT motif (green), STS motif (purple), and Arg/His (orange) C. A close up view of IFS highlighting the SPN interacting loops, SIL1 and SIL2 (cyan). D. A close up view of SPNct showing secondary structural elements and the ADase signature motifs. The N-terminal linker region (red) between the catalytic domain and secretory domain of SPN (not present in the crystal structure) is a unique adaptation of SPN's RTase-like fold. The conserved ARTT motif (green), STS motif (purple), and Arg/His motif (orange) of the RTase superfamily are present in SPNct. See also Figures S1 and S2.
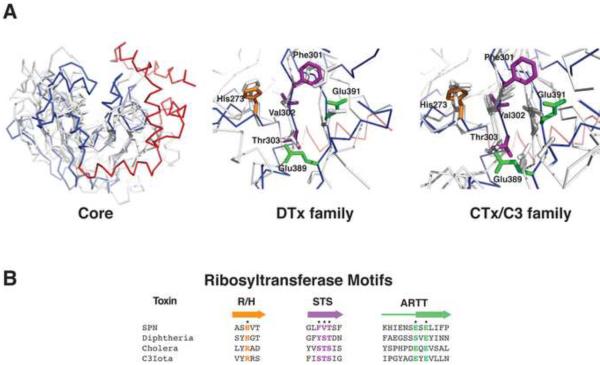
Figure 2.
SPNct Has a RTase-like Fold. A. Structural alignment of SPNct with ribosyltransferases: diphtheria toxin (1dtp), cholera toxin (2a5f), C3bot (1gzf), C3tau (1ojz), Vip2 (1qs2). In the left panel, SPNct is shown in blue (RTase domain) and red (helical linker) with the other RTases shown in gray. The helical linker of SPNct is not present in other RTase superfamily enzymes. Conserved active residues of the DTx (center) and CTx/C3 (right) families of RTases are present in SPNct . SPNct residues corresponding to the ARTT motif (green), the Arg/His motif (orange), and STS motif (purple) are shown as sticks (cf. Figure 1C). B. Aligned sequences of RTase motifs colored according to the scheme in panel (A).
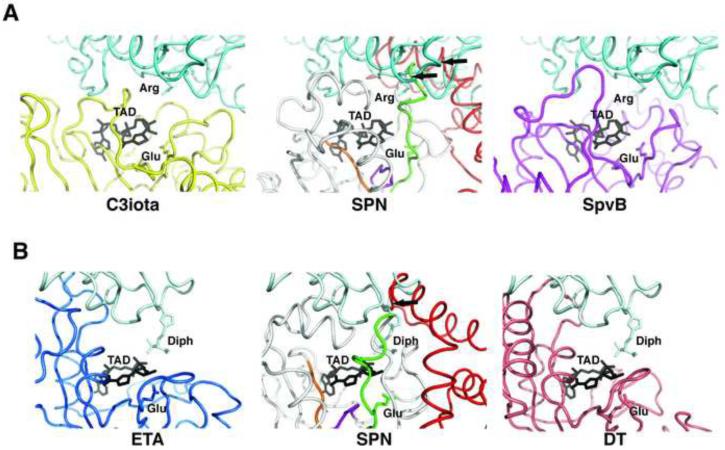
Figure 3.
The Binding Site for Protein Substrates of Other RTases is Occluded in SPNct. A. The structures of SPNct and the RTase SpvB (2gwl) were superimposed onto the structure of C3iota in complex with its Actin substrate (3buz; left panel)(Tsuge et al., 2008). The β-NAD+ mimic TAD (β-methylene-thiazole-4-carboxamide adenine dinucleotide; grey) identifies the active site of C3iota in the complex. The helical linker of SPNct (red) clashes with the docked actin protein (middle panel), suggesting that SPN does not accommodate protein substrates in the canonical mode of other RTases. The docking model for SpvB readily accommodates actin, which is a known substrate for ADP-ribosylation by this enzyme. B. In a similar manner as in panel (A), the structures of SPNct and DTx were superimposed onto the RTase ETA in complex with its substrate eEF2. The left panel is the crystal structure of ETA-eEF2 complex (Jorgensen et al., 2005). The model of SPNct docked against eEF2 reveals a clash with the helical linker of SPNct (red). In contrast, the docking model for DTx does not show significant clashes with eEF2, a substrate for ADP-ribosylation by DTx .
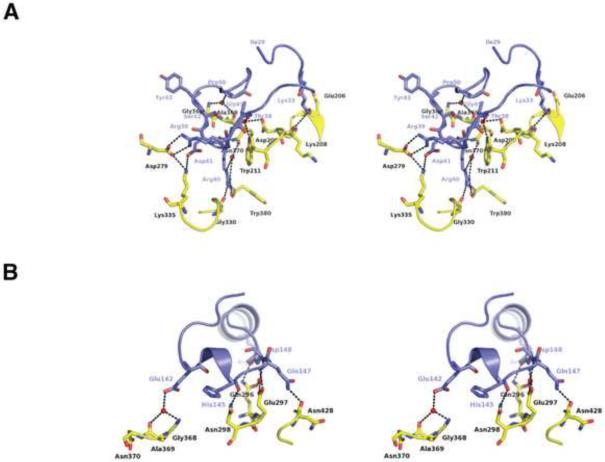
Figure 4.
Polar Interactions Stabilize the SPNct-IFS Complex. A. SIL1 inserts into the active site pocket of SPN and make numerous hydrogen-bonding and electrostatic interactions. B. SIL2 makes hydrogen-bonding interactions on the rim that forms part to the active site of SPN.
Figure 5.
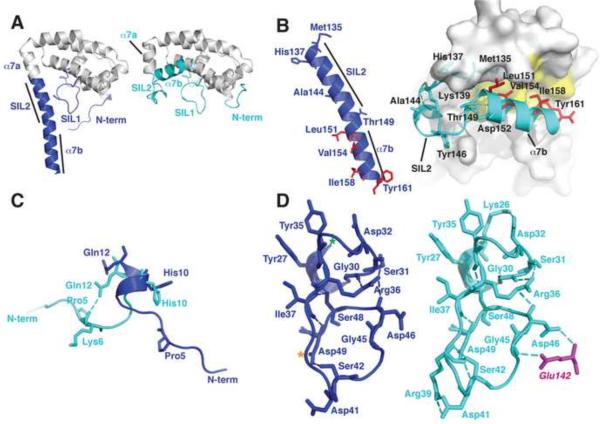
IFS Changes Conformation in Complex with SPN. A. The unbound IFS structure (left panel) shows an extended conformation of the C-terminal region (blue) whereas this region adopts a compact structure (cyan, right panel) in IFS bound to SPN (cf. Figure 1B). B. Hydrophobic residues (red) that are exposed in unbound IFS (left panel) become buried in the fold of bound IFS (right panel). Refolding of the C-terminus breaks helix α7 into two helical segments α7a and α7b. C. The N-terminal region of IFS is flexible and adopts different conformations in the free and bound structures. D. A network of hydrogen bonds stabilizes the structure of the SIL1 loop in free and bound structures of IFS (see text for details).
RESULTS
Overall Structure of the SPNct-IFS complex
The full-length, mature SPN protein (residues 38–451) in complex with IFS (161 amino acids) did not crystallize, so a C-terminal enzymatically active domain of SPN (SPNct; residues 191–451) was identified by limited proteolysis, and overexpressed in E. coli as a recombinant protein in complex with IFS. The deleted N-terminal CMT domain (residues 1–190) is predicted to have a β-strand structure (Rost et al., 2004) and the ProDom database (Servant et al., 2002) predicts a “jelly-roll” fold for this region. Although the N-terminal CMT domain is required for translocation of SPN into host cells and biological activity (Ghosh and Caparon, 2006), it has no significant effect on enzymatic activity in vitro (Ghosh and Caparon, 2006). The SPNct-IFS complex produced well diffracting crystals (Figures 1A and S2; Table 1) and the structure was determined at 2.8 Å resolution using phasing information from a selenomethionyl-labeled derivative by a multiple wavelength anomalous diffraction X-ray experiment (Table 1).
Table 1.
Crystallographic Data and Model Refinement Statistics
| Crystal | SPN/IFS | IFS | ||||
|---|---|---|---|---|---|---|
| Peak | Inflection | Remote | Peak | Inflection | Remote | |
| Resolution (Å) | 42.72–2.8 (2.90–2.80)a | 42.64–2.8 (2.90–2.80) | 42.72–2.8 (2.90–2.80) | 46.8–2.5 (2.59–2.5) | 46.8–2.5 (2.59–2.5) | 46.8–2.5 (2.59–2.5) |
| λ | 0.97892 | 0.97938 | 0.99510 | 0.97901 | 0.97932 | 0.99504 |
| mosaicity | 0.86 | 0.83 | 0.86 | 0.43 | 0.42 | 0.41 |
| Total measurements | 181424 | 181118 | 182378 | 470532 | 471606 | 547619 |
| Unique measurements | 46783 | 46736 | 46783 | 66240 | 66415 | 66456 |
| Redundancy | 3.88 (3.83) | 3.88 (3.84) | 3.88 (3.83) | 7.1 (7.05) | 7.1 (7.07) | 8.24 (8.15) |
| Completeness (%) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) |
| <I/σI> | 9.2 (2.6) | 7.2 (2.2) | 10.4 (3.4) | 12.6 (3.3) | 12.8 (3.0) | 14.3 (4.3) |
| Rsym | 0.079 (0.328) | 0.091 (0.391) | 0.069 (0.265) | 0.077 (0.427) | 0.077 (0.457) | 0.069 (0.363) |
| MAD phasing | ||||||
| Resolution (Å) | 41.7–2.8 | 41.7–2.8 | 46.8–2.5 | 46.8–2.5 | ||
| Number of sites | 26 | 26 | 26 | 26 | ||
| Phasing power (anom) | 1.05 (1.02) | 0.92 (0.64) | 1.25 (1.24) | 2.07 (1.22) | ||
| Rcullis (anom) | 0.70 (0.83) | 0.73 (0.91) | 0.56 (0.71) | 0.51 (0.72) | ||
| Overall FOM | 0.286 | 0.301 | ||||
| Refinement statistics | ||||||
| # of reflections | ||||||
| working set | 23069 | 32775 | ||||
| test set | 1240 | 1739 | ||||
| Rwork | 0.212 | 0.259 | ||||
| Rfree | 0.287 | 0.289 | ||||
| rmsd bond length | 0.010 | 0.0071 | ||||
| rmsd bond angles | 1.193 | 1.109 | ||||
Values in parenthesis are for the highest resolution shell
The SPNct domain has a mixed α/β fold (Figure 1B) with a shape reminiscent of an open bowl. IFS serves as a lid covering the active site pocket of SPN (Figure 1A). The active site motifs and core β-sheet structure of the RTase family are conserved in the SPNct protein (Figure 3) as described in more detail below. The core structure of IFS comprises a single domain of eight helices that are connected by several extended loops (Figure 1C). Two of these loops make intimate contacts with SPNct and we have named them the SPN Interaction Loops, SIL1 (residues 29–50) and SIL2 (residues 137–150; Figures 1C and 5). SIL1 is preserved as a loop in the bound and free conformations of IFS, whereas SIL2 rearranges in the unbound form of IFS and is incorporated into a continuous helix projecting from the protein core (figure 5A). This extended helix is incompatible with the binding interactions seen in the SPNct-IFS complex.
SPNct is Structurally Homologous to Ribosyltransferases
We compared the structure of SPNct with those of related β-NAD+-dependent enzymes. There are no structures of strict GHases in the Protein Data Bank and the amino acid sequence of SPN has limited homology with other proteins, except for SPN orthologs from other Streptococcus spp. (e.g., (Kimoto et al., 2005)). The SPN sequence and structure are unrelated to those of multi-functional GHase/ACases, including human CD38 and a GHase/ACase from Aplysia californica {Love, 2004 #212}, which contain multiple disulfide bonds that stabilize the protein fold. Furthermore, the eukaryotic GHase/ACases are homodimers, whereas SPNct is a monomer (Meehl et al., 2005). Based on these criteria, we determined that SPNct is not structurally homologous to these enzymes.
However, the SPNct structure superimposes well onto several different RTases, including two universally conserved motifs in the active site (figure 3). Although the sequences of bacterial RTases are divergent, these proteins share a conserved core structure consisting of seven β-strands arranged in two perpendicular β-sheets that bracket the β-NAD+ binding pocket (Han and Tainer, 2002). This core β-sheet sandwich is decorated with different configurations of helices, forming a variety of mixed α/β structures in various RTase family members. A helix or a variable sized loop lies on one side of the β-NAD+ binding pocket in various RTase family members and is hypothesized to be important for substrate recognition (Han et al., 2001). A structure-based alignment of the SPN amino acid sequence with several structurally homologous RTases did not reveal this element, or any significant sequence homology outside of a few active site motifs described below.
All RTases harbor an ADP-ribosyl-turn-turn (ARTT) motif in their active site, which includes a catalytically essential glutamic acid (Holbourn et al., 2006). SPN's ARTT motif features Glu391, which is required for enzymatic activity in vitro (Ghosh et al., 2010). A second highly conserved residue of the ARTT motif is a glutamic acid or glutamine (SPN Glu389) located two residues N-terminal to the essential catalytic glutamic acid. The conformation of SPN's ARTT loop is different from other RTase proteins because of packing interactions involving SPN's unique α-helical linker subdomain (figure 1D). Nonetheless, ARTT residues Glu389 and Glu391 in SPN superimpose well with the analogous residues of other RTases (figure 3A). The presence of a typical ARTT motif in the SPN active site is additional evidence for SPN's inclusion as a member of the RTase superfamily. However, the high level of sequence divergence between SPN other RTases suggests that SPN represents an atypical member of the superfamily.
Another universal feature of RTase active sites is the R/H motif, consisting of an arginine or histidine preceded by two hydrophobic residues, which are typically an aliphatic residue followed by an aromatic residue. These residues support binding to β-NAD+ and/or maintaining the structure of the active site (Holbourn et al., 2006). The signature histidine (His273; Figure 2A middle and right panels) of SPN's R/H motif is located on β-strand 1, although His273 is preceded by an alanine (Ala271) and serine (Ser272) instead of the aliphatic/aromatic residue pair that is typical of a canonical R/H motif (Figure 2B). It remains to be determined if His273 contributes to the enzymatic activity of SPN.
Atypical β-NAD+ binding pocket
The Diphtheria Toxins (DTx) and Cholera Toxins (CTx) constitute two large families within the RTase superfamily that are distinguished by different motifs within the β-NAD+ binding pocket and active site. The DTx family is defined by a Y-X10-Y motif, consisting of a pair of tyrosines separated by a 10-residue spacer that stack against the aromatic nicotinamide ring of β-NAD+ {Domenighini, 1996 #289}. In the CTx family, the Y-X10-Y motif is replaced by a Ser-Thr-Ser (STS) motif in an analogous location, and CTx family members have either an “active site loop” or an “α3 helix” that participates in binding to polypeptide substrates (Holbourn et al., 2006). The STS and Y-X10-Y motifs of the CTx and DTx proteins, respectively, are located in the vicinity of strand β2 on the core β-sheet structure of the RTase superfamily (Figure 1B).
The SPNct active site lacks the Y-X10-Y (not shown) and STS motifs (Figure 2A) and there is no obvious “active site loop” or the “α3 helix” found in CTx-type enzymes. Consistent with its role as a pure GHase, SPN lacks these protein substrate-binding motifs, although other RTase signature residues are present in the active site. The NAD+ cofactor binds in slightly different orientations within the active sites of RTases crystallized in complex with NAD+. A docking model based on superposition of these structures onto SPNct suggests that SPN's active site can easily accommodate NAD+, with several residues in hydrogen-binding distance of the modeled NAD+ ligand. Arg295 is positioned for hydrogen bonding to the adenine phosphate of β-NAD+. Lys335 is also in close proximity and could potentially hydrogen bond to the ribose oxygen of adenine. Arg289 lies close to the amino group of the adenine base. The NAD+ binding pocket is capped by IFS, which appears to prevent binding.
SPNct modeled with known RTase protein substrates
Purified SPN does not exhibit RTase activity (Ghosh et al., 2010), and we superimposed the structure onto various RTase-protein complexes to examine if SPN could potentially bind to protein substrates. The RTase-substrate complexes show different extents and modes of interacting with proteins targeted for ADP-ribosylation (Figure 3), yet the amino acid side chain targeted for ADP-ribosylation is positioned similarly in the β-NAD+ binding pocket and the protein substrate binds on the same aspect of the conserved RTase fold. However, our substrate docking models reveal a clash with the α-helical subdomain that joins the SPN catalytic domain to an N-terminal domain (Figure 3, in red). The SPN interdomain linker occludes the usual substrate-binding site present in the RTases, as shown by two examples in Figure 3. In one example, we superimposed the crystal structure of the CTx family member Clostridium botulinum C3iota in complex with its G-actin substrate (Tsuge et al., 2008) onto the SPNct structure (Figure 3A). C3iota binds to G-actin in a different orientation than that of the ETA-eEF2 complex, but the superimposed SPNct structure still clashes with G-actin in the docking model (Figure 3A, center panel). Specifically, the ARTT loop and helices α1–α3 of SPN's linker subdomain both clash with G-actin. Notably, the G-actin residue Arg177 targeted for ADP-ribosylation by C3iota poses a severe clash with SPNct residues Trp380 and Leu372. The Salmonella enterica SpvB protein is a RTase that posttranslationally modifies G-actin, and superposition of SpvB onto C3iota complexed to G-actin shows a good fit of actin in the docking model (Figure 3A, right panel). We conclude that SPN cannot bind to a protein substrate in the same mode as these RTases without a rearrangement of SPN's α-helical linker subdomain.
In a second example of RTase-substrate interactions, the crystal structure of Pseudomonas aeruginosa enterotoxin A (ETA, a DTx family member) complexed with the protein substrate eEF2 (Jorgensen et al., 2005) was superimposed onto SPNct. Helix α2 within SPNct's linker subdomain (figure 1B) severely clashes with the docked eEF2 substrate (Figure 3B, center panel) and additional clashes are observed with SPN's ARTT loop and helix α5. In contrast to SPN, the structure of the Corynebacterium diptheriae DTx protein, which posttranslationally modifies the eEF2 protein, is well accommodated by a docking model based on the DTx-eEF2 complex (Figure 3B, right panel). These docking studies reveal that SPN is unable to accommodate protein substrates in the modes utilized by other RTases, supporting the conclusion that SPN's catalytic activity is limited to GHase activity despite having many features of an RTase toxin.
The presence of the linker subdomain also increases the depth of SPN's β-NAD+ binding pocket in comparison to the bona fide RTase proteins, which could further interfere with activity towards protein substrates. In fact, the IFS subunit in the SPNct-IFS complex structure does not directly contact the catalytic residues of the SPN active site. SPN's α-helical linker also contributes interactions with IFS, supporting immunity to SPN-based toxicity. This protective function may be a strong evolutionary driver for maintaining the linker as a protein interaction surface that gives SPN a unique structural identity among RTase family proteins.
Structure of the IFS inhibitory factor
In the complex with SPNct, the protein fold of IFS is a compact helical bundle with a loosely structured N-terminal region. Helices α4–α6 constitute the core of the protein, which is decorated by helices α1–α3. In the N-terminal region of IFS, the twenty-two residue SIL1 loop connects helices α2 and α3, and a 10 residue loop connects helices α5 and α6; both loops pack against helices α3 and α4. At the C-terminal end of IFS, helices α7a and α7b are joined together by SIL2 and pack against the core of the protein. Helix α7a makes hydrophobic and polar interactions with the helical head domain, whereas helix α7b makes hydrophobic interactions with the hydrophobic patch created from helices α5 and α6.
Structural homology searches using SSM (Krissinel and Henrick, 2004) and DALI (Holm and Sander, 1996) reveal similarities between IFS and several other alpha-helical proteins sharing 5–12% sequence identity, including the ubiquitin addition domain of RanGAP1 (Hillig et al., 1999) and VHS domain of ADP-ribosylating factor (ADF) interaction proteins (Boman et al., 2002; Mullins and Bonifacino, 2001; Shiba et al., 2003; Shiba et al., 2002; Zhu et al., 2003). However, these similarities are limited and appear to be fortuitous—the functional surfaces of IFS contacting SPN are not conserved with these other proteins, which function as adaptors that bind to signaling and vesicular proteins. These findings suggest that these proteins are functionally unrelated although they are likely to have evolved from a common ancestral fold.
Interactions Between SPNct and IFS
The extensive interaction between SPNct and IFS buries more than 3200 Å2 of protein surface area, forming a seal around the entire rim of SPNct's concave active site (Figure 1A). IFS does not directly contact the catalytic residues in SPN's active site but instead blocks access to β-NAD+ by covering over the active site pocket. Numerous hydrogen bonds and electrostatic interactions secure the complex between SPNct and IFS, which features a striking complementarity of electrostatic potential. The surface of IFS is predominately negatively charged whereas the opposing surface of SPNct is positively-charged (supplementary figure S1). Charge-charge interactions are likely to contribute to the long-lived complex with IFS that effectively blocks SPN enzymatic activity. In the absence of IFS, this hydrophilic surface of SPNct would be exposed to solvent and could mediate interactions with substrates.
IFS contacts SPN mainly through two extended loops, SIL1 and SIL2 (Figure 1C). SIL1 is a highly convoluted loop connecting helices α2 and α3 of IFS. The beginning of the SIL1 loop (residues 29–38) contacts another small loop located between the first two helices of SPNct (Figure 4A). Lys33 of IFS is within hydrogen-bonding distance of the carbonyl oxygens of Glu206 and Lys208 in the SPN protein. Thr38 makes water-mediated interactions with SPN Asp209 and Trp211 (Figure 4A). The middle of SIL1 is poised over the β-NAD+ binding pocket where IFS residues 39–43 make electrostatic and hydrogen-bonding interactions with SPN residues on either side of the pocket. Arg39 forms a salt bridge with Asp279 (Figure 4A). Arg40 forms a hydrogen bond with the carbonyl of Gly330 and Trp380. Asp41 makes an electrostatic interaction with Lys335. The carbonyls of Ser42 and Tyr43 are within hydrogen-bonding distance of the Nε atom of Arg295 in SPNct (figure 4A). The last section of the SIL1 (residues 44–50) winds around the back of the β-NAD+ binding pocket and makes its interactions with loop between β2 strand and α6 helix (residues 285–300) of SPNct. The amide of Gly45 of IFS makes a water-mediated hydrogen bonding interaction with the carbonyl oxygen of Ala369 and the amide nitrogen of Gly368 (Figure 4A).
The SIL2 loop, located between helices α7a and α7b, is a second major region of IFS interaction with SPN. SIL2 contacts two loops on the rim of SPN's β-NAD+ binding pocket (Figure 4B), one that is located between the SPNct β4 strand and ARTT loop (see below; residues 361–380) and the other between the α6 helix and β2 strand (residues 285–300). In a manner analogous to Gly45 of SIL1, the SIL2 residue Glu142 makes water-mediated hydrogen-bonding interactions with the carbonyl oxygens of Ala369 and amide of Gly368 (Figure 4B). The amide nitrogen of Asp148 makes water-mediated interactions with Gln296 and Glu297 while the Asp148 side chain makes interactions with Gln296 (Figure 4B). Auxiliary interactions in this region occur through helix α7b of IFS. Arg156 makes interactions with the carbonyl of Ala 287 and Thr157 makes a water-mediated hydrogen bond with Asp286 (Figure 4B). This network of polar interactions between IFS and SPNct is suggestive of a stable protein complex. However, the hydrophilic character of this interface is also consistent with SPN's stand-alone role as a soluble toxin that is injected into the host cytosol.
Structural Dynamics of IFS
The crystal structure of IFS in the absence of SPN reveals a different conformation of the C-terminal helices α7a and α7b (Figure 5), which in the absence of SPN fold into a continuous α-helix (α7) that projects from the core of the IFS protein and resembles the letter “P” (figure 5A). This rearrangement eliminates the SIL2 interaction loop (Table 1; Figure 5B) and presumably is inactive for binding to SPN. It is likely this extended conformation of IFS is energetically favorable and populated in solution. Four crystallographically independent molecules of IFS exhibit the same extended conformation of helix α7, despite different crystal packing environments (not shown). These IFS molecules differ only in the number of disordered residues at their C-termini (the ordered region of monomer A spans residues 1–156, monomer B spans residues 1–173, monomer C spans residue 1–164, and monomer D: 1–159). The elongated shape of unliganded IFS is also consistent with its anomalous behavior on gel filtration and large apparent molecular mass (31,500 Da including the c-Myc tag and His6 affinity tag, versus the calculated molecular mass of 21,960 Da) that was estimated by dynamic light scattering. Furthermore, deuterium exchange studies of IFS support the conclusion that the C-terminal residues spanning helix α7 are exposed and freely exchangeable in the unbound state (Justin Sperry and Michael Gross, Washington University, unpublished observations).
The core structure of IFS spanning helices α1–α6 is unchanged in bound and free forms of IFS, except for localized rearrangements in three regions of interaction with SPNct. The transformation of α7 buries ~200 Å of hydrophobic surface area that is exposed on the surface of α5 and α6 in the unbound IFS conformation (involving C-terminal residues Leu151, Val154, Ile158, and Tyr161; shown in red in Figure 5B).
N-terminal residues 1–9 are in an extended conformation in the uncomplexed IFS and, when bound to SPNct, the protein backbone is twisted by two rotations of approximately 180° (Figure 5C). This twisted conformation is further stabilized by the side chain of IFS Gln12, which donates a bifurcated hydrogen bond to the main chain carbonyl oxygens of Pro5 and Gly7 (Figure 5C). This change upon binding to SPNct redirects the polypeptide chain of IFS and extends the surface of contact with SPNct.
The conformation of the SIL1 loop changes significantly in the bound and unbound structures of IFS. The conformation of SIL1 in the SPN-IFS complex is established by a series of internal hydrogen bonds, including a hydrogen bond between Lys26 and Asp32 within the beginning of the loop (Figure 5D, right panel). Arg36 interacts with the carbonyls of Gly30 and Ser31, while Ser48 hydrogen bonds to the carbonyl of Tyr35 and the amide of Ile37. The middle of the SIL1 loop is conformationally constrained by two hydrogen bonds from Ser42 to the main chain amide of Arg39 and the side chain of Asp49. The C-terminal end of SIL1 interacts with SIL2 via several hydrogen bonds involving SIL1 residues Asp46 and Gly45 and SIL2 residue Glu142. In the unbound IFS structure, the middle of the SIL1 loop moves closer to helix α7, requiring Ser42 to forego its interaction with Arg39. This apparent increase in the flexibility of SIL1 is consistent with an absence of electron density for the Ser42 side chain in two of the four crystallographically independent molecules of unbound IFS.
DISCUSSION
SPN and IFS constitute a protein complex that enhances bacterial virulence while preventing self-destruction of the pathogen. Similar toxin-antitoxin pairs have a wide phylogenetic distribution and include Staphylococcal proteases and their endogenous inhibitors (Potempa et al., 2005), toxin-antitoxins that ensure carriage of plasmids {Hayes, 2003 #220} and the chromosomally-encoded toxin-antitoxin pairs that modulate growth of prokaryotes in response to nutritional stress {Gerdes, 2005 #218}. Since these antitoxins typically block toxins affecting bacterial cell growth or viability, chemical inhibitors of antitoxin function could prove useful as antimicrobials. Inhibitor development would benefit from structural information about the antitoxin and its mode of binding to the toxin, and knowledge of the mechanism of toxin activity and its blockade by the antitoxin.
SPN is an atypical RTase with the conserved protein fold and active site ARTT motif of the RTase superfamily including the catalytic residue Glu391 (Ghosh et al., 2010)(Figure 3). However, SPNct does not accommodate protein substrates for ADP-ribosylation and lacks the Y-X10-Y and STS motifs contributing to substrate interactions by diphtheria toxin and cholera toxin family proteins, respectively (Holbourn et al., 2006). A unique α-helical linker in SPNct (Figure 1) creates a clash with protein substrates docked onto SPNct in the orientation of other RTase-substrate complexes (Figure 4). The linker contributes interactions with IFS (Figure 4A), which may explain the selection for this feature of SPN. SPN is an efficient GHase in vitro and causes a significant depletion of β-NAD+ stores in epithelial cells exposed to SPN-producing strains of S. pyogenes or in yeast expressing recombinant SPN {Michos, 2006 #213}. These findings indicate that SPN's biological function(s) as a virulence factor are significantly coupled with GHase activity, depleting β-NAD+ stores instead of modifying proteins through ADP-ribosylation.
The RTase-type fold of the SPN protein may have evolved to catalyze efficient GHase activity through two types of genetic alterations. The first alteration may have been the addition of the α–helical linker region to couple SPN's catalytic activity to its cellular import mediated by SPN's N-terminal domain. This architectural addition would have the secondary consequence of interfering with binding of protein substrates to SPN, therefore limiting enzymatic activity to the reaction of β-NAD+ with water. Subsequent modifications of the SPN active site may have occurred by positive selection to increase the catalytic efficiency of the GHase reaction in order to enhance bacterial virulence and provide a growth advantage. Mutations in other RTase enzymes have been reported to change relative rates of RTase and GHase activities; however, the mutations that strongly suppress RTase activity typically cause a concomitant reduction in GHase activity (Holbourn et al., 2006). Thus, understanding the mechanistic basis for selective β-NAD+ hydrolysis in the absence of ADP-ribosylating activity would benefit from comparative structural analyses of dedicated bacterial GHases. Several instances of bacterial GHase activity have been reported {Davis, 1980 #214;Everse, 1975 #172;Mather, 1972 #216}, but the genes encoding these enzymes have not been isolated and their enzymatic activities remain to be characterized.
Some clues about residues contributing to efficient GHase activity have come from clinical isolates of S. pyogenes collected during the past 20 years. Recent isolates express SPN proteins with higher specific activity than strains isolated prior to 1989 (Tatsuno et al., 2007). Reduced GHase activity is associated with a polymorphism at SPN residue 330 in which an aspartic acid replaces the glycine found in the native enzyme (Tatsuno et al., 2007). Residue 330 lies near the surface of the β-NAD+-binding pocket (figure 4A) where the larger size and negative charge of the aspartic acid may inhibit access of β-NAD+ to the binding pocket. It seems unlikely that the high activity Gly330 allele evolved from the lower activity Asp330 allele, which is consistently found in association with a nonfunctional, truncated IFS allele suggestive of a lack of selection for IFS activity when the SPN Asp330 variant is present (Meehl et al., 2005). Thus, it seems more likely that the high activity Gly330 allele is ancestral to the low activity Asp330 allele, which consequently relieved selective pressure for maintaining a functional IFS.
Bacterial expression of a GHase toxin requires coexpression of a cognate antitoxin like IFS to prevent depletion of β-NAD+ {Everse, 1975 #215;Davis, 1980 #214;Mather, 1972 #216}. The occurrence of toxin-antitoxin pairs may argue for a different evolutionary model in which the GHase enzymes are progenitors of RTases and evolve by first decreasing GHase activity to eliminate the need for the protein inhibitor before acquiring efficient RTase activity through additional mutational drift. The extant bacterial RTases are expressed without a cognate inhibitory protein and their enzymatic activity is contingent upon interaction with protein substrates in host cells that are either absent from the bacterium or not essential for bacterial growth. The transition from GHase to RTase activity could have occurred by adaptations that changed the binding site for a protein inhibitor of the GHase to a site that binds the protein substrate of a RTase.
A structural analysis of the interaction between SPN and IFS is informative in this regard. The crystal structure of the SPNct-IFS complex reveals that IFS binds to SPNct in a 1:1 complex and that the SIL1 loop would sterically clash with β-NAD+ in the active site, providing the molecular basis for the known ability of IFS to act as a competitive inhibitor of β-NAD+ (Meehl and Caparon, 2004). It is noteworthy that a second IFS binding site has been described corresponding to SPN residues 38–166 (Tatsuno et al., 2007), which are not present in the SPNct protein (residues 191–451) that was crystallized in complex with IFS. However, since this site is not required to suppress the toxic effects of SPNct in vivo (Ghosh and Caparon, 2006), its biological significance is uncertain.
A comparison of IFS structures, alone and in complex with SPNct, reveals features that could be exploited for the development of small molecule inhibitors of IFS that block interaction with SPN, unleashing its auto-toxic activity against S. pyogenes. The α-helical head domain of unbound IFS is appended by an elongated C-terminal helix extending away from the head domain. Consistent with the conformational flexibility revealed in the crystal structure, the C-terminal region of purified IFS is susceptible to proteolysis, and deuterium isotope exchange studies reveal high rates of exchange in this region relative to the head domain (J. Sperry and M. Gross, manuscript in preparation). In order to bind to SPNct, IFS must undergo extensive conformational changes, including an adjustment of the N-terminal SIL1loop, and the repacking of residues from the C-terminal helix α7 to form two helices (α7a, α7b ; figure 1C) interrupted by the SIL2 interaction loop.
An inhibitor of IFS might be developed to lock the C-terminal helix in the extended conformation that cannot refold to bind and inhibit SPN. Consistent with this, bacterial growth is attenuated by overexpression of SPN-IFS and this effect can be mitigated by C-terminal truncations of IFS, which include SIL2 and α7b (S. Chandraeskaran and M. Caparon, unpublished data). C-terminal deletions may relieve a barrier to conformational rearrangement of the C-terminal residues of IFS that enables binding to SPN. An antibacterial agent that binds to the hydrophobic surface of IFS serving as the docking site for helices α7a and α7b might stabilize IFS in an inactive, extended conformation to unleash the cellular toxicity of SPN, causing depletion of β-NAD+ within the bacteria and cell death.
EXPERIMENTAL PROCEDURES
Construction of SPNct-IFS expression plasmid
The SPNct-IFS expression plasmid was constructed from the plasmid pMAM 3.18 (Meehl et al., 2005) in two steps. First the nucleotides coding the gIIIB signal sequence, 6XHis and SPN residues 38–190 were removed from this plasmid to create an intermediate plasmid pJOY114 using an inside-out PCR strategy and two 5' phosphorylated primers (oJOY187: /5Phos/CAT CAT CAT ACT TTT GGC AAT ATG GAA CGC GAT CTT TTT GAA AAA AAG TTT AAA G and oJOY155: /5Phos/ATG ATG ATG CAT GGT TAA TTC CTC CTG TTA GCC CAA AAA ACG GGT ATG GAG AAA CAG). Then nucleotides for two stop codons (underlined in the primer sequence below) were introduced in pJOY114 between the coding sequences of the final IFS residue (Phe 161) and its carboxy-terminal c-myc epitope tag using the QuikChange site-directed mutagenesis kit (Stratagene) and two complementary primers (IFS stopF: GCT TGA TAT GGT CGA AAG AAC AAT AGA AAC ATT TTA GTG AAG CTT TCT AGA ACA AAA ACT CAT CTC AGA AGA GG and IFS StopR). The final plasmid pJOY126 expresses SPNct (Met - 6XHis - SPN Thr191 to Lys451) and full length IFS (residues 1 to161) from an L-arabinose-inducible araBAD promoter and these proteins form the SPNct-IFS complex in the cytoplasm of the E. coli hosts.
Purification and crystallization of SPNct-IFS complex
The SPNct protein representing the C-terminal glycohydrolase domain of SPN (residues 191–451), and full length IFS (residues 1–161) were co-expressed in E. coli DL41 (methionine auxotroph) using the plasmid pJOY126 as described previously (Ghosh and Caparon, 2006; Meehl and Caparon, 2004). Cells were grown in a medium consisting of Luria broth and Bovine Heart Infusion broth with 100 μg/ml of ampicillin at 37 °C. When the culture reached an OD600nm of 2.5 expression was induced by the addition of 0.2% arabinose and growth continued for additional 1–1.5 hours at which time cells were harvested by centrifugation, and immediately processed, or the pellets were stored at −20 °C. For expression of selenomethionine protein, cells were grown in M9 media supplemented with all 20 amino acids except methionine, which was replaced in selenomethionine. For processing, cells were resuspended in phosphate buffered saline (PBS), pH 7.4 and protease inhibitor cocktail (Sigma Aldrich) then lysed by sonication while chilled on ice and the cellular debris removed by centrifugation at 20,000g for 20 minutes. The resulting lysate was subjected to metal affinity chromatography using a TALON Superflow column (Clontech). The SPNct-IFS complex was eluted using a step gradient of PBS supplemented with 0.3 M imidazole. Fractions containing the complex were pooled and dialyzed three times for two hours each against 20 mM Tris pH 8.0 at 4 °C and then subjected to ion exchange chromatography using Q Sepharose FF (GE Healthcare) eluted with a step gradient of 20 mM Tris, pH 8.0 and 0.5 M NaCl. Fractions containing the complex were pooled, concentrated and subjected to a size-exclusion chromatography over a S-200 column (GE Heathcare). Fractions were analyzed for purity by SDS-PAGE, pooled, dialyzed against 20 mM Tris pH 8.0, 50 mM NaCl, 0.1 mM EDTA and stored at 4 °C. For crystallization, drops containing 1 μl of 15–20 mg/ml of protein and 1 μl of precipitant containing 20–25% PEG 8K and 100mM Tris-HCl pH 8.0 were equilibrated against the same precipitant by vapor diffusion at 22 °C. Crystals appeared in 1–2 days and grew to full size in 3–5 days. Crystals were in space group C2 with cell dimensions a=198.42, b=57.81, c=89.52; β=107.14 with two molecules per asymmetric unit.
Structure Determination of SPNct-IFS complex
Before data collection, crystals in stabilizing solution (25% PEG 8K and Tris-HCl pH 8.0) were transferred to the same solution containing 20% glycerol for 3–5 minutes and frozen with cold nitrogen gas from an X-Stream system (Rigaku). X-ray diffraction data were collected at Lawrence Berkeley Laboratory beamline 4.2.2 (Molecular Biology Consortium). Images were indexed, integrated, and scaled using D*TREK {Pflugrath, 1999 #242}. Initial phases were calculated from a three-wavelength (peak, inflection, and low remote) selenomethione MAD experiment. Structure factor calculation and scaling between sets were done using the CCP4 suite {, 1994 #248}. Of the twenty-eight expected selenium sites, nineteen of the strongest sites were located using SHELXD {Schneider, 2002 #237} and AutoSHARP {Vonrhein, 2007 #230} was used to locate an additional 8 sites, refine the selenium sites, and calculate initial phases. Maps were improved by density modification using DM {Cowtan, 1998 #249}. Maps were interpretable as we were able to quickly locate the helical head structure of IFS. The atomic model was built using COOT {Emsley, 2004 #228}. Initial refinement was done by simulated annealing protocols in CNS {Brunger, 1998 #238}. The model was further refined using REFMAC which included TLS refinement {Murshudov, 1997 #273}. Ramachandran analysis showed that 93.6% of the main chain dihedral angles are in the most favorable regions, 6.27 in additional regions, and 0.12% in disallowed regions. The final model includes SPNct (residues 196–445) and the entire IFS molecule (residues 1–161). Residues 191–195 and 446–451 of SPNct were not seen in the electron density map and assumed to be disordered. Figures were produced using PyMOL {DeLano, 2002 #250}.
Purification and Crystallization of IFS
IFS alone with a carboxy terminal c-myc- and 6XHis-tag was expressed from the plasmid pMAM 3.19 (Meehl et al., 2005) in E. coli DL41 under the same culture conditions described above. When the OD600nm reached 3.0 expression was induced with 0.2% arabinose and cells harvested following an hour of additional growth. Selenomethionine-incoporated protein was expressed as described for the SPNct-IFS complex. –Lysates were prepared and subjected to metal affinity chromatography as described above. Collected fractions were dialyzed three times against 20 mM Tris pH 8.0 then subjected to a Source 15Q column eluted with a salt gradient (0–0.5 M) in 20 mM Tris pH 8.0 and fractions analyzed by SDS-PAGE. Fractions containing greater than 95% IFS were pooled, dialyzed against 20mM Tris pH 8.0, 50 mM NaCl, and concentrated to 12.8 mg/ml. Crystals were grown using the vapor diffusion method with hanging drops comprised 2 μl of IFS mixed with 2 μl of precipitant solution containing 1.6–2.0 M ammonium sulfate and Tris pH 8.0 and equilibrated over the same precipitant solution. Crystals were of space group P3121 will cell dimensions a=108.03 Å, c=147.05 Å with four molecules per asymmetric unit.
Structure Determination and Refinement of IFS
Prior to freezing, the crystals were transferred from a stabilizing solution containing 2.5 M ammonium sulfate and 20 mM Tris pH 8.0 and the same solution supplemented with 15% glycerol and frozen with cold nitrogen gas as described above. X-ray diffraction data were collected at Lawrence Berkeley Laboratory beamline 4.2.2 (Molecular Biology Consortium). Diffraction data were indexed, integrated, and scaled using D*TREK {Pflugrath, 1999 #242}. Initial phases were calculated from a three-wavelength (peak, inflection, and low remote) selenomethione MAD experiment. The structure factor calculation and scaling between sets were done using the CCP4 suite {, 1994 #248}. Sixteen of the strongest selenium sites (of the expected twenty-eight) were located from the dataset collected at the peak wavelength using SHELXD {Schneider, 2002 #237}. AutoSHARP {Vonrhein, 2007 #230} was used to locate an additional 10 sites, refine the selenium sites, and calculate initial phases. Maps were improved by density modification using DM {Cowtan, 1998 #249} and allowed for unambiguous tracing of the electron density. The atomic model was built using O {Jones, 1991 #252}. The initial model was subjected to refinement using CNS {Brunger, 1998 #238} and further refined using REFMAC {Murshudov, 1997 #273}. Interestingly, two molecules include contain the entire IFS (1–161) plus a portion of the c-myc tag (B: 1–173; C: 1–166) and the other molecules were truncated (A:1–155; D:1–159). Thus, residues 156–161 in molecule A and residues 160–161 in molecule D were not seen in electron density and were assumed to be disordered.
E-TOC PARAGRAPH.
The Streptococcus pyogenes SPN protein is injected into host cells where it hydrolyzes β-NAD+ and enhances infectivity. The bacterium is protected from SPN toxicity by an immunity factor known as IFS (immunity factor for SPN), which functions as a competitive inhibitor of SPN enzymatic activity by binding to the active site. We report a crystal structure of the SPN-IFS complex that shows an extensive and convoluted protein interaction surface characterized by many polar interactions. A crystal structure of IFS alone reveals a different conformation that may inform the rational design of small molecule inhibitors of IFS with the capability of unleashing SPN toxicity in the bacterium.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jay Nix (Molecular Biology Consortium beamline 4.2.2 at the Lawrence Berkeley Laboratory) with help with X-ray diffraction experiments. This study was supported in part by the UNCF/Merck Science Initiative Postdoctoral Fellowship awarded to C.L.S. and grants GM52504 (T.E.E.) and AI064721 (M.G.C.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Boman AL, Salo PD, Hauglund MJ, Strand NL, Rensink SJ, Zhdankina O. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol Biol Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol. 2002;44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Anderson PJ, Chandrasekaran S, Caparon MG. Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis. J Biol Chem. 2010;285:5683–5694. doi: 10.1074/jbc.M109.070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Caparon MG. Specificity of Streptococcus pyogenes NAD(+) glycohydrolase in cytolysin-mediated translocation. Mol Microbiol. 2006;62:1203–1214. doi: 10.1111/j.1365-2958.2006.05430.x. [DOI] [PubMed] [Google Scholar]
- Grushoff PS, Shany S, Bernheimer AW. Purification and properties of streptococcal nicotinamide adenine dinucleotide glycohydrolase. J Bacteriol. 1975;122:599–605. doi: 10.1128/jb.122.2.599-605.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Arvai AS, Clancy SB, Tainer JA. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J Mol Biol. 2001;305:95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- Han S, Tainer JA. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int J Med Microbiol. 2002;291:523–529. doi: 10.1078/1438-4221-00162. [DOI] [PubMed] [Google Scholar]
- Hillig RC, Renault L, Vetter IR, Drell T.t., Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Merrill AR, Yates SP, Marquez VE, Schwan AL, Boesen T, Andersen GR. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- Karasawa T, Takasawa S, Yamakawa K, Yonekura H, Okamoto H, Nakamura S. NAD(+)-glycohydrolase from Streptococcus pyogenes shows cyclic ADP-ribose forming activity. FEMS Microbiol Lett. 1995;130:201–204. doi: 10.1111/j.1574-6968.1995.tb07720.x. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Fujii Y, Hirano S, Yokota Y, Taketo A. Genetic and biochemical properties of streptococcal NAD-glycohydrolase inhibitor. J Biol Chem. 2006;281:9181–9189. doi: 10.1074/jbc.M506879200. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Fujii Y, Yokota Y, Taketo A. Molecular characterization of NACasestreptolysin O operon of hemolytic streptococci. Biochim Biophys Acta. 2005;1681:134–149. doi: 10.1016/j.bbaexp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- Meehl MA, Caparon MG. Specificity of streptolysin O in cytolysin-mediated translocation. Mol Microbiol. 2004;52:1665–1676. doi: 10.1111/j.1365-2958.2004.04082.x. [DOI] [PubMed] [Google Scholar]
- Meehl MA, Pinkner JS, Anderson PJ, Hultgren SJ, Caparon MG. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog. 2005;1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS. Structural requirements for function of yeast GGAs in vacuolar protein sorting, alpha-factor maturation, and interactions with clathrin. Mol Cell Biol. 2001;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J, Golonka E, Filipek R, Shaw LN. Fighting an enemy within: cytoplasmic inhibitors of bacterial cysteine proteases. Mol Microbiol. 2005;57:605–610. doi: 10.1111/j.1365-2958.2005.04714.x. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein Server. Nucleic Acids Research 32 (Web Server issue):W321-W326. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant F, Bru C, Carrere S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, Wakatsuki S. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature. 2002;415:937–941. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- Stevens DL, Salmi DB, McIndoo ER, Bryant AE. Molecular epidemiology of nga and NAD glycohydrolase/ADP-ribosyltransferase activity among Streptococcus pyogenes causing streptococcal toxic shock syndrome. J Infect Dis. 2000;182:1117–1128. doi: 10.1086/315850. [DOI] [PubMed] [Google Scholar]
- Tatsuno I, Sawai J, Okamoto A, Matsumoto M, Minami M, Isaka M, Ohta M, Hasegawa T. Characterization of the NAD-glycohydrolase in streptococcal strains. Microbiology (Reading, Engl) 2007;153:4253–4260. doi: 10.1099/mic.0.2007/009555-0. [DOI] [PubMed] [Google Scholar]
- Tsuge H, Nagahama M, Oda M, Iwamoto S, Utsunomiya H, Marquez VE, Katunuma N, Nishizawa M, Sakurai J. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc Natl Acad Sci U S A. 2008;105:7399–7404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, He X, Zhai P, Terzyan S, Tang J, Zhang XC. Crystal structure of GGA2 VHS domain and its implication in plasticity in the ligand binding pocket. FEBS Lett. 2003;537:171–176. doi: 10.1016/s0014-5793(03)00095-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.