Abstract
Purpose
The vitreous gel is a highly hydrated extracellular matrix containing many proteins. These proteins are likely accumulated in the vitreous by local secretion, filtration from the blood, or diffusion from the surrounding tissues and vasculature, and may be altered in disease state. In the last several years, several reports of large-scale profiling of vitreous proteins have been published; however, there is little information on the characterization of the phosphoproteome of vitreous. Here, we sought to identify phosphopeptides and their phosphorylation sites from vitreous.
Experimental design
We used titanium dioxide (TiO2) to enrich phosphopeptides from vitreous and identified them by liquid chromatography coupled tandem mass spectrometry (LC-MS/MS). Results: We identified 85 unique phosphopeptides and the phosphorylation sites from 44 proteins.
Conclusions and clinical relevance
We present a method for characterization of phosphoproteome from vitreous samples using current mass spectrometry technologies and yielded an initial assessment of the phosphoprotein/peptide content of human vitreous, thus providing important biological information toward further understanding of the post-translational modifications of vitreous proteins and their functional significance in disease.
Keywords: Titanium dioxide, phosphopeptide, vitreous, crystallin, LTQ-Orbitrap, mass spectrometry
1. Introduction
The vitreous gel is a highly hydrated extracellular matrix whose structure is maintained by a network of collagen fibrils and filled with glycosaminoglycan hyaluronan. It has been demonstrated that the vitreous has certain physiological functions, such as providing a reservoir of nutrients for the metabolic requirements of the lens, excluding cells and large macromolecules from the vitreous cavity in order to maintain transparency [1], coordinating eye growth [2, 3], and protecting the eye during mechanical trauma [4, 5]. The proteins in the vitreous are likely accumulated there by local secretion, filtration from the blood, or diffusion from the surrounding tissues and vasculature [6, 7]. Recently, several lines of evidence revealed that the proteins retained in the closed vitreous chamber may be altered in disease state: (1) significant changes in human vitreous humour were observed following cataract extraction [8]; (2) higher abundance of pigment epithelium-derived factor, a potent inhibitor of angiogenesis, was detected in vitreous with diabetes [9]; (3) elevated levels of vascular endothelial growth factor (VEGF) was found in vitreous from patients with diabetic retinopathy and retinal-vein occlusion [10]; (4) increased amount of extracellular carbonic anhydrase-I was characterized in vitreous from individuals with diabetic retinopathy [11]; (5) levels of phosphorylated VEGF receptor in the vitreous of eyes with wet age-related macular degeneration was noted to correlate with response to anti-VEGF therapy [12].
Despite the sophisticated imaging technology and treatment modalities available to ophthalmologists, many eye diseases cannot be diagnosed until they are at a stage where irreversible, vision-limiting changes have occurred. Ocular diseases are increasing in prevalence. In part this is because many ocular diseases arise as a consequence of the normal aging process, with many retinal disorders caused by vascular changes associated with diabetes as well as sclerotic processes. Thus, there is a tremendous unmet need for developing new diagnostic technologies to identify protein molecules or their modified forms in the vitreous that could be early warning for pathologies or markers of a particular disease stage that cause severe vision loss. In the last several years, proteomics studies by mass spectrometry technique identified more than 500 proteins in vitreous, including multiple post-translationally modified forms of metabolic, antioxidant, angiogenic, and immune modulatory proteins [13–18], however, there is little information on the characterization of the phosphoproteome of vitreous. Post-translational phosphorylation is a common and important mechanism of acute and reversible regulation of protein function in mammalian cells. Dynamic phosphorylation of proteins on serine, threonine and tyrosine residues is recognized as a key mode of regulating cell cycle, cell growth, cell differentiation and metabolism [19, 20]. In this present work, we report an approach to identify a large number of phosphopeptides and their phosphorylation sites using titanium dioxide (TiO2)-based phosphopeptide enrichment followed by liquid chromatography coupled nano-electrospray tandem mass spectrometry (LC-MS/MS) analysis. The result yielded an initial assessment of the phosphoprotein/peptide content of human vitreous, which can serve as a launch-point for further exploration and analysis.
2. Materials and methods
2.1. Materials
Dithiothreitol (DTT), iodoacetamide, urea, ammonium bicarbonate, 2,5-dihydroxybenzoic acid (DHB), trifluoroacetic acid (TFA), and phosphoprotein bovine beta-casein were from Sigma-Aldrich; Coomassie (Bradford) protein assay kit was from Pierce; trypsin was from Promega; Human angiotensin I (AngI) peptide DRVYIHPFHL, and human tyrosine phosphorylated Angiotensin II (AngII-P) DRVpYIHPF were from Calbiochem; phosphopeptide DRFpSVNLDVK was synthesized from Peptide 2.0 Inc.
The clinical vitreous samples were obtained from 11 patients undergoing total vitrectomy under IRB approval (Istituto Superiore di Sanit, Italy) and patient consent (Table 1). Immediately after collection, samples were diluted with Lactated Ringer’s solution (the solution is isotonic with blood and intended for intravenous administration, containing 130 mM sodium ion, 109 mM chloride ion, 28 mM lactate, 4 mM potassium ion, and 1.5 mM calcium ion from sodium chloride, sodium lactate, calcium chloride and potassium chloride), and centrifuged at 5000 g for 7 minutes at 4 °C to eliminate insoluble material. The supernatant was transferred to another tube, and the protein concentration was measured by Bradford Assay (BioRad). The samples were stored at −80 °C until use.
Table 1.
The vitreous samples used for the study.
| Patient ID | Protein concentration (mg/mL) | Eye disease | Age | Gender |
|---|---|---|---|---|
| 1 | 3.70 | hemovitreous, no diabetes | 59 | female |
| 2 | 3.04 | retinal detachment, no diabetes | 74 | male |
| 3 | 1.57 | hemovitreous, no diabetes | 73 | female |
| 4 | 5.23 | retinal detachment, no diabetes | 78 | female |
| 5 | 4.69 | retinal detachment, no diabetes | 70 | female |
| 6 | 12.71 | diabetes with hemovitreous | 69 | female |
| 7 | 3.41 | diabetes with hemovitreous | 69 | female |
| 8 | 1.73 | diabetes with ischemic cardiopathy | 59 | male |
| 9 | 9.29 | diabetes with hemovitreous | 77 | female |
| 10 | 17.67 | diabetes with hemovitreous | 82 | female |
| 11 | 4.97 | diabetes with retinopathy | 59 | male |
2.3. Trypsin digestion and desalting
Proteins from 1 mg vitreous, pooled from several patients, were spiked with the phosphoprotein bovine beta-casein as an internal standard, reduced by 10 mM DTT in the presence of 8 M urea for 30 minutes at 37 °C, and then alkylated by 50 mM iodoacetamide at room temperature. The concentrated urea in the sample was diluted to a final concentration of 2 M, and the proteins were digested by trypsin at 37 °C for 6 hours in a buffer containing ammonium bicarbonate (50 mM, pH 9). The digestion mixture was then acidified by adding glacial acetic acid to a final concentration of 2% and desalted by SepPak C18 column (Waters, catalogue number WAT054955).
2.3. TiO2 enrichment of phosphopeptides
Phosphopeptides were enriched from the mixture of all tryptic peptides using TiO2 column (200 μm × 2 cm) packed in-house [21]. 1 pmol of standard phosphopeptide angiotensin II phosphate was added to the SepPak-cleaned sample. The sample then was mixed with an equal volume of Loading Buffer (200 mg/mL DHB, 5% TFA, 80% acetonitrile), and loaded into the TiO2 column using the Pressure Cell (Brechbühler Inc.) with flow rate of 3 μL per minute. The column was washed by 200 μL of Wash Buffer 1 (40 mg/mL DHB, 2% TFA, 80% acetonitrile) and 2 × 200μL of a second Wash Buffer 2 (2% TFA, 50% acetonitrile) to remove non-phosphopeptides. Phosphopeptides were eluted from the column with the Elution Buffer (5% ammonia solution). Ammonia in the eluate was removed by lyophilization (~3 minutes), and the sample was acidified by adding glacial acetic acid to a final concentration of 2%, and desalted by ZipTip (Millipore, catalogue number ZTC18S960). 100fmol of standard peptide angiotensin I were spiked into the enriched sample as an internal standard.
2.4. Mass spectrometry for phosphopeptide identification
The purified phosphopeptides were analyzed by reversed-phase liquid chromatography coupled nanospray tandem mass spectrometry (LC-MS/MS) using an LTQ-Orbitrap mass spectrometer (Thermo Fisher) [21]. The reversed-phase LC column was slurry-packed in-house with 5 μm, 200 Å pore size C18 resin (Michrom BioResources, CA) in a 100 μm i.d. × 10 cm long piece of fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a laser-pulled tip. After sample injection, the column was washed for 5 minutes with mobile phase A (0.1% formic acid), and peptides were eluted using a linear gradient of 0% mobile phase B (0.1% formic acid, 80% acetonitrile) to 45% mobile phase B in 120 minutes at 200 nL/minute, then to 100% B in an additional 5 minutes. The LTQ-Orbitrap mass spectrometer was operated in a data-dependent mode in which each full MS scan (60,000 resolving power) was followed by eight MS/MS scans where the eight most abundant molecular ions were dynamically selected and fragmented by collision-induced dissociation (CID) using a normalized collision energy of 35%. The “FT master scan preview mode”, “Charge state screening”, “Monoisotopic precursor selection”, and “Charge state rejection” were enabled so that only the 2+ and 3+ ions were selected and fragmented by CID. Tandem mass spectra were searched against the NCBI human protein database using SEQUEST (Bioworks 3.3.1, ThermoFisher) with full tryptic cleavage constraints, static cysteine alkylation by iodoacetamide, and variable phosphorylation of Ser/Thr/Tyr and methionine oxidation. Mass tolerance for precursor ions was 10 ppm and mass tolerance for fragment ions was 0.25 Da. Confident phosphopeptide identifications were determined using stringent filter criteria for database match scoring followed by manual evaluation of the results.
3. Results
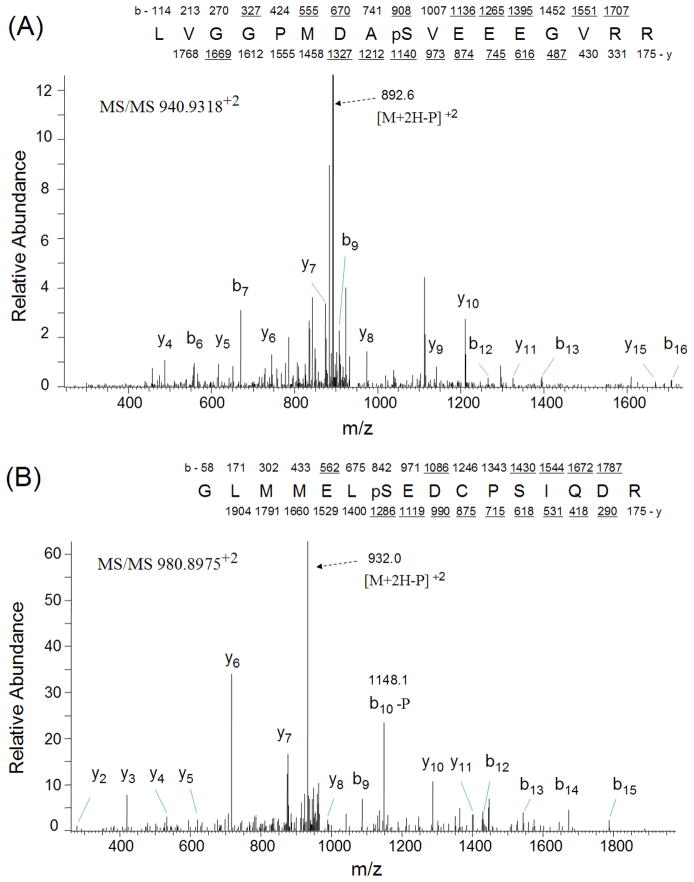
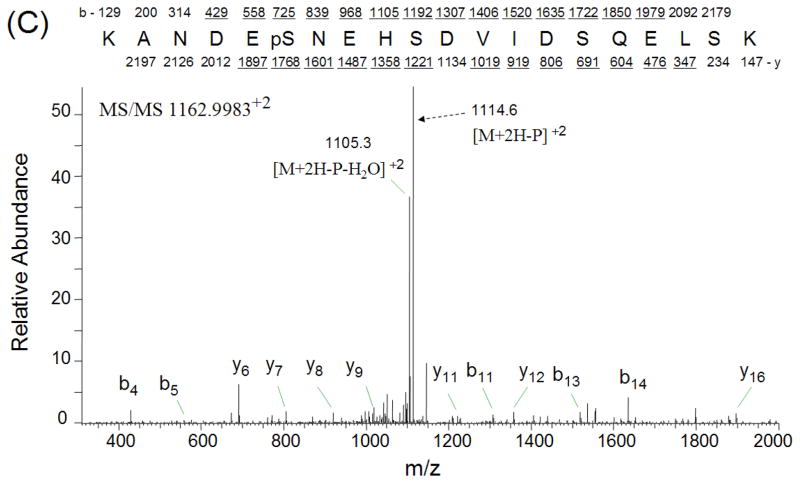
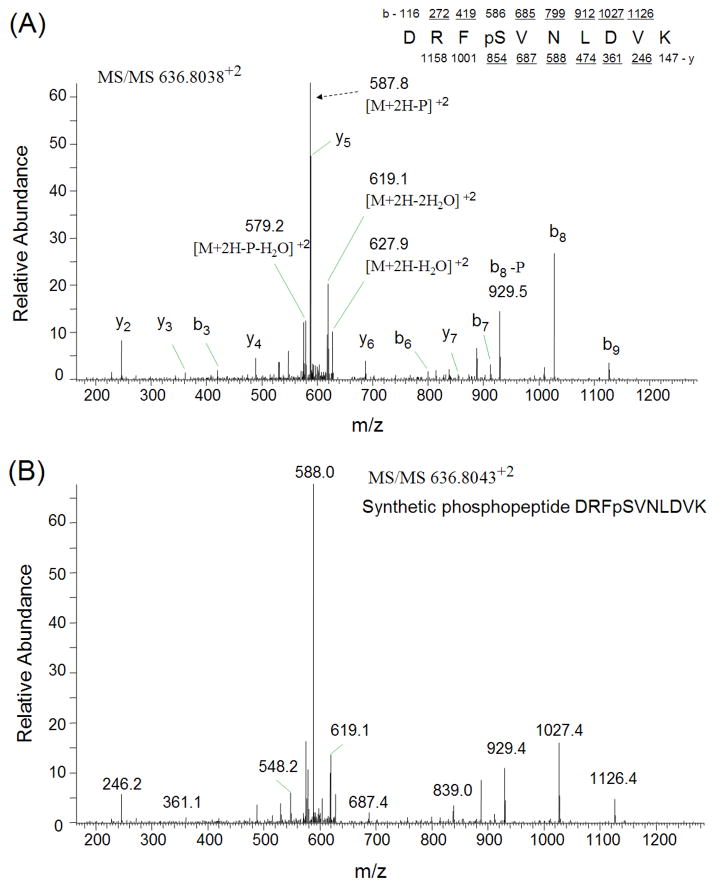
The phosphopeptides from trypsinized vitreous (~ 1 mg) were enriched by TiO2 column and analyzed by LC-MS/MS using LTQ-Orbitrap mass spectrometer. The SEQUEST search results were filtered using the following criteria: “ranked top #1; Xcorr versus charge 1.8, 2.5 for 2+, 3+ ions; mass accuracy 3 ppm; probability of randomized identification of peptide < 0.05”, which yielded 548 matched MS2 spectra. Among these, 253 (46.2%) spectra were matched to non-phosphopeptides, and 295 (53.8%) spectra were matched to phosphopeptides. A total of 85 unique phosphopeptides was identified from 44 proteins (Table 2). The estimated false discovery rate (FDR) was lower than 1% by searching a combined forward-reversed database as described by Elias [22]. Example CID spectra from three of the identified phosphopeptides, LVGGPMDApSVEEEGVRR from cystatin C, GLMMELpSEDCPSIQDR from crystallin, gamma C, and KANDEpSNEHSDVIDSQELSK from secreted phosphoprotein 1 isoform b, are shown in Figures 1. In addition to the matched b-ions and y-ions, the obtained CID spectra exhibited neutral loss of phosphate group from parent ions, which enabled the confident identification of these phosphopeptides. To confirm the validity of the spectral findings, one of these identified phosphopeptides, DRFpSVNLDVK from crystallin, alpha B, was obtained as synthetic peptides, and its MS2 spectrum was acquired using the LTQ-Orbitrap. The MS2 spectrum of this phosphopeptide obtained from TiO2 enriched vitreous was almost identical to the MS2 spectrum of synthesized peptide (Figures 2), indicating accurate identification of this phosphopeptide.
Table 2.
The identified vitreous phosphopeptides by LTQ-Orbitrap from TiO2 enrichment.
| Protein description | Positiona | Phosphopeptides sequence |
|---|---|---|
| crystallin, alpha A | 120–145 | LPpSNVDQSALSCSLSADGMLTFCGPK |
| crystallin, alpha B | 12–22 | RPFFPFHpSPSR |
| 57–69 | APpSWFDTGLSEMR | |
| 73–82 | DRFpSVNLDVK | |
| crystallin, beta A3 | 46–64 | MEFTpSSCPNVSERSFDNVR |
| 46–64 | MEFpTSSCPNVSERSFDNVR | |
| crystallin, beta A4 | 49–71 | VLpSGAWVGFEHAGFQGQQYILER |
| crystallin, beta B1 | 93–110 | SIIVSAGPWVAFEQpSNFR |
| crystallin, gamma C | 100–115 | GLMMELpSEDCPSIQDR |
| albumin precursor | 76–88 | TCVADEpSAENCDK |
| transferrin | 385–399 | IECVpSAETTEDCIAK |
| 684–698 | CSTSSLLEACpTFRRP | |
| alpha 1 globin | 129–140 | FLApSVSTVLTSK |
| 129–140 | FLASVpSTVLTSK | |
| beta globin | 42–60 | FFEpSFGDLSTPDAVMGNPK |
| 42–60 | FFESFGDLSpTPDAVMGNPK | |
| delta globin | 42–60 | FFESFGDLSpSPDAVMGNPK |
| 68–83 | VLGAFpSDGLAHLDNLK | |
| secreted phosphoprotein 1 isoform a | 52–70 | QNLLAPQNAVpSpSEETNDFK |
| secreted phosphoprotein 1 isoform b | 21–35 | QADSGSpSEEKQLYNK |
| 190–206 | AIPVAQDLNAPSDWDpSR | |
| 190–206 | AIPVAQDLNAPpSDWDSR | |
| 190–206 | AIPVAQDLNAPpSDWDpSR | |
| 207–227 | GKDSYETSQLDDQpSAETHSHK | |
| 207–227 | GKDpSYETSQLDDQpSAETHSHK | |
| 235–254 | KANDEpSNEHSDVIDSQELSK | |
| 235–254 | KANDESNEHSDVIDpSQELSK | |
| 235–254 | KANDESNEHpSDVIDpSQELSK | |
| 235–254 | KANDESNEHSDVIDSQELSKVpSR | |
| 235–254 | KANDEpSNEHSDVIDSQELpSKVpSR | |
| 286–300 | FRISHELDSApSSEVN | |
| 286–300 | FRIpSHELDSASSEVN | |
| 286–300 | FRIpSHELDpSASSEVN | |
| 286–300 | FRIpSHELDpSApSSEVN | |
| secreted phosphoprotein 1 isoform c | 31–50 | QNAVpSSEETNDFKQETLPSK |
| 31–50 | QNAVpSpSEETNDFKQETLPSK | |
| chromogranin A precursor | 319–331 | GGKpSGELEQEEER |
| chromogranin B precursor | 134–153 | ADEPQWSLYPSDSQVpSEEVK |
| 179–202 | GEDpSSEEKHLEEPGETQNAFLNER | |
| 255–277 | GQPRpSQEEpSEEGEEDATSEVDK | |
| 259–277 | SQEEpSEEGEEDATSEVDKR | |
| 372–387 | APRPQpSEESWDEEDKR | |
| 397–409 | MAHGYGEEpSEEER | |
| transmembrane protein 132A isoform a | 515–536 | VPGPAEGPAEPAAEApSDEAERR |
| glycophorin A precursor | 120–150 | KSPSDVKPLPSPDTDVPLSSVEIENPETpSDQ |
| inter-alpha globulin inhibitor H2 polypeptide | 55–76 | SLPGEpSEEMMEEVDQVTLYSYK |
| myocilin | 194–216 | AVPPGpSREVSTWNLDTLAFQELK |
| clusterin isoform 1 | 438–460 | VTTVASHTSDpSDVPSGVTEVVVK |
| serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | 62–78 | ATEDEGpSEQKIPEATNR |
| kininogen 1 | 325–343 | ETpTCSKESNEELTESCETK |
| 325–343 | ETTCSKEpSNEELTESCETK | |
| 325–343 | ETTCpSKEpSNEELTESCETK | |
| 416–427 | AGAEPApSEREVS | |
| SPARC-like 1 | 71–89 | SSVLKpSKEEpSHEQSAEQGK |
| 187–220 | DQGNQEQDPNIpSNGEEEEEKEPGEVGTHNDNQER | |
| 227–255 | EHANpSKQEEDNTQSDDILEESDQPTQVSK | |
| 256–286 | MQEDEFDQGNQEQEDNpSNAEMEEENASNVNK | |
| 287–299 | HIQETEWQpSQEGK | |
| 409–426 | KAENSpSNEEETSSEGNMR | |
| alpha-2-HS-glycoprotein | 132–143 | CDSSPDpSAEDVR |
| 318–337 | HTFMGVVSLGSPpSGEVSHPR | |
| apolipoprotein A-II preproprotein | 52–62 | VKpSPELQAEAK |
| apolipoprotein E precursor | 138–152 | GEVQAMLGQpSTEELR |
| apolipoprotein L1 isoform a precursor | 306–320 | VTEPISAEpSGEQVER |
| 306–320 | VTEPIpSAESGEQVER | |
| 306–320 | VTEPIpSAEpSGEQVER | |
| secretory granule, neuroendocrine protein 1 (7B2 protein) | 199–211 | SVPHFpSDEDKDPE |
| fibrinogen, alpha polypeptide isoform alpha-E preproprotein | 354–367 | PGSTGTWNPGpSSER |
| natrium-phosphate cotransporter IIa C-terminal-associated protein 2 | 19–46 | FNPKLGIDNPVLpSLAEDHDPpYDPWpSLER |
| chondroitin sulfate proteoglycan 2 (versican) | 2108–2124 | QEIESETTpSEEQIQEEK |
| ankyrin 1 isoform 1 | 777–787 | LGYIpSVTDVLK |
| 804–828 | MSFPETVDEILDVpSEDEGEELISFK | |
| 1683–1695 | ITHSPpTVSQVTER | |
| 1683–1695 | ITHpSPTVSQVTER | |
| spectrin beta isoform b | 2122–2137 | LSpSSWESLQPEPSHPY |
| 2122–2137 | LSpSpSWESLQPEPSHPY | |
| cystatin C precursor | 35–51 | LVGGPMDApSVEEEGVRR |
| ATP-binding cassette, sub-family B, member | 6744–763 | APGIILLDEATpSALDTSNER |
| frizzled-related protein | 295–313 | HLGLSKpSDSSNSDSTQSQK |
| amyloid precursor-like protein 1 isoform 1 precursor | 348–369 | QALNEHFQSILQTLEEQVpSGER |
| PREDICTED: similar to retinitis pigmentosa 1-like 1 | 169–183 | PDAGSEEPDpSEGGRR |
| tenascin XB isoform 1 | 3678–3702 | LGPLSAEGTTGLAPAGQTSEEpSRPR |
| nuclear ubiquitous casein kinase and cyclin-dependent kinase substrate | 9–28 | KVVDYSQFQEpSDDADEDYGR |
| vitronectin precursor | 311–324 | DpSWEDIFELLFWGR |
| glyceraldehyde-3-phosphate dehydrogenase | 163–186 | VIHDNFGIVEGLMTTVHAITApTQK |
Position of the initial and final peptide amino acids in the protein sequence.
Figure 1.
CID spectra of three phosphopeptides. CID spectra were labeled to show singly-charged b and y ions, as well as ions corresponding to neutral losses of the phosphate group and water. A) phosphopeptide LVGGPMDApSVEEEGVRR from cystatin C; B) phosphopeptide GLMMELpSEDCPSIQDR from crystallin, gamma C; C) phosphopeptide KANDEpSNEHSDVIDSQELSK from secreted phosphoprotein 1 isoform b.
Figure 2.
Verification of identified phosphopeptide DRFpSVNLDVK from crystallin, alpha B by MS2 spectra comparison. A) CID spectrum of the phosphopeptide obtained from TiO2 enriched vitreous phosphopeptides; B) CID spectrum of synthetic phosphopeptide DRFpSVNLDVK.
As shown in Table 2, some of these identified phosphopeptides were from abundant serum proteins such as albumin, apolipoproteins, kininogen 1, serine (or cysteine) proteinase inhibitor, secreted phosphoprotein 1, and alpha-2-HS-glycoprotein. These proteins have also been shown to be phosphorylated in our previous characterization of serum phosphoproteome [21]. In addition, several cellular phosphopeptides such as ITHSPpTVSQVTER from ankyrin 1 isoform 1, LSpSSWESLQPEPSHPY from spectrin beta isoform b, HIQETEWQpSQEGK from SPARC-like 1 protein, were identified as well. Interestingly, 9 phosphopeptides from three types of crystallins—i.e., LPpSNVDQSALSCSLSADGMLTFCGPK from alpha A, RPFFPFHpSPSR, APpSWFDTGLSEMR, and DRFpSVNLDVK from alpha B, MEFTpSSCPNVSERSFDNVR and MEFpTSSCPNVSERSFDNVR from beta A3, VLpSGAWVGFEHAGFQGQQYILER from beta A4, SIIVSAGPWVAFEQpSNFR from beta B1, and GLMMELpSEDCPSIQDR from gamma C, were also identified from vitreous. It is well-known that crystallins are the principal components of eye lens, contributing about 20–60% of total wet weight [23].
We next examined the phosphopeptides from patients with different diseases. For this purpose, vitreous from patient 2 and 4 who had retinal detachment without diabetes were pooled to generate Group 1, and vitreous from patient 6 and 10 who had hemovitreous with diabetes were pooled to generate Group 2. Using similar approach, phosphopeptides were enriched from two groups of trypsinized vitreous by TiO2 column, analyzed by LC-MS/MS, and compared with each other. As a result, phosphopeptides from abundant serum proteins such as albumin, apolipoproteins, kininogen 1, and secreted phosphoprotein 1, were identified from both groups, whereas the 9 phosphopeptides from three types of crystallins were found only in Group 2, indicating that the phosphorylation of crystallins were involved in the disease of hemovitreous with diabetes.
Discussions
Protein phosphorylation can affect catalytic activity, localization of a protein in the cell, protein stability, and the ability of a protein to dimerize or form a stable complex with other molecules. Protein phosphorylation and dephosphorylation function together in signal transduction pathways to induce rapid changes in response to hormones, growth factors, and neurotransmitters [19, 20]. To understand exactly why a particular protein becomes phosphorylated, it may be necessary to identify precisely which amino acid residues are phosphorylated. These residues then can be changed by site-directed mutagenesis, and the mutated protein can be examined for changes in activity, intracellular localization, and association with other proteins in the cell. In addition, identification of phosphorylation sites could reveal which protein kinase regulates the protein and thereby help to understand the biological function and significance of signal transduction pathway. Today, mass spectrometry (MS)-based methods are the most commonly used technique for global profiling of phosphoproteins and phosphorylation sites [24]. However, due to the low stoichiometry of most phosphorylation, enrichment of phosphopeptides by immobilized metal ion affinity chromatography (IMAC), or titanium dioxide (TiO2) chromatography, is advantageous or required before MS analysis [25].
While there have been reports that bovine crystallin alpha B was a phosphoprotein [26–28], and phosphorylation of rat alpha B was involved in uveitis progression and inflammatory responses [29], the large scale identification of phosphoproteins and mapping their phosphorylation sites in vitreous, to our knowledge, has never been reported. The vitreous proteome, containing more than 500 proteins based on recent proteomics studies [13–18], is a very complex matrix predominated by high-abundance resident proteins, such as albumin and apolipoproteins, together with proteins that are shed or secreted by key ocular structures. The complexity of vitreous protein mixture and the high background level of the most abundant proteins make MS-based phosphoprotein identification and localization of phosphorylation sites a challenge. Here, in order to obtain an improved characterization of vitreous phosphoproteome, we conducted the first attempt and successfully identified a large number of phosphopeptides and their phosphorylation sites using TiO2-based phosphopeptide enrichment followed by LC-MS/MS analysis using an LTQ-Orbitrap instrument which combines high sensitivity, high resolution and high mass accuracy.
In this study, phosphopeptides from trypsinized clinical vitreous samples (~ 1mg), pooled from several patients, were enriched by TiO2 column and analyzed by LTQ-Orbitrap mass spectrometer. Due to a) the high concentrations of resident proteins such as albumin and apolipoproteins in vitreous, b) the low abundance stoichiometry of phosphorylated proteins in the biological samples, c) the lack of sufficient volume and amount of clinical vitreous samples, we chose to analyze the pooled vitreous (~ 1 mg) instead of vitreous from a single patient in order to yield the highest number of identified phosphopeptides and provide the most comprehensive view of the vitreous phosphoproteome. This effort was not meant to serve as a complete description of the vitreous phosphoproteome nor identify disease-specific biomarker candidates, but as an initial effort to begin the description of this potentially important information archive.
In addition to the phosphopeptides from cellular proteins and abundant serum proteins, we identifed several phosphopeptides from all three types of crystallins in this pilot study. Among these, Ser19 of phosphopeptide RPFFPFHpSPSR and Ser59 of phosphopeptide APpSWFDTGLSEMR from crystalline alpha B are the homologous residues of phosphorylated rat alpha B that was involved in uveitis progression and inflammatory responses [29]. As mentioned previously, the vitreous proteins, shed or secreted by surrounding ocular structures such as the retina and the lens, provide a record of the health or disease state of the ocular tissue. We therefore compared the identified phosphopeptides from patients who had retinal detachment without diabetes and patients who had hemovitreous with diabetes, and found that phosphopeptides from crystallins were identified only in patients who had hemovitreous with diabetes. The preliminary findings from this comparison, as usual, require further study to determine their relevant role in the disease.
It is not clear what kinds of kinases are responsible for the phosphorylation of these proteins and what is the function of these vitreous phosphoproteins, however, these modified forms of proteins are definitely a reflection of ongoing physiological and pathological events, and could be early warnings for diseases or stages of diseases such as diabetic retinopathy or macular degeneration. Further investigation of these phosphoproteins may lead to a better understanding of their roles in both normal and disease conditions. We believe that vitreous phosphoproteome may be a future source of candidate biomarkers for individualized therapy of age-related macular degeneration and other ocular diseases.
Acknowledgments
We thank Virginia Espina, and Lindsay Wescott from George Mason Univeristy for helping with this study. The research is supported by funding from the College of Science at George Mason University, NIH grant 1R21EY018942-01 and Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanit, Italy.
References
- 1.Fatti I. Hydraulic flow conductivity of the vitreous gel. Invest Ophthalmol Vis Sci. 1977;16:565–568. [PubMed] [Google Scholar]
- 2.Coulombre AJ. The role of intraocular pressure in the development of the chicken eye. J Exp Zool. 1956;133:211–223. [Google Scholar]
- 3.Arciniegas A, Amaya LE. Bio-structural model of the human eye. Ophthalmologica. 1980;180:207–211. doi: 10.1159/000308975. [DOI] [PubMed] [Google Scholar]
- 4.Foulds WS. Is your vitreous really necessary? The role of the vitreous in the eye with particular reference to retinal attachment, detachment and the mode of action of vitreous substitutes. Eye. 1987;1:641–664. doi: 10.1038/eye.1987.107. [DOI] [PubMed] [Google Scholar]
- 5.Bishop PN. Structural macromolecules and supramolecular organisation of the vitreous gel. Progress in Retinal and Eye Research. 2000;19:323–344. doi: 10.1016/s1350-9462(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 6.Haddad A, de Almeida JC, Laicine EM, Fife RS, Pelletier G. The origin of the intrinsic glycoproteins of the rabbit vitreous body: an immunohistochemical and autoradiographic study. Exp Eye Res. 1990;50:555–561. doi: 10.1016/0014-4835(90)90045-v. [DOI] [PubMed] [Google Scholar]
- 7.Haddad A, Laicine EM, de Almeida JC, Costa MS. Partial characterization, origin and turnover of glycoproteins of the rabbit vitreous body. Exp Eye Res. 1990;51:139–143. doi: 10.1016/0014-4835(90)90065-3. [DOI] [PubMed] [Google Scholar]
- 8.Neal RE, Bettelheim FA, Lin C, Winn KC, Garland DL, Zigler JS., Jr Alteration in human vitreous humor following cataract extraction. Exp Eye Res. 2005;80:337–347. doi: 10.1016/j.exer.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Nakanishi T, Koyama R, Ikeda T, Shimizu A. Catalogue of soluble proteins in the human vitreous humor: comparison between diabetic retinopathy and macular hole. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;776:89–100. doi: 10.1016/s1570-0232(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 10.Aiello LP, Avery RL, Arrigg PG, et al. Vascular endothelial growth factor in ocular fluid of patients with diabetic retinopathy and other retinal disorders. N Engl J Med. 1994;331:1480–1487. doi: 10.1056/NEJM199412013312203. [DOI] [PubMed] [Google Scholar]
- 11.Gao BB, Chen X, Timothy N, Aiello LP, Feener EP. Characterization of the vitreous proteome in diabetes without diabetic retinopathy and diabetes with proliferative diabetic retinopathy. Journal of Proteome Research. 2008;7:2516–2525. doi: 10.1021/pr800112g. [DOI] [PubMed] [Google Scholar]
- 12.Davuluri G, Espina V, Petricoin EF, Ross M, Deng J, Liotta L, Glaser BM. Activated VEGF receptor shed into the vitreous in eyes with wet AMD. Arch Ophthalmol. 2009;127:613–621. doi: 10.1001/archophthalmol.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyama R, Nakanishi T, Ikeda T, Shimizu A. Catalogue of soluble proteins in human vitreous humor by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrospray ionization mass spectrometry including seven angiogenesis-regulating factors. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;792:5–21. doi: 10.1016/s1570-0232(03)00133-8. [DOI] [PubMed] [Google Scholar]
- 14.Yamane K, Minamoto A, Yamashita H, Takamura H, Miyamoto-Myoken Y, Yoshizato K, Nabetani T, Tsugita A, Mishima HK. Proteome analysis of human vitreous proteins. Mol Cell Proteomics. 2003;2:1177–1187. doi: 10.1074/mcp.M300038-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Wu CW, Sauter JL, Johnson PK, Chen CD, Olsen TW. Identification and localization of major soluble vitreous proteins in human ocular tissue. Am J Ophthalmol. 2004;137:655–661. doi: 10.1016/j.ajo.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Ouchi M, West K, Crabb JW, Kinoshita S, Kamei M. Proteomic analysis of vitreous from diabetic macular edema. Exp Eye Res. 2005;81:176–182. doi: 10.1016/j.exer.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Kim T, Kim SJ, Kim K, Kang UB, Lee C, Park KS, Yu HG, Kim Y. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics. 2007;7:4203–4215. doi: 10.1002/pmic.200700745. [DOI] [PubMed] [Google Scholar]
- 18.Mukai N, Nakanishi T, Shimizu A, Takubo T, Ikeda T. Identification of phosphotyrosyl proteins in vitreous humours of patients with vitreoretinal diseases by sodium dodecyl sulphate-polyacrylamide gel electrophoresis/Western blotting/matrix-assisted laser desorption time-of-flight mass spectrometry. Ann Clin Biochem. 2008;45(Pt 3):307–312. doi: 10.1258/acb.2007.007151. [DOI] [PubMed] [Google Scholar]
- 19.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 20.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Ross MM, Tessitore A, Ornstein D, VanMeter A, Liotta LA, Petricoin EF. An Initial Characterization of the Serum Phosphoproteome. Journal of Proteome Research. 2009;8:5523–5531. doi: 10.1021/pr900603n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 23.Wistow G, Piatigorsky J. Recruitment of enzymes as lens structural proteins. Science. 1987;236:1554–1556. doi: 10.1126/science.3589669. [DOI] [PubMed] [Google Scholar]
- 24.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 25.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 26.Chieasa R, Gawinowicz Kolks MA, Kleiman NJ, Spector A. The phosphorylation sites of the B2 chain of bovine alpha-crystallin. Biochem Biophys Res Commun. 1987;144:1340–1347. doi: 10.1016/0006-291x(87)91457-4. [DOI] [PubMed] [Google Scholar]
- 27.Voorter CE, de Haard Hoekman WA, Roersma ES, Meyer HE, Bloemendal H, de Jong WW. The in vivo phosphorylation sites of bovine alpha B-crystallin. FEBS Lett. 1989;259:50–52. doi: 10.1016/0014-5793(89)81491-7. [DOI] [PubMed] [Google Scholar]
- 28.Smith JB, Sun Y, Green B. Identification of the posttranslational modifications of bovine lens alpha B-crystallins by mass spectrometry. Protein Sci. 1992;1:601–608. doi: 10.1002/pro.5560010506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahk SC, Jang JU, Choi CU, Lee SH, Park ZY, Yang JY, Kim JD, Yang YS, Chung HT. Post-Translational Modification of Crystallins in vitreous body from experimental autoimmune uveitis of rats. Journal of Proteomic Research. 2007;6:3891–3898. doi: 10.1021/pr070133k. [DOI] [PubMed] [Google Scholar]