Abstract
We developed a general approach that combines Chromosome Conformation Capture Carbon Copy with the Integrated Modeling Platform to generate high-resolution three-dimensional models of chromatin at the Mb scale. We applied this approach to the ENm008 domain on human chromosome 16 containing the α-globin locus, which is expressed in K562 cells and silenced in lymphoblastoid cells (GM12878). The models accurately reproduce the known looping interactions between the α-globin genes and their distal regulatory elements. Further, we find that the domain folds into a single globular conformation in GM12878 cells, whereas two globules are formed in K562 cells. The central cores of these globules are enriched for transcribed genes, whereas non-transcribed chromatin is more peripheral. We propose that globule formation represents a higher-order folding state related to clustering of transcribed genes around shared transcription machineries, as observed by microscopy.
Currently, efforts are directed at producing high-resolution genome annotations where the positions of functional elements or specific chromatin states are mapped onto the linear genome sequence1. However, these linear representations do not indicate functional or structural relationships between distant elements. For instance, recent insights suggest that widely spaced functional elements cooperate to regulate gene expression by engaging in long-range chromatin looping interactions. The three-dimensional (3D) organization of chromosomes is thought to facilitate compartmentalization2,3, chromatin organization4, and spatial sequestration of genes and their regulatory elements5–7, all of which may modulate the output and functional state of the genome. A general approach to determine the spatial organization of chromatin can aid in the identification of long-range relationships between genes and distant regulatory elements as well as in the identification of higher-order folding principles of chromatin in general.
Chromosome conformation capture (3C)-based assays use formaldehyde cross-linking followed by restriction digestion and intra-molecular ligation to study chromatin looping interactions7–12. 3C-based assays have been used to show that specific elements such as promoters, enhancers and insulators are involved in the formation of chromatin loops5,7,13–16. The frequencies by which loci interact reflect chromatin folding7,17 and thus comprehensive chromatin interaction datasets can help building spatial models of chromatin. Previously, chromatin conformation has been modeled using polymer models8,18 and molecular dynamics simulations19, which have proven valuable for understanding general features of chromatin fibers including flexibility and compaction20,21. However, such methods only partially leverage the current wealth of experimental data on chromatin folding. Recently, experiment-driven approaches, in combination with computational modeling, have resulted in low-resolution models for the topological conformation of the immunoglobulin heavy-chain22, the HoxA23 loci and the yeast genome24. However, those methods were limited by the resolution and completeness of the input experimental data22, by insufficient model representation, scoring and optimization23 or limited analysis of the 3D models24. To overcome such limitations, we developed a new approach that couples high-throughput 3C-carbon copy (5C) experiments9 with the Integrative Modeling Platform (IMP)25. We applied this approach to determine the higher-order spatial organization of a 500 Kb gene dense domain located near the left telomere of human chromosome 16 (Fig. 1a). Embedded in this cluster of ubiquitously expressed house keeping genes is the tissue-specific α-globin locus that is only expressed in erythroid cells. This 500 Kb domain corresponds to the ENm008 region extensively studied by the ENCODE pilot project1.
Figure 1.
ENCODE region ENm008 on human chromosome 16. (a) Map of ENm008 including the ζ, μ, α2, α1, and θ globin genes. Genes are indicated by grey lines above the linear representation. Vertical black lines indicate HindIII restriction sites. Colored restriction fragments contain annotated genes. Red, orange and green circles localize the HS40, other α-globin related HS sites and CTCF sites, respectively. (b) ENCODE annotations for the ENm008 region. RNA expression data, CTCF data, Histone modification data (H3K4me3) and DNAse I sensitivity data 56,57 are generated by the ENCODE project (http://genome.ucsc.edu/ENCODE/).
The α-globin locus has been widely used as a model to study the mechanism of long-range and tissue-specific gene regulation15,26–30. The α-globin genes are up regulated by a set of functional elements, characterized by the presence of DNAse I hypersensitive sites (HSs) located 33 to 48 Kb upstream of the ζ gene. One of these elements, HS40, is considered to be of particular importance31,32. This element can act as an enhancer in reporter constructs and its deletion severely impacts activation of the α-globin genes33. HS40 is bound by several erythroid transcription factors including GATA factors and NF-E234. Importantly, previous 3C studies have demonstrated direct long-range looping interactions between some of these distant functional elements (i.e., HS48, HS46 and HS40) and the α-globin genes upon gene activation in mouse and human erythroid cells15,30. Major unanswered questions revolve around the higher-order folding of multi-gene domains like ENm008, and how long-range interactions involved in regulation of each of the resident genes are accommodated.
We have obtained comprehensive interaction maps of the α-globin locus by performing 5C analysis of the ENm008 region in GM12878 and K562 cells. These two cell lines, which differ in the expression of the α-globin genes, are studied by the ENCODE consortium and therefore extensive chromatin structural and functional information for ENm008 is publicly available. We developed a general approach to generate 3D chromatin models based on chromatin interaction data. Our models of the ENm008 domain in GM12878 cells show that it forms a single compact structure, which we refer to as a chromatin globule. We find that active genes and promoters tend to be located at the center of the globule, whereas inactive genes are more peripherally positioned. Interestingly, in cells that express high levels of α-globin (K562) the chromatin is broken into two globules separated by an extended chromatin segment. We propose that sets of neighboring active genes cluster to form chromatin globules, perhaps analogous to transcription factories, and that a given globule can accommodate only a limited number of active genes.
RESULTS
Our approach to determine the 3D conformation of genomic domains consists of four steps (Supplementary Fig. 1): (i) data collection by 5C experiments, (ii) data translation into points and spatial restraints between them, (iii) model building by optimization of the imposed restraints, and (iv) ensemble analysis of the optimal 3D solutions. The following sections describe the results of each of these key steps in our approach to 3D structure determination of the ENm008 region. A summary and further details of the methods are provided in the Online Methods and Supplementary Methods, respectively.
5C analysis of ENm008
5C, described in detail before9,35, employs highly multiplexed ligation-mediated amplification to detect sets of 3C ligation products. 5C primers were designed at HindIII sites using computational algorithms through our online My5C software package (http://my5C.umassmed.edu)36. In total, 30 forward primers and 25 reverse primers were designed throughout the 500 Kb ENm008 region with the capability of detecting 750 unique pair-wise chromatin interactions (Supplementary Table 1). The quantitative number of 5C ligation products, which corresponded to pairs of interacting fragments, was determined by paired-end Solexa sequencing. Consistent with previous analyses9,37, the 5C interaction maps display prominent signals between sites located near each other. Further, GM12878 displays more abundant long-range interactions suggesting a more compact conformation compared to K562 (Fig. 2).
Figure 2.
5C analysis of the 500 Kb ENCODE region ENm008. (a) 5C experimental data for GM12878 cell lines. Upper plot shows 5C count matrix colored yellow to blue to indicate low to high counts. For an easy inspection, the axis labels are substituted by the linear representation of the forward and reverse fragments of the ENm008 region. Lower plots show 5C interaction profiles for fragments containing HS48, HS46, HS40, HBM, HBA2, HBA1, and 3’ end of LUC7L, respectively. The plots show the 5C counts and their associated standard error of interactions between the anchor fragment (indicated by vertical arrows) and the rest of queried fragments in the ENm008 region (colored bars indicate the positions of HS elements (red), globin genes (green) and LUC7L gene (blue)). Blue solid lines show the average and standard error expected relationship between interaction frequency (5C counts) and genomic distance (Kb) determined by LOESS smoothing of the complete dataset (Supplementary Fig. 1). Red circles show the observed 5C counts for each of the queried fragments. (b) 5C experimental data for K562 cell lines. Data are represented as in panel a.
We determined the average relationship between genomic distance (in Kb) and interaction probability (average read count) using the entire 5C data set (blue lines in 5C interaction profiles of Fig. 2). This is important because this relationship can be used as an estimate for the expected random collision frequency for pairs of loci in the absence of specific looping interactions37 (Supplementary Fig. 2). In K562 cells we detected all previously known long-range looping interactions between the active α-globin genes and the upstream distant regulatory elements (i.e., HS48, HS46, and HS40), which interacted up to 6-fold more frequently than the estimated expected frequency (Fig. 2b). Such frequent interactions were not present in GM12878 cells with a repressed α-globin domain (Fig. 2a). Therefore, K562 can serve as a model cell line to study the conformation of the active α-globin locus, despite the fact that i) these cells are transformed and can be variable in karyotype and gene expression profile, and ii) primary erythroid cells could have a different conformation of this region.
Interestingly, novel long-range interactions were identified. For example, in both cell types, HS46 interacted very frequently with a locus located just downstream of the α-globin genes (3’ end of LUC7L, which encodes a RNA-binding protein similar to the yeast Luc7p). This downstream locus in turn interacted more frequently than expected with a region located within the more distant Axin1 gene. The nature of the elements involved in these interactions is currently unknown, although it is noteworthy that all these interacting fragments contain sites bound by the CTCF protein (Fig. 1) often involved in long-range interactions13.
From 5C data to points and restraints
Chromatin interaction frequencies can be used as a proxy for spatial distance between interacting fragments12. Thus, our first step was to translate the 5C experimental data into a set of distances dependent on the observed interactions. IMP represents a genomic domain as a set of points (one per restriction fragment) and the spatial restrains (or springs) between them whose distances are proportional to the observed frequency of interaction. The type and force of the restraints that place each of the 70 points representing the ENm008 region were defined by the “IMP calibration”, which was carried out in two steps. First, 5C counts were normalized by log10 transformation and Z-score computation based on the average and standard deviation of all log10 values in the interaction matrix. A Z-score indicates how many standard deviations a measure is above or below the mean of the measure. Second, two linear relationships relating 5C Z-scores to spatial distances for restraining pairs of fragments were defined: (i) two neighbor fragments (i.e., i to i+1‥2) were restrained based on the linear relationship between the 5C Z-scores and the sum of the excluded volume occupied by the nucleotides between the centers of the two fragments (Supplementary Table 1), and (ii) two non-neighbor fragments (i.e., i to i+3‥n) were restrained based on the relationship bound by a empirically determined closest possible distance between two non-interacting fragments and the excluded volume of a canonical 30 nm fiber (Supplementary Fig. 3). These two linear relationships between 5C Z-score and spatial distances rely on the following assumptions: (i) the different 5C Z-scores distribution between neighbor and non-neighbor fragments reflected their different response in 5C experiments37; (ii) consecutive fragments were spatially restrained proportionally to the occupancy of their chromatin fragments with a relationship of 0.01 nm per base pair, assuming a canonical 30 nm fiber38; and (iii) two non-neighbor fragments could not get closer in space than 30 nm, which corresponds to the diameter of the chromatin fiber. Even though the precise diameter of the chromatin fiber in vivo is unknown and likely fluctuates, it has been shown that the observed looping frequencies by 5C experiments in human cells are consistent with a 30 nm fiber39. Moreover, the assumption that chromatin adopts a 30nm fiber only affects the final scale of the resulting 3D models, which is controlled by the excluded volume assigned to the fragments. Based on the results from our FISH experiments (below), the use of 0.01 nm per base pair resulted in models of the appropriate scale. Finally, the values of two Z-scores cut-offs were also optimized and defined the type of restraint imposed between two non-neighbor fragments. The optimal parameters found were: 500 nm for the lowest Z-score, a Z-score of −0.2 for the lower-bound cut-off, and a Z-score of 0.1 for the upper-bound cut-off for GM12878 cells; 400 nm for the lowest Z-score, a Z-score of −0.1 for the lower-bound cut-off, and a Z-score of 0.9 for the upper-bound cut-off for K562 cells (Supplementary Methods).
All 70 fragments representing the studied region were restrained with a total of 1,520 and 1,049 restraints for GM12878 and K562 cells, respectively (Supplementary Fig. 3). The forces applied to the defined restraints were also set proportional to the absolute value of the 5C Z-score observed between a pair of fragments. That is, the more extreme the Z-score the stronger the force constant applied to the restraint. By making the harmonic forces proportional to the variability of the Z-score we ensured that restraints between pairs of points with extreme Z-score values were stronger than those between pairs of points with average frequencies. An exception to this rule was applied to neighbor fragments. In such cases, the forces were set to a value of 5.0, which was large enough to maintain connectivity between neighbor fragments.
Generation of spatial models of ENm008
Once the restraints have been defined, IMP generated a 3D model of the ENm008 region by searching for a spatial arrangement of all points that minimizes the violation of the imposed restraints (Supplementary Fig. 3cd). Thus, IMP expressed the problem of determining the chromatin structure as an optimization problem, assuming that the conformation of the locus is largely determined by chromatin interactions within the locus. The absence of strong interactions outside the locus comparable in frequency to the ones we observe within the locus was recently confirmed by Hi-C, a method that couples proximity-based ligation with massively parallel sequencing to probe the three-dimensional architecture of whole genomes12.
Starting from a random position of all points within a cube of side length of 1 µm, IMP iteratively moved all points so as to force them to a conformation that minimally violated the imposed restraints. Given the population-averaged nature of the 5C analyses, the 3D models generated by IMP can only represent the macroscopic state of the system and thus result in an ensemble of solutions reflecting the variable nature of chromatin conformation40. It is important to note that the 3D positions obtained by IMP correspond to points representing the center of the ligation positions designed as part of the 5C experiments. The path between points shown in our 3D models does not necessarily correspond to the path that chromatin may follow in vivo.
A total of 50,000 models were generated for each cell type, which ensured a fair coverage of the searching space. We then selected 10,000 models with the least number of violated restraints to be clustered based on their structural similarity (Supplementary Methods). GM12878 models clustered in a total of 4 different conformations (Fig. 3a). The first and second most populated clusters contained the conformations with the lowest IMP objective function, indicating that a minimum in the search space was found for most of the independent runs. This shows that: (i) 5C data are sufficient for uniquely identifying a set of dominant conformations; and (ii) the top two clusters represent topological mirror solutions, providing further confidence in the results.
Figure 3.
Ensemble of solutions. (a) Cluster analysis for the GM12878 selected 10,000 models. Upper plot shows the number of models per cluster plotted against the cluster number. Points are colored proportional to the lowest IMP objective function in the cluster. IMP mirroring is illustrated by the superimposition of the centroids (i.e., the solution closest to the center of the cluster) of clusters one (red) and two (blue). Lower plot shows the structural relationship between the top cluster centroids. The tree was generated based on the structural similarity between each of the centroids. The branch thickness is proportional to the number of solutions at each branch point. Each centroid, colored as in its linear representation (Fig. 1a), is vertically placed proportional to the lowest IMP objective function within the cluster their represent. (b) Cluster analysis for the K562 selected 10,000 models. Data are represented as in panel a. (c) Model consistency for the ensemble of solutions in cluster 1 of GM12878 models (blue) and cluster 2 of K562 models (red).
Models obtained for K562 cells form a more variable set of solutions with a total of 393 different structure clusters including ten large clusters with more than 150 solutions each and 194 clusters with less than 10 solutions each (Fig. 3b). This result suggests that the large number of clusters with few members, represent a diverse set of local minima conformations that partly satisfy the K562 5C interaction data. Such diverse solutions could reflect a higher variability of chromatin conformation of the domain in K562 cells, perhaps related to variable karyotypes and gene expression in individual cells in this cancer-derived cell line. It is important to note that even though we selected representative clusters to describe key properties of the α-globin locus structure (below), only the ensemble of all solutions from the top clusters reflected the range of multiple distinct conformations that may be present in the cell population.
We studied whether the different conformations we observed between individual models within a cluster of solutions could be considered locally consistent (Fig. 3c). Such analysis allowed us to identify local regions in the structures that were conserved for most of the pair-wise structure alignments between the models in the selected cluster. Clearly, GM12878 models were locally consistent and only one fragment (reverse 21) of the models did not have a consistent local conformation (i.e., not superimposable within 150 nm for more than 75% of the models). In K562 cells as many as 82% of the fragments were consistent across the models. This analysis shows that even in the more variable K562 models most of the region contains conserved local features, and that the diversity is the result of variable positioning of only a small minority of fragments (18%).
3D models reproduce known long-range interactions
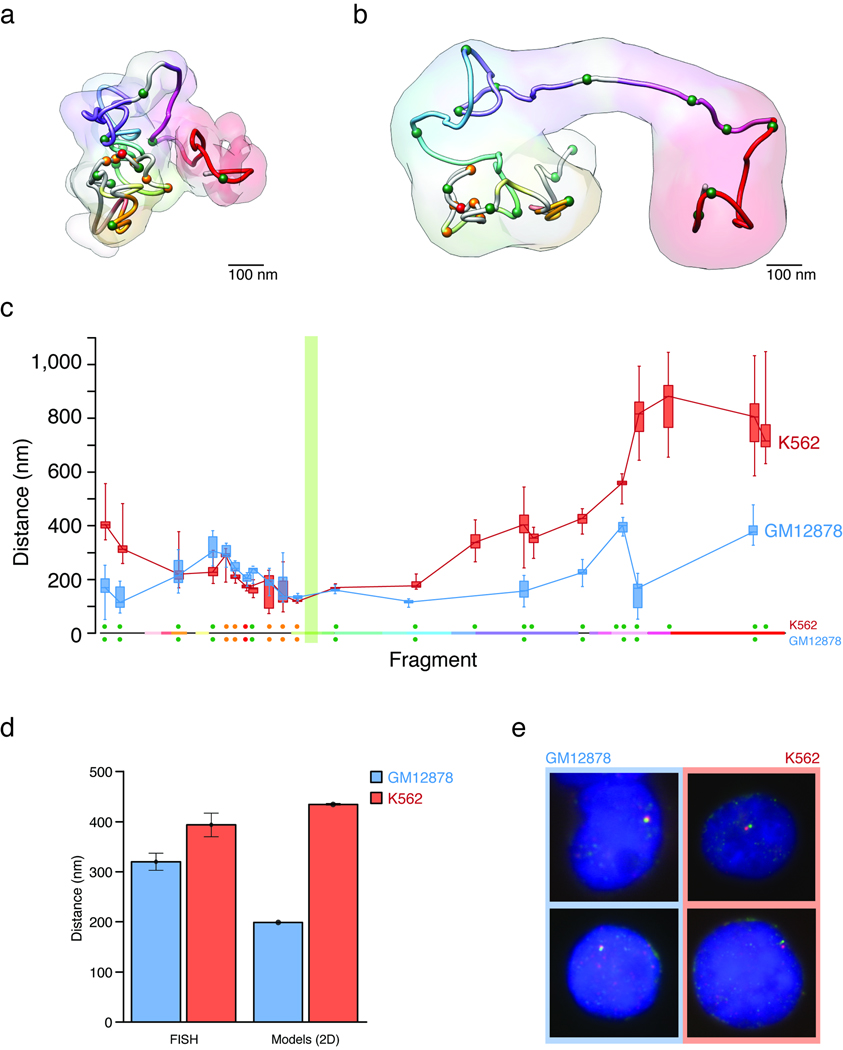
We determined whether the 3D models reflected the known long-range interactions involving the α-globin genes (Fig. 4). We used the selected cluster of models to calculate the average distance between the restriction fragment containing the α-globin genes and other restriction fragments in ENm008 in both GM12878 and K562 cells. Restriction fragments containing the enhancer (HS40) and α-globin genes were closely juxtaposed in K562 cells (159.1 ± 13.3 nm). Conversely, HS40 was the only fragment that was located farther from the α-globin genes in the inactive GM12878 cells (228.2 ± 17.3 nm) as compared to K562 cells, whereas all other fragments were located closer to the α-globin genes (Fig. 4c). These observations are consistent with previous 3C experiments that have shown that strong interaction between HS40 and the α-globin genes is only observed when the genes are expressed.
Figure 4.
3D models of the ENm008 ENCODE region containing the α-globin locus. (a) 3D structure of the GM12878 models represented by the centroid of cluster number 1. The 3D model is colored as in its linear representation (Fig. 1a). Regulatory elements are represented as spheres colored in red (HS40), orange (other HSs), and green (CTCFs). (b) 3D structure of the K562 models represented by the centroid of cluster number 2. Data are represented as in panel a. (c) Distances between the α-globin genes (restriction fragments 31–32) and other restriction fragments in ENm008. The plot shows the distribution and standard deviation of the mean of distances for GM12878 models in cluster 1 (blue) and K562 models in cluster 2 (red). (d) Average distances and their standard error between a pair of loci located on either end of the ENM008 domain as determined by FISH with two fosmid probes (see Methods) and from a 2D representation of the IMP-generated models in both cell lines. (e) Example images obtained with FISH of GM12878 and K562 cell lines. The images show smaller distances between the probes in GM12878 than in K562 cell lines.
FISH experimental validation
We employed an entirely independent method, Fluorescence In Situ Hybridization (FISH), to validate a particular aspect of our 3D models for the ENm008 region. For small genomic domains, such as the one studied here, determining the spatial positions of individual restriction fragments within this domain by FISH is not straightforward given the resolution of light microscopy, which is limited to ~200 nm. However, the models of the ENm008 domain predict that the locus is in a more extended conformation in K562 cells than in GM12878 cells, which would predict a greater average 2D interphase distance between the ends of the 500 kb locus. Prior work has demonstrated that this is large enough to be measured by interphase mapping with FISH41. We find that in GM12878 these loci are on average 318.8±17.0 nm apart, whereas in K562 cells they are 391.9±23.4 nm apart. These differences, which are statistically significant (p-value <0.011), show that in K562 cells the locus is in a more extended conformation consistently with the models generated by IMP where the 2D distances (that is, without considering the orientation of the model) resulted 198.9±0.7 and 434.6±1.4 nm apart for GM12878 and K562 models, respectively (Fig. 4de).
Formation of chromatin globules
Interestingly, a feature observed in both cell lines is the formation of compact chromatin clusters, which we termed “chromatin globules”. In GM12878 cells the ENm008 region forms a single chromatin globule whereas in K562 cells the locus forms two chromatin globules (Fig. 4ab, and Supplementary Videos 1 and 2). This large-scale difference in conformation between the two cell lines is also illustrated by the contact map differences between GM12878 and K562 models (Fig. 5a). The heat map shows that most distances in GM12878 are smaller than in K562 cells, consistent with formation of a single compact chromatin globule. However, and also consistent with the 5C data, the α-globin genes and the distant regulatory elements are closer in space in K562 cells than in GM12878 cells (red areas in Fig. 5a).
Figure 5.
Analysis of chromatin globules. (a) Frequency contact map differences between models in cluster 1 of GM12878 cells and cluster 2 of K562 cells. Differential expression levels are shown next to the 1D representation of the ENm008 in the axis of the plot. (b) Relative abundance of different ENm008 fragment types to the center of their chromatin globules for GM12878 (upper plot) and K562 (lower plot). Plots show cumulative relative abundance of annotations vs. radial position in the globule. Active genes and promoters are enriched in the center. (c) Observed loops in the centroids of selected cluster for GM12878 (upper) and K562 (lower) models. The loops are placed over the 1D representation of the ENm008 region. Loop height is proportional to the path length of the loop. Loops are colored proportional to the distance between the anchor points (dark = near and light = far). Loop sizes in Kilobases (Kb) are indicated at the tip of the loop. (d) Chromatin density for the ensemble of solutions in cluster 1 of GM12878 models (blue) and cluster 2 of K562 models (red). DNAse I hypersensitive sites are shown next to the 1D representation of the ENm008 in the x-axis of the plot.
To explore whether these globules display some level of internal organization, we determined the locations of genes and putative regulatory elements within the chromatin globules. We measured the radial positions of active genes, gene promoters, HSs, sites bound by CTCF as well as sites marked by H3K4Me3 by calculating the average distance between the corresponding restriction fragments and the geometrical center of the globules. Strikingly, we found that in both cell types active genes and gene promoters are enriched near the center of the globule, whereas inactive genes and restriction fragments that do not contain genes are more peripheral (Fig. 5b). In contrast, HSs and CTCF-bound and sites marked by H3K4Me3, are not preferentially located in the center, but are found throughout the globules.
In GM12878 cells we visually identified 9 loops with an average length of ~50 Kb, ranging from about 20 to 70 Kb, average distance between anchors of 102.8 ± 5.1 nm, and average path length of 547.9 ± 96.9 nm (Fig. 5c). In K562 cells the locus forms two chromatin globules (5 loops and 2 loops, respectively) with an average length of ~60 Kb, ranging from about 30 to 70 Kb, average distance between anchors of 231.2 ± 129.2 nm (190.6 ± 43.5 nm not considering loop number 6 connecting the two globular domains), and average path length of 600.1 ± 90.2 nm. Our experiments, which only covered the ENm008 region, prevented us to determine whether the second chromatin globule observed in K562 cells contained additional genes beyond the LOC100134368, DECR2 and RAB11FIP3 genes. Overall, the models suggest that chromatin is organized around chromatin globules with rosettes of 50–60 Kb chromatin loops and centers enriched with active genes and their promoters.
Estimates of chromatin compaction based on spatial models
Chromatin across the ENm008 region was not uniformly dense as determined by the contour length of the chromatin fiber (Fig. 5d). As expected, the average chromatin path was much denser than for naked DNA, which is about 3 bp nm−1. We found that the telomere proximal end of ENm008, which contains the highest density of active genes as well as most of the regulatory elements (as estimated based on the density of DNAse I hypersensitive sites; Fig. 1b), has a chromatin fiber compaction level that corresponds to ~50 bp nm−1. Conversely, the telomere-distal region displays a denser chromatin region (~100 bp nm−1). Interestingly, GM12878 cell models result on average in a less dense chromatin fiber, despite folding into a single chromatin globule. However, the region containing the HS40 enhancer of α-globin genes is more compact on average in GM12878 cells compared to K562 cells, consistent with predicted relationships between transcription and formation of more open chromatin.
Local chromatin features and three-dimensional folding
The analysis of chromatin compaction illustrates how our models can reveal new insights into spatial relationships between distant 1D annotations and their 3D conformation. To further illustrate this, we have generated tracks for the UCSC Genome Browser42 showing the interaction frequency maps resulting from our 5C experiments and 3D models (Supplementary Fig. 4). These tracks allow direct visualization of spatial relationships between widely spaced genomic elements in the context of all publicly available 1D genome annotations. For instance, we find that the α-globin genes are spatially close to a region containing the genes POLR3K and MPG near the left end of the region. Both interacting regions are transcriptionally active and marked by histone modification associated with open chromatin (e.g., H3K4Me2, H3K4Me2, H3Ac, and H4Ac). This is consistent with our observation that active genes tend to form the cores of the chromatin globules, which has been identified here as well as in previous work showing association between active genes10,15,43.
DISCUSSION
Here, we have combined high-throughput in vivo chromatin interaction mapping with the Integrative Modeling Platform to characterize the higher-order chromatin conformation of the ENm008 region containing the α-globin domain in cells that do or do not express the globin locus. The 5C data and the 3D models derived from them accurately reflect the known long-range interactions between the α-globin genes and their distant regulatory elements, thereby validating our approach. Furthermore, we identify a higher-order chromatin folding motif in which groups of adjacent genes cluster to form “chromatin globules”. Analysis of the internal architecture of these globules revealed that active genes are enriched in the cores of these structures. These observations suggest that chromatin globules may represent sub-nuclear structures dedicated to gene expression, perhaps related to the clustering of shared transcription machineries.
Formation of chromatin globules
Chromatin globules result in a rosette-like structure with loops of ~50–60 Kb, an average path length of 500–600 nm and a distance between anchors of 100–200 nm. Such spatial organization would agree with the Multi-Loop-Subcompartment (MLS) model, which proposes that chromatin is folded into rosettes of small loops, connected by linkers of variable size22,44. Importantly, FISH experiments have also revealed that chromatin domains can form strings of globular domains of around the same size (in Kb) as the globules identified here45. The type and function of proteins involved in maintaining these chromatin globules are unknown.
It has been proposed that active genes interact at discrete sites (also called transcription factories) where several RNA polymerases are concentrated46. It is still unresolved whether such transcription machineries are a consequence or cause of transcription of gene clusters in the nucleus47. However, it has been observed by electron microscopy that these sites of transcription can range from 45 to 100 nm in diameter46,48–51 and include a limited number of active RNA polymerases (~8) as estimated by the number of nascent RNA molecules46. Our models agree with these estimates. The first chromatin globule in our K562 models, which includes the α-globin genes as well as other near-by house-keeping genes, wraps around a cavity with an average diameter of ~100–110 nm, which would fit a hypothetical transcription factory. Of particular interest is our observation that the ENm008 region forms a single large chromatin globule in GM12878, but two smaller globules in K562 cells. The major difference between these two cell lines is the expression the α-globin gene cluster, which is actively transcribed in K562 cells. In GM12878 cells there are 6 to 7 genes being actively transcribed, which would all fit within a single transcription factory. However, in K562 cells, the activity of the α-globin genes appears to exceed the capacity of a single transcription factory having about 10 genes actively transcribed. We entertain the idea that the number of active genes that can cluster to form a chromatin globule may be limited to only around 8 genes, which would agree with the elongated-beaded structures observed by light microscopy of active chromatin regions45,52. Clearly, this is a highly speculative idea, and it is also possible that the extended conformation in K562 cells is related to the transformed state of this cell line. Further experiments are needed to shed light on the determinants of globule formation.
From our models, we cannot say whether these chromatin globules would self-assemble around genes sharing common transcription machineries, actively assemble on demand or already exist as a complex fixed to a yet unknown underlying nuclear substructure. It has been proposed that transcriptionally active regions may attain increased chromatin mobility6. It is interesting that the K562 models have higher variability and lower consistency compared to models from GM12878 cells, which could correspond to the fact that the region is broken in two globules or to the fact that the region is overall more transcriptionally active.
Chromatin density
Even for transcriptionally active regions, chromatin is about 400–1,000 fold more compact than the “30-nm” fiber53. Therefore, de-condensation of chromatin may be transient54. We have observed for both cell lines that transcriptionally inactive regions are, on average, about twice as dense as regions containing either transcribed genes or their regulatory elements. Interestingly, the region including HS40, HS46 and HS48 was on average denser in GM12878 than in K562 cells, while the remaining studied regions were on average denser in K562 than in GM12878 cells. Our results indicate that chromatin undergoes a certain level of de-condensation when genes are expressed55. Thus, 5C experiments, reflected by our models, are able to capture such subtle differences.
A “chromatin globule” model
Our 3D structures suggest a model for higher-order chromatin folding based on the formation of “chromatin globules” (Fig. 6). Chromatin globules would be spatially separated and would form by clustering of a limited number of actively transcribed genes. Within the context of the chromatin globules, our analysis identified specific long-range interactions between genes and their regulatory elements, as well as novel interactions between sites bound by CTCF. The potential roles of such regulatory elements in globule formation are currently unknown.
Figure 6.
Diagram of the proposed chromatin globule model for higher-order chromatin folding of actively transcribed genomic regions.
The identification of chromatin globules indicates how our models can point to the presence of novel higher-order features of chromosome architecture. Our 3D models for the Enm008 region are in agreement with: i) our own FISH experiments validating their overall size and shape; ii) previously described biological phenomena such as the clustering of active genes; and iii) local chromatin structural features from the ENCODE consortium such as DNAse I sensitivity. Our approach has the potential to further leverage large-scale efforts for annotating genes and their regulatory elements along the linear genome by revealing their relative spatial arrangements.
METHODS
Methods and associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank the IMP community (http://www.integrativemodeling.org/) specially Daniel Russell, Ben Webb and Andrej Sali as well as the Chimera developers (http://www.cgl.ucsf.edu/chimera/) specially Thomas Goddard and Tom Ferrin. We also thank Mark Umbarger, Matthew Wright, George Church, M.S. Madhusudhan, Marian Walhout, and Dekker lab members for fruitful discussions. MAM-R acknowledges support from the Spanish Ministerio de Ciencia e Innovación (BIO2007/66670; BFU2010/19310). JD acknowledges support from NIH (HG003143) and the Keck Foundation. Finally, we are grateful to the ENCODE project (funded by the National Institutes of Health and the National Human Genome Research Institute) for providing annotations of the ENm008 region. In particular, we thank the ENCODE groups led by Tom Gingeras (expression data, Cold Spring Harbor), Greg Crawford (DNAse I data, Duke University) and Bradley Bernstein (CTCF data, H3K4Me3 data, Broad Institute of Harvard and MIT). ENCODE data are publicly available through the ENCODE Data Coordination Center at the University of California, Santa Cruz (http://genome.ucsc.edu/ENCODE/).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
B.R.L performed the bioinformatics design and analysis of the 5C experiments. A.S. performed the 5C experiments. D.B., E.C. and M.A.M-R. carried out the IMP computational modeling. M.B. and J.B.L. performed the FISH experiments. D.B., B.R.L, A.S., J.D. and M.A.M-R wrote the manuscript. J.D. and M.A.M-R conceived the work.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 3.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 5.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 6.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 7.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 12.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhou GL, et al. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 17.Ohlsson R, Gondor A. The 4C technique: the 'Rosetta stone' for genome biology in 3D? Curr Opin Cell Biol. 2007;19:321–325. doi: 10.1016/j.ceb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Mateos-Langerak J, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci U S A. 2009;106:3812–3817. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wedemann G, Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–34540. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: Balancing stability and plasticity. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Jhunjhunwala S, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser J, et al. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10:R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 26.Hughes JR, et al. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci U S A. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs DR, Vernimmen D, Hughes J, Gibbons R. Using genomics to study how chromatin influences gene expression. Annu Rev Genomics Hum Genet. 2007;8:299–325. doi: 10.1146/annurev.genom.8.080706.092323. [DOI] [PubMed] [Google Scholar]
- 28.Higgs DR, Wood WG. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr Opin Hematol. 2008;15:176–183. doi: 10.1097/MOH.0b013e3282f734c4. [DOI] [PubMed] [Google Scholar]
- 29.Lower KM, et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci U S A. 2009;106:21771–21776. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs DR, et al. A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Lowrey CH, Stamatoyannopoulos G. Analysis of enhancer function of the HS-40 core sequence of the human alpha-globin cluster. Nucleic Acids Res. 1997;25:2917–2922. doi: 10.1093/nar/25.14.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernet A, et al. Targeted inactivation of the major positive regulatory element (HS-40) of the human alpha-globin gene locus. Blood. 1995;86:1202–1211. [PubMed] [Google Scholar]
- 34.De Gobbi M, et al. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007;110:4503–4510. doi: 10.1182/blood-2007-06-097964. [DOI] [PubMed] [Google Scholar]
- 35.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 36.Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekker J. The three 'C' s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 38.Gerchman SE, Ramakrishnan V. Chromatin higher-order structure studied by neutron scattering and scanning transmission electron microscopy. Proc Natl Acad Sci U S A. 1987;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa A, Becker NB, Everaers R. Looping probabilities in model interphase chromosomes. Biophys J. 2010;98:2410–2419. doi: 10.1016/j.bpj.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voss TC, Hager GL. Visualizing chromatin dynamics in intact cells. Biochim Biophys Acta. 2008;1783:2044–2051. doi: 10.1016/j.bbamcr.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence JB, Singer RH, McNeil JA. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science. 1990;249:928–932. doi: 10.1126/science.2203143. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn RM, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munkel C, et al. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 45.Muller WG, et al. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol Cell Biol. 2004;24:9359–9370. doi: 10.1128/MCB.24.21.9359-9370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin S, Pombo A. Transcription factories: quantitative studies of nanostructures in the mammalian nucleus. Chromosome Res. 2003;11:461–470. doi: 10.1023/a:1024926710797. [DOI] [PubMed] [Google Scholar]
- 47.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 48.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription "factories' in human nuclei. J Cell Sci. 1996;109(Pt 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 49.Pombo A, et al. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. Embo J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eskiw CH, Rapp A, Carter DR, Cook PR. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J Cell Sci. 2008;121:1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 51.Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem Soc Trans. 2008;36:585–589. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- 52.Goetze S, et al. The 3D Structure of Human Interphase Chromosomes is Related to the Transcriptome Map. Mol Cell Biol. 2007 doi: 10.1128/MCB.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gheldof N, Tabuchi TM, Dekker J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc Natl Acad Sci U S A. 2006;103:12463–12468. doi: 10.1073/pnas.0605343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crawford GE, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7 Suppl 1:S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tam R, Shopland LS, Johnson CV, McNeil J, Lawrence JB. Applications of RNA FISH for visualizing gene expression and nuclear architecture. In: Beatty BG, Mai S, Squire J, editors. FISH: A Practical Approach. New York: Oxford University Press; 2002. pp. 93–118. [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.