Abstract
Chemical reactions in living cells are under strict enzyme control and conform to a tightly regulated metabolic program. However, uncontrolled and potentially deleterious endogenous reactions occur, even under physiological conditions. Aging, in this chemical context, could be viewed as an entropic process, the result of chemical side reactions that chronically and cumulatively degrade the function of biological systems. Mitochondria are a main source of reactive oxygen species (ROS) and chemical sidereactions in healthy aerobic tissues and are the only known extranuclear cellular organelles in animal cells that contain their own DNA (mtDNA). ROS can modify mtDNA directly at the sugar-phosphate backbone or at the bases, producing many different oxidatively modified purines and pyrimidines, as well as single and double strand breaks and DNA mutations. In this scenario, natural selection tends to decrease the mitochondrial ROS generation, the oxidative damage to mtDNA, and the mitochondrial mutation rate in long-lived species, in agreement with the mitochondrial oxidative stress theory of aging.
1. Introduction
Living cells emerged as open systems, exchanging matter and energy with their surrounding, extracting and channelling energy to maintain themselves in a dynamic steady state distant from equilibrium. One of the properties that distinguish living organisms is, among others, a high degree of chemical complexity and molecular organization, defined functions for each of their components and regulated interactions among them. The overall network of interconnected reaction sequences that interconvert cellular metabolites constitutes cellular metabolism [1]. Metabolism is adapted to achieve balance and economy [2]. For this reason, chemical reactions in living cells are under strict enzyme control and conform to a tightly regulated metabolic program. However, uncontrolled and potentially deleterious endogenous reactions occur, even under physiological conditions [3]. Aging, in this chemical context, could be viewed as an entropic process, the result of chemical side-reactions that chronically and cumulatively degrade the function of biological systems. Death, the endpoint in the kinetic process of aging, represents the threshold of damage sufficient to compromise the normal function of an essential physiological subsystem or its ability to survive a challenge to that function [4].
Although aging seems to be a multicausal process (other causal factors involved in the aging process are, e.g., membrane unsaturation [5], Maillard reaction [6], proteasome [7], autophagy [8]), it is likely due to a relatively small number of main causes with major effects. This would make it possible to study the main endogenous mechanisms of aging. Reactive oxygen species (ROS), especially those of mitochondrial origin, are most likely among those main causes [9–11]. Mitochondria are a main source of ROS in healthy aerobic tissues and are the only known extra-nuclear cellular organelles in animal cells that contain their own DNA (mtDNA). Mitochondrial ROS generation occurs continuously throughout life at a animal species-specific rate [12] independent of the rate of mitochondrial oxygen consumption but related to the longevity of each species and, consequently, with their rate of aging [11, 13].
In spite of the existence of many kinds of cellular antioxidants, as adaptive response to ROS generation, there is always a certain steady-state level of oxidative damage to macromolecules even in healthy unstressed animals [14, 15]. ROS can modify mtDNA directly at the sugar-phosphate backbone or at the bases, producing many different oxidatively modified purines and pyrimidines, including the most commonly measured 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), as well as single and double strand breaks and DNA mutations [16, 17] (Figure 1). Reactive carbonyl compounds derived from carbohydrates and lipid oxidation reactions can also react at the exocyclic amino groups of deoxyguanosine, deoxyadenosine, and deoxycytosine to form various alkylated products [18, 19] (Figure 1). Some common carbonyl compounds that can potentially cause mtDNA damage are malondialdehyde, acrolein and 4-hydroxynonenal, among others. The most common adducts arising from enals are exocyclic adducts such as malondialdehyde-deoxyguanosine (M1G).
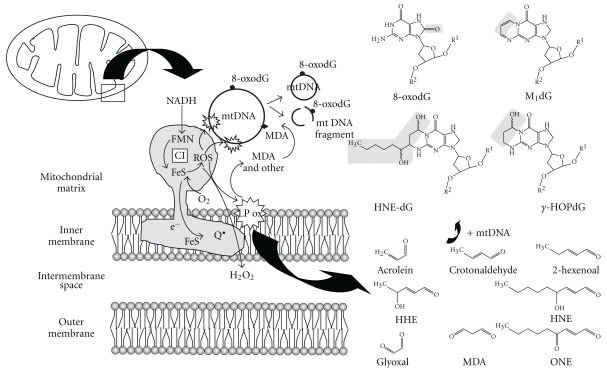
Figure 1.
Schematic diagram of mitochondrial processes that are important for aging and longevity. The schematic shows that mitochondrial complex I is the main ROS generator. Several physiological mechanisms influencing the rate of mitochondrial ROS generation include: (i) the relative concentration of the respiratory complexes, (ii) the degree of electronic reduction of these generators, (iii) the uncoupling proteins, and (iv) specific chemical modifications. Oxygen radicals attack lipids, carbohydrates, proteins, and DNA. The products of lipid peroxidation include highly reactive molecules that can also cause lipoxidative damage to mitochondrial DNA. The scheme highlights two main characteristics of long-lived animal species likely contributing to their slow aging rate and superior longevity: highly resistant macromolecular components and low rate of generation of endogenous damage. Abbreviations: 8-oxodG, 8-oxo-7,8-dihydro-2′-deoxyguanosine; M1dG, Malondialdehyde-deoxyguanosine; HNE-dG, 4-hydroxy-2-nonenal-deoxyguanosine; HOPdG, Acrolein-deoxyguanosine.
Many investigations have found that tissue steady-state levels of 8-oxodG show a moderate increase during aging in brain, heart, or liver nuclear (nDNA) or mitochondrial DNA (mtDNA) in rodents and humans [20–23], while such increases were not detected in other cases [23–25]. While 8-oxodG is normally present both in nDNA and in mtDNA, its level is several folds higher in mtDNA than in nDNA [12, 23, 26]. This has been attributed to various reasons like the lack of protective histones and polyamines or to a lower repair of mtDNA in relation to nDNA. However, while mtDNA lacks some forms of DNA repair, its capacity to repair 8-oxodG seems to be similar to that of nDNA [27]. Thus, the most probable reason for the higher level of 8-oxodG in mtDNA than in nDNA is its closeness to the main ROS generator of healthy cells, the inner mitochondrial membrane (Figure 1). Other factors, like the lack of introns in mtDNA, can amplify the detrimental consequences of the relatively higher level of oxidative damage of mtDNA. The genetic information is tightly packed on the mtDNA molecule and, therefore, most mutational changes on this genome should have injurious effects.
The observed increases in oxidative damage to mtDNA with aging likely reflect the net flux of oxidative damage resulting from the balance between prooxidant sources and antioxidant/detoxifying/repair systems that would contribute to age-related deterioration at all levels of the biological organization, possibly due to its capacity to generate mtDNA mutations [28]. The mitochondrial genome principally suffers from two types of mutations: point mutations, which are changes of one or a few nucleotides; and large deletions, which involve the removal of large portions of the genome (from a few hundred base pairs to almost the entire genome). The exact sources of mtDNA mutations are matter of debate, but might be similar to those causing nuclear DNA mutations, that is, errors during DNA damage processing or spontaneous polymerase errors [28, 29].
2. Rate of Generation of Mitochondrial Reactive Oxygen Species and Animal Longevity
2.1. Generation of Mitochondrial ROS
Inside mitochondrial respiratory chain, electrons from reduced substrates are passed from complexes I and II of the electron transport chain through complexes III and IV to oxygen, forming water and causing protons to be pumped across the mitochondrial inner membrane. The proton motive force set up by proton pumping drives protons back through the ATP synthase in the inner membrane, forming ATP from ADP and phosphate [30]. In this context, a major side reaction is that electrons may leak from the respiratory chain and react with oxygen to form the free radical superoxide. Superoxide anion, the product of a one-electron reduction of oxygen, is the precursor of most ROS and a mediator in oxidative chain reactions [15]. There is, however, a lack of stoichiometric coupling of ROS production to oxygen consumption [13].
It is well known that mitochondrial ROS (mtROS) generation occurs at complex I and at complex III (reviewed in [11, 14]). Concerning the electron transport component responsible for mtROS generation within complex I, flavin mononucleotide, ubisemiquinone species, or iron-sulphur clusters have been proposed (reviewed in [31–35]). Interestingly two of these, the flavin and all the FeS clusters, are located in the hydrophilic complex I domain facing the mitochondrial matrix, which would facilitate close proximity, or even the contact between mtDNA and the ROS generator (see Figure 1). In contrast, complex III produces ROS directed to the cytosolic side of the inner membrane, although recent studies suggest that part of the production could also occur towards the matrix side [36].
Are there biological adaptations determining the rate of mitochondrial free radical generation? Available evidence seems to suggest that this is the case. Among these adaptations the following can be included (reviewed in [14, 33, 37]): (i) to adapt the amount of the respiratory complex/es responsible for ROS generation. Thus, a decrease in the amount of complex I protein will lead to a decreased rate of ROS generation; (ii) to adjust the amount of uncoupling proteins. The mitochondrial superoxide production is very sensitive to the proton motive force, so it can be strongly decreased by mild uncoupling. In this scenario, an ancestral function of uncoupling proteins has been proposed: to cause mild uncoupling and so diminish mitochondrial superoxide production, hence protecting against oxidative damage; (iii) to regulate the degree of electronic reduction of these generators: the higher their degree of reduction, the higher will be their rate of ROS production; and (iv) to modify by enzymatic and nonenzymatic pathways such as S-nitrosation, acetylation, and glutathionylation specific peptides of the ROS generators.
2.2. Mitochondrial ROS Production and Animal Longevity
Several works have evaluated the relationship between mitochondrial ROS production and animal longevity. These studies found that the rate of mtROS production is lower in the tissues of long-lived than in those of short-lived animal species [11, 12, 14]. Early studies, however, only included animal species following the “rate of living theory”—the inverse relationship between longevity and metabolic rate-, so the results obtained could also be interpreted as a correlate of that phenomenon. Species with short longevity could show high mitochondrial ROS production simply because their rates of mitochondrial oxygen consumption are higher. Fortunately, comparative physiology offers more possibilities to test the hypothesis that mitochondrial ROS production is involved in the rate of aging and, consequently, in determining longevity. Three groups of homoeothermic vertebrates have an extraordinarily high longevity in relation to their body size and metabolic rate: birds, bats, and primates. Birds can be a strong test because if a low rate of ROS production contributes to slowing of the aging rate, the mitochondria of birds should show a low rate of ROS generation in spite of the high rate of oxygen consumption of these animals. Thus, in spite of their high rates of whole body O2 consumption, parakeets, canaries, and pigeons (with longevities of 21, 24, and 35 years, resp.) have lower rates of mtROS generation than mice and rats (3.5 and 4 years of longevity, resp.), even though their metabolic rates and body sizes are of a similar magnitude. The difference in ROS production between these species occurred at only one of the electron transport complexes: complex I. The low rate of mtROS production of birds, in spite of their high rates of oxygen consumption, is possible because (a) the percent of total electron flow in the respiratory chain directed to ROS production (the % free radical leak, %FRL) is lower [12], and (b) the total amount of mitochondrial complex I is also lower [38, 39]. This means that their respiratory chain transports electrons more efficiently avoiding univalent electron leaks to oxygen upstream of cytochrome oxidase. All the comparative investigations performed in mammals and birds show that mitochondrial ROS generation is lower in long- than in short-lived animals. Interestingly, this finding is even more significant after correction for effects of body size and phylogeny [40]. Furthermore, other interspecies comparisons between rodents with widely different longevities but similar body size (see [12]), or between rodents and bats [41], also showed that the longest-lived species had lowest mitochondrial ROS production and leak. Recent studies have also found much lower rates of ROS generation in human (primates—and especially humans—also live much longer than expected for their body size and metabolic rate) than in rat brain mitochondria [42]. If mitochondrial oxygen radical production controls aging rate, the rate of ROS production of animals with different metabolic rates and body sizes but similar longevity should be equal. This is exactly what happened in another interspecies comparison when mitochondrial ROS production of mouse (longevity, 3.5 yrs) and rat (longevity, 4 yrs) were compared [43, 44]. The longevity of these species are similar (3.5 and 4 years), whereas metabolic rate is threefold higher and body size is around 15 fold lower in the mouse than in the rat.
The relevance of ROS production in determining longevity has also been recently reinforced by using Drosophila as experimental model [45]. In this study, transgenic strains of Drosophila were created that express yeast NDI1 (in yeast, the single-subunit NADH dehydrogenase Ndi1 serves as a nonproton-translocating alternative enzyme that replaces complex I, bringing about the reoxidation of intramitochondrial NADH) ubiquitously. NDI1 expression mitigated the aging-associated decline in respiratory capacity and the accompanying increase in mitochondrial ROS production, and resulted in decreased accumulation of markers of oxidative damage in aged flies, resulting in an increased median, mean, and maximum longevity. These results support a central role of mitochondrial complex I in influencing longevity via oxidative stress.
In accordance with the low mitochondrial free radical production of long-lived animal species, there is an adaptive response concerning endogenous cellular antioxidant systems (reviewed in [14, 32]). Thus, long-lived vertebrates, including mammals, constitutively have lower (instead of higher) tissue levels of antioxidant enzymes and low molecular weight endogenous antioxidants, as well as repair systems [46], than short-lived ones. This observation has been recently extended to invertebrate models [47]. That characteristic can thus explain why endogenous tissue antioxidants correlate negatively with longevity across species: long-lived animals have constitutively low levels of antioxidants because they produce ROS at a low rate. If long-lived animals had high rates of ROS production together with their very low levels of endogenous antioxidants, their tissue cells would not be able to maintain oxidative stress homeostasis. Decreasing mitochondrial ROS production instead of increasing antioxidants or repair systems makes sense when considered from the point of view of evolution of longevity among species. It would be very inefficient to generate large amounts of ROS and, afterwards, try to intercept them before they reach cellular components like DNA, or even worse, try to repair DNA after heavily damaging it. This makes even more sense taking into account the high energetic cost of continuously maintaining high levels of antioxidant and repair molecules in cells. Interestingly, there is a close proximity or even contact between main sources of ROS at the inner mitochondrial membrane and mtDNA. So, lowering the rate of mtROS generation near mtDNA decreases its damage much more efficiently and at much lower cost [12].
In agreement with the adaptive response that represents antioxidant defences, and with the ability of all kinds of animals (short- or long-lived) to transitorily induce or repress these protective molecules when necessary in the needed amounts, the outcomes of two experimental paradigms deserve also special mention (reviewed in [11, 48, 49]): (i) experimentally increasing tissue antioxidants through dietary supplementation, pharmacological induction, or transgenic techniques sometimes moderately increases mean longevity (life expectancy) but does not change maximum longevity; and (ii) animals in which genes coding for particular antioxidant enzymes are knocked out can show different pathologies but their rates of aging do not seem to be affected. It is proposed that these findings are probably due to the activation of negative feedback mechanisms in order to maintain the redox balance, which is cell and species specific. It is this low endogenous rate of generation of highly damaging substances that can importantly contribute to a slower rate of accumulation of DNA damage and mutations, and then to a longer life, in long-lived species.
3. Mitochondrial DNA Damage and Longevity
Mitochondrial DNA oxidation is one of the natural consequences of aerobic life. Among different kinds of molecular damage caused by ROS, that to mtDNA is important for longevity because it can lead to irreversible loss or alteration of its coded information, which will be especially deleterious in postmitotic tissues. Determinant mechanisms of the steady-state level of mtDNA damage include (i) because long-lived animal species have low rates of mtROS generation, this should be reflected in the steady-state level of oxidative damage and the accumulation of somatic mutations in their mtDNA; (ii) mtDNA is located very close to or even in contact with the site/s of mitochondrial ROS production (see Figure 1); and (iii) the structural susceptibility of the mtDNA molecule to damage.
The susceptibility of mtDNA to damage could be related to the simplest property of a DNA sequence, the proportion of A, C, G, and T in the mtDNA molecule. In a recent study [50], analyzing the “light strand” mtDNA sequence of 94 animals species (including invertebrates, birds, and mammals), it was found that short-lived species have higher A and T abundances and lower C abundances than the long-lived species, while G is almost uniformly low in all species. The light strand was given this name because it is unusually deficient in G, and data from this study shows that this characteristic low G abundance carries through to all of these species. A conclusion is that there is some strong evolutionary pressure to keep the G abundance of the light strand low. The standard hypothesis is that the asymmetric mode of replication of the mtDNA molecule causes an increase in the G to A transition mutations on the light strand, depleting it of G nucleotides. In addition to this finding, it was verified that this pattern of nucleotides affects a DNA sequence property, the free energy (a physical property of the double-stranded DNA molecule related to the binding energy between the two DNA strands; the more negative the free energy is, the less likely is the spontaneous separation of the two strands through thermal fluctuations, conferring to the mtDNA a greater structural stability and lesser susceptibility to damage) [50]. The data showed a strong relationship between the mtDNA free energy and life span. Thus, the longer the maximum longevity of a species, the lower is the free energy of mtDNA [50]. Considering that of the four nucleobases, guanine has the lowest oxidation potential and is thus generally most easily oxidized [16], it is proposed that these mtDNA base sequence pattern and derived properties represent an evolutionary adaptation of long-lived species to increase mtDNA resistance to endogenous damage by chemical side-reactions and so to decrease the susceptibility of mtDNA to damage and mutation. A similar evolutionary adaptation has been demonstrated for proteins (by the methionine content) and membrane unsaturation (by the polyunsaturated fatty acids content) in long-lived species [5, 14].
In agreement with their high resistance to endogenous damage and low rates of mitochondrial ROS production, long-lived animal species have lower steady-state levels of 8-oxodG in their mtDNA [51, 52] and lower rates of urinary excretion of 8-oxo-7,8-dihydroguanine [53] than short-lived ones. Long-lived birds also usually show lower oxidative damage to mtDNA in their tissues than short-lived rodents of similar body size [54]. It has been also described that the rate of accumulation of mtDNA mutations with age is also much slower in humans than in mice [55]. In this line, in a recent study [56], analyzing the lineage-specific mitochondrial mutation rate across 1,696 mammalian species and comparing it with the nuclear rate, is reported a selected decrease of substitution rate in long-lived species, in agreement with the evidence for a causal role of mtDNA mutations in aging (reviewed in [57]), suggesting that natural selection tends to decrease the mitochondrial mutation rate in long-lived species. In addition, oxidative damage is around 5–9 fold higher in mtDNA than in nDNA in the brain and heart of all the 11 mammalian and bird species studied [51, 54], a difference similar to that generally observed for spontaneous mutations when comparing both DNAs [58].
In order to maintain mitochondrial genomic integrity and to avoid the gradual accumulation of oxidative DNA base lesions, mtDNA damage is repaired by a number of mechanisms (for review see [59]). In general, oxidized mtDNA bases are removed by base excision repair (BER) mechanisms [59, 60] and although these correcting mechanisms show high fidelity, DNA lesions can still accumulate with age. DNA polymerase enzymes (as component of the BER machinery) have a proofreading error-correcting capacity, and when mitochondrial DNA polymerase proofreading activity was eliminated in mice there was a 3–5 fold increase in mitochondrial mutations and a decreased longevity in such mice [61, 62]. In support for a relevant role of mtDNA mutations in the rate of aging, in a recent study it has been demonstrated that mutations in mtDNA profoundly affect the assembly of fully functional ETC complexes, leading to mitochondrial dysfunction and ultimately, to the activation of a mitochondrial-mediated apoptotic program, as mechanism underlying the accelerated aging process of this experimental model [63].
All that might suggest that the flux rates of both ROS attack on and repair of DNA are much higher in the mtDNA of short-lived than in that of long-lived animals, and are also much higher in the mtDNA than in the nDNA of all species [64]. In addition, the higher rate of mtROS production and mtDNA susceptibility to oxidative damage of short-lived animals may be an important cause of their much faster rate of accumulation of mtDNA mutations during aging [55, 65].
4. mtDNA Mutations and Rate of Aging
Aging is irreversible in nature. Thus, it could be caused in part by the long-life progressive accumulation of nonrepairable macromolecular damage. This occurs at the level of the main information-containing macromolecule of postmitotic cells, the DNA. As mentioned above, it is known that somatic mtDNA mutations can accumulate with age reaching very high levels in postmitotic tissues [66, 67], and some studies in mutant mice suggest that increasing the level of mtDNA mutations increases the aging rate [61–63].
It has been argued that the high copy number of mtDNA molecules per cell is contradictory with a possible role of mtDNA in aging, since loss of some mtDNA molecules would not be deleterious to the cell because the remaining wild type ones would be enough for the continuation of cell function. However, it is known that clonal expansion of mutated mtDNA molecules can bring them near to homoplasmy in old tissues [68]. Furthermore, the reliability theory of aging [69] points out that aging appears in complex systems thanks to the presence of redundancy of vital system components. Such redundancy also occurs at subcellular level in the case of mtDNA which is present in multiple copies per cell. If a single mtDNA molecule would exist per postmitotic cell instead of many, its deletion could easily result in cell death. According to Gavrilov and Gavrilova (2006) [69], some cases of the so-called “negliglible senescence” in simple organisms would not be cases of very slow aging as it is frequently assumed. Instead, they would be the simple result of a lack of redundancy in their vital component parts. They would just die when they are seriously damaged for the first time, and thus they could not tell us anything interesting about slow aging and longevity in mammals [69]. This concept of reliability and aging due to high redundancy could be also applied to mtDNA. Cells containing large amounts of mutated mtDNA molecules seem to survive many years or even decades in human postmitotic tissues, and their wrong functioning likely contributes to the age-related deterioration of tissue functions. Large accumulations of mtDNA mutations have been recently reported in substantia nigra [67, 70] as well as in human skeletal muscle in which 31% of the fibers of old individuals showed electron transport abnormalities and the abnormal fibers showed accumulations of deleted mtDNA molecules to detrimental (>90% of the total mtDNA) levels [71, 72].
On the other hand, ROS have mutagenic capacity, and the rate of accumulation of DNA mutations is known to be higher when the cellular pO2 is increased, suggesting a role for ROS in their formation [73]. mtROS generation can be among the causes of such accumulation in vivo and thus of aging. Also agreeing with this possibility, both 8-oxodG [51, 74] and somatic mutations [58] are present in mtDNA at levels around 10 folds higher than in nuclear DNA (nDNA) [75]. Since it is now known that the capacity of the mitochondria to repair 8-oxodG is similar or even higher (instead of lower) than that of the nucleus [59], the difference in steady-state oxidative DNA damage between both compartments should be mainly due to the much higher capacity of the mitochondria for continuous ROS generation, not to differences in 8-oxodG repair [64], and possibly to the closeness or even the contact between mtDNA and the inner mitochondrial membrane [76–78].
5. Rate of Generation of mtROS-Induced Damage and Longevity: The Dietary Restriction Approach
The comparative studies described above are strongly suggestive of a causal relationship between mtROS production and longevity via mtDNA damage. But correlation does not necessarily mean that a cause-effect relationship exists, and experimental studies are needed for confirmation. Dietary restriction (DR) is the best described experimental intervention that slows down aging and increases longevity in different animal species such as C. Elegans [79], Drosophila [45], rodents (mice and rats) [80], and monkeys [81]. The effect of DR on mtROS production has been studied intensively in rodents and especially in rats [80]. These investigations, usually applying 40% DR, consistently demonstrated that long-term dietary restriction (between 1 year of DR and life-long DR) significantly decreases the rate of mtROS generation in rat tissues including heart, brain, skeletal muscle, liver, and kidney. It was also found that 6-7 weeks of dietary restriction are enough to decrease mtROS production and 8-oxodG in mtDNA and nDNA in rat liver [82]. It was also found that the decrease in mtROS generation in DR rats specifically occurs at complex I in all the organs studied so far (heart, liver, and brain), along with a decrease in the amount of the mtROS generators, and that it takes place together with lack of changes in mitochondrial O2 consumption and with a decrease in %FRL, indicating that the mitochondria of DR animals are more efficient in avoiding ROS production per unit electron flow [82–84]. These characteristics were also observed in long-lived animal species when compared to those of short-lived ones. The lack of changes in mitochondrial O2 consumption also agreed with the lack of variations in basal metabolic rate in DR [85, 86]. In addition, the decrease in mtROS generation observed in DR rats was accompanied by significant decreases in 8-oxodG levels in mtDNA alone, or in mtDNA and nDNA, depending on the tissue studied [82, 84, 87].
On the other hand, the repair of 8-oxodG in mtDNA through the mitochondrial base excision repair pathway does not increase and even decreases during DR [88]. Thus, the decrease in 8-oxodG steady-state level in mtDNA during DR must be due to a decrease in 8-oxodG formation. This is further supported by the fact that the three implicated parameters, mtROS generation, steady state 8-oxodG levels in mtDNA, and 8-oxodG repair through mitochondrial base excision repair decrease by a similar quantitative extent during 40% DR: around 30–40%. This agrees with the previously proposed model indicating that there is a decrease in the flow of oxidative damage through the mtDNA in DR [64], similar to what occurs in long-lived compared to short-lived species. In other words, mitochondrial 8-oxodG repair, like tissue antioxidants in long-lived animals, is lower (instead of higher) in tissues of DR rodents because their rate of mtROS production is lower than in those fed ad libitum. Neither 8-oxodG repair nor endogenous antioxidants should be continuously maintained at levels higher than required because that would be energetically costly. Both the antioxidants and the mtDNA repair systems are inducible. Thus, they can be transitorily increased at moments of higher than normal oxidative damage to quickly return to their basal levels when the situation is normalized. For instance, the repair of 8-oxodG is maximally induced in the rat kidney after only 6 hours of exposure to potassium bromate [89]. The lower steady-state oxidative damage in the mtDNA of long-lived and DR animals is much better obtained by decreasing the rate of mtROS generation than by increasing antioxidants and repair because this is much less costly and much more efficient and simple [11, 33].
In summary, lowering the rate of mtROS production seems to be a highly conserved mechanism developed during the evolution to be used both within (DR) and between species. It allows both long-lived species and DR animals to decrease steady-state oxidative damage to mtDNA, and likely the rate of accumulation of mtDNA mutations and the aging rate. In this scenario, it is proposed that natural selection tends to decrease the mitochondrial ROS generation, the oxidative damage to mtDNA and the mitochondrial mutation rate in long-lived species, in agreement with the mitochondrial oxidative stress theory of aging.
Acknowledgments
Investigations of the author of this review have been supported by grants ref. BFI2003-01287, BFU2006-14495/BFI, and BFU2009-11879/BFI; Reticef RD06/0013/0012, and 2005SGR00101 and 2009SGR735 from the Ministry of Science and Innovation, Ministry of Health and the Autonomous Government of Catalonia, respectively.
References
- 1.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York, NY, USA: W. H. Freeman; 2005. [Google Scholar]
- 2.Meléndez-Hevia E, Montero-Gómez N, Montero F. From prebiotic chemistry to cellular metabolism—the chemical evolution of metabolism before Darwinian natural selection. Journal of Theoretical Biology. 2008;252(3):505–519. doi: 10.1016/j.jtbi.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Golubev AG. The other side of metabolismIznanka metabolizma. Biokhimiia. 1996;61(11):2018–2039. [PubMed] [Google Scholar]
- 4.Baynes JW. From life to death—the struggle between chemistry and biology during aging: the Maillard reaction as an amplifier of genomic damage. Biogerontology. 2000;1(3):235–246. doi: 10.1023/a:1010034213093. [DOI] [PubMed] [Google Scholar]
- 5.Pamplona R. Membrane phospholipids, lipoxidative damage and molecular integrity: a causal role in aging and longevity. Biochimica et Biophysica Acta. 2008;1777(10):1249–1262. doi: 10.1016/j.bbabio.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Monnier VM, Sell DR, Nagaraj RH, Miyata S. Mechanisms of protection against damage mediated by the Maillard reaction in aging. Gerontology. 1991;37(1–3):152–165. doi: 10.1159/000213256. [DOI] [PubMed] [Google Scholar]
- 7.Merker K, Grune T. Proteolysis of oxidised proteins and cellular senescence. Experimental Gerontology. 2000;35(6-7):779–786. doi: 10.1016/s0531-5565(00)00140-6. [DOI] [PubMed] [Google Scholar]
- 8.Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: theimportance of exquisite quality control. doi: 10.1016/j.arr.2010.02.001. Ageing Research Reviews. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harman D. The biologic clock: the mitochondria? Journal of the American Geriatrics Society. 1972;20(4):145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 10.Miquel J, Economos AC, Fleming J, Johnson JE. Mitochondrial role in cell aging. Experimental Gerontology. 1980;15(6):575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- 11.Sanz A, Pamplona R, Barja G. Is the mitochondrial free radical theory of aging intact? Antioxidants and Redox Signaling. 2006;8(3-4):582–599. doi: 10.1089/ars.2006.8.582. [DOI] [PubMed] [Google Scholar]
- 12.Barja G. Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biological Reviews of the Cambridge Philosophical Society. 2004;79(2):235–251. doi: 10.1017/s1464793103006213. [DOI] [PubMed] [Google Scholar]
- 13.Barja G. Mitochondrial oxygen consumption and reactive oxygen species production are independently modulated: implications for aging studies. Rejuvenation Research. 2007;10(2):215–223. doi: 10.1089/rej.2006.0516. [DOI] [PubMed] [Google Scholar]
- 14.Pamplona R, Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Research Reviews. 2007;6(3):189–210. doi: 10.1016/j.arr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- 16.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutation Research. 2003;531(1-2):37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 17.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chemical Research in Toxicology. 2006;19(2):173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 18.Marnett LJ, Plastaras JP. Endogenous DNA damage and mutation. Trends in Genetics. 2001;17(4):214–221. doi: 10.1016/s0168-9525(01)02239-9. [DOI] [PubMed] [Google Scholar]
- 19.Dutta U, Cohenford MA, Dain JA. Nonenzymatic glycation of DNA nucleosides with reducing sugars. Analytical Biochemistry. 2005;345(2):171–180. doi: 10.1016/j.ab.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-Hydroxy-2’-deoxyguanosine in rat organ DNA and urine. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(12):4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mecocci P, MacGarvey U, Kaufman AE, et al. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Annals of Neurology. 1993;34(4):609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 22.Asunción JG, Millan A, Pla R, et al. Mitochondrial glutathione oxidation correlates with age-associated oxidative damage to mitochondrial DNA. FASEB Journal. 1996;10(2):333–338. doi: 10.1096/fasebj.10.2.8641567. [DOI] [PubMed] [Google Scholar]
- 23.Herrero A, Barja G. Effect of aging on mitochondrial and nuclear DNA oxidative damage in the heart and brain throughout the life-span of the rat. Journal of the American Aging Association. 2001;24(2):45–50. doi: 10.1007/s11357-001-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirano T, Yamaguchi R, Asami S, Iwamoto N, Kasai H. 8-Hydroxyguanine levels in nuclear DNA and its repair activity in rat organs associated with age. Journals of Gerontology. Series A. 1996;51(5):B303–B307. doi: 10.1093/gerona/51a.5.b303. [DOI] [PubMed] [Google Scholar]
- 25.Davies SMK, Poljak A, Duncan MW, Smythe GA, Murphy MP. Measurements of protein carbonyls, ortho- and meta-tyrosine and oxidative phosphorylation complex activity in mitochondria from young and old rats. Free Radical Biology and Medicine. 2001;31(2):181–190. doi: 10.1016/s0891-5849(01)00576-7. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton ML, Van Remmen H, Drake JA, et al. Does oxidative damage to DNA increase with age? Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anson RM, Croteau DL, Stierum RH, Filburn C, Parsell R, Bohr VA. Homogenous repair of singlet oxygen-induced DNA damage in differentially transcribed regions and strands of human mitochondrial DNA. Nucleic Acids Research. 1998;26(2):662–668. doi: 10.1093/nar/26.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khrapko K, Vijg J. Mitochondrial DNA mutations and aging: devils in the details? Trends in Genetics. 2009;25(2):91–98. doi: 10.1016/j.tig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijg J. Aging of the Genome. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- 30.Scheffler IE. Mitochondria. New York, NY, USA: Wiley-Liss; 1999. [Google Scholar]
- 31.Pamplona R, Barja G. Aging rate, free radical production, and constitutive sensitivity to lipid peroxidation: insights from comparative studies. In: Van Zglinicki T, editor. Biology of Aging and Its Modulation Series. Vol. 1. New York, NY, USA: Kluwer Academic Publisher; 2003. pp. 47–64. [Google Scholar]
- 32.Brand MD, Affourtit C, Esteves TC, et al. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radical Biology and Medicine. 2004;37(6):755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 33.Pamplona R, Barja G. Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochimica et Biophysica Acta. 2006;1757(5-6):496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Koopman WJH, Nijtmans LGJ, Dieteren CEJ, et al. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxidants and Redox Signaling. 2010;12(12):1431–1470. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- 35.Lenaz G, Fato R, Genova ML, Bergamini C, Bianchi C, Biondi A. Mitochondrial Complex I: structural and functional aspects. Biochimica et Biophysica Acta. 2006;1757(9-10):1406–1420. doi: 10.1016/j.bbabio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Brand MD. The sites and topology of mitochondrial superoxide production. Experimental Gerontology. 2010;45(7-8):466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. Life and death: metabolic rate, membrane composition, and life span of animals. Physiological Reviews. 2007;87(4):1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 38.Pamplona R, Portero-Otín M, Sanz A, Ayala V, Vasileva E, Barja G. Protein and lipid oxidative damage and complex I content are lower in the brain of budgerigar and canaries than in mice. Relation to aging rate. Age. 2005;27(4):267–280. doi: 10.1007/s11357-005-4562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert AJ, Buckingham JA, Boysen HM, Brand MD. Low complex I content explains the low hydrogen peroxide production rate of heart mitochondria from the long-lived pigeon, Columba livia. Aging Cell. 2010;9(1):78–91. doi: 10.1111/j.1474-9726.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 40.Lambert AJ, Boysen HM, Buckingham JA, et al. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6(5):607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 41.Brunet-Rossinni AK. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mechanisms of Ageing and Development. 2004;125(1):11–20. doi: 10.1016/j.mad.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Kudin AP, Bimpong-Buta NYB, Vielhaber S, Elger CE, Kunz WS. Characterization of superoxide-producing sites in isolated brain Mitochondria. Journal of Biological Chemistry. 2004;279(6):4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 43.Herrero A, Barja G. Sites and mechanisms responsible for the low rate of free radical production of heart mitochondria in the long-lived pigeon. Mechanisms of Ageing and Development. 1997;98(2):95–111. doi: 10.1016/s0047-6374(97)00076-6. [DOI] [PubMed] [Google Scholar]
- 44.Herrero A, Barja G. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: sites of free radical generation and mechanisms involved. Mechanisms of Ageing and Development. 1998;103(2):133–146. doi: 10.1016/s0047-6374(98)00035-9. [DOI] [PubMed] [Google Scholar]
- 45.Sanz A, Soikkeli M, Portero-Otín M, et al. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salway KD, Page MM, Faure PA, Burness G, Stuart JA. Enhanced protein repair and recycling are not correlated with longevity in 15 vertebrate endotherm species. doi: 10.1007/s11357-010-9157-5. Age. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanz A, Fernández-Ayala DJ, Stefanatos RK, Jacobs HT. Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila. Aging. 2010;2(4):220–223. doi: 10.18632/aging.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez VI, Bokov A, Remmen HV, et al. Is the oxidative stress theory of aging dead? Biochimica et Biophysica Acta. 2009;1790(10):1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmon AB, Richardson A, Pérez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radical Biology and Medicine. 2010;48(5):642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuels DC. Life span is related to the free energy of mitochondrial DNA. Mechanisms of Ageing and Development. 2005;126(10):1123–1129. doi: 10.1016/j.mad.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB Journal. 2000;14(2):312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 52.Sanz A, Barja G, Pamplona R, Leeuwenburgh C. Free radicals and mammalian aging. In: Jacob C, Winyard PG, editors. Redox Signaling and Regulation in Biology and Medicine. Weinheim, Germany: Wiley-VCH; 2009. pp. 433–472. [Google Scholar]
- 53.Foksinski M, Rozalski R, Guz J, et al. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radical Biology and Medicine. 2004;37(9):1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Herrero A, Barja G. 8-oxo-deoxyguanosine levels in heart and brain mitochondrial and nuclear DNA of two mammals and three birds in relation to their different rates of aging. Aging. 1999;11(5):294–300. doi: 10.1007/BF03339803. [DOI] [PubMed] [Google Scholar]
- 55.Wang E, Wong A, Cortopassi G. The rate of mitochondrial mutagenesis is faster in mice than humans. Mutation Research. 1997;377(2):157–166. doi: 10.1016/s0027-5107(97)00091-2. [DOI] [PubMed] [Google Scholar]
- 56.Nabholz B, Glémin S, Galtier N. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Molecular Biology and Evolution. 2008;25(1):120–130. doi: 10.1093/molbev/msm248. [DOI] [PubMed] [Google Scholar]
- 57.Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genetics. 2007;3(2, article e24) doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanchard JL, Lynch M. Organellar genes. Why do they end up in the nucleus? Trends in Genetics. 2000;16(7):315–320. doi: 10.1016/s0168-9525(00)02053-9. [DOI] [PubMed] [Google Scholar]
- 59.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radical Biology and Medicine. 2002;32(9):804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 60.Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging—an update. Experimental Gerontology. 2010;45(7-8):478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 62.Kujoth CC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 63.Hiona A, Sanz A, Kujoth GC, et al. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS ONE. 2010;5(7, article e11468) doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barja G. The flux of free radical attack through mitochondrial DNA is related to aging rate. Aging. 2000;12(5):342–355. doi: 10.1007/BF03339859. [DOI] [PubMed] [Google Scholar]
- 65.Melov S, Coskun PE, Wallace DC. Mouse models of mitochondrial disease, oxidative stress, and senescence. Mutation Research. 1999;434(3):233–242. doi: 10.1016/s0921-8777(99)00031-2. [DOI] [PubMed] [Google Scholar]
- 66.Crott JW, Choi SW, Branda RF, Mason JB. Accumulation of mitochondrial DNA deletions is age, tissue and folate-dependent in rats. Mutation Research. 2005;570(1):63–70. doi: 10.1016/j.mrfmmm.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nature Genetics. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 68.Kraytsberg Y, Nekhaeva E, Bodyak NB, Khrapko K. Mutation and intracellular clonal expansion of mitochondrial genomes: two synergistic components of the aging process? Mechanisms of Ageing and Development. 2003;124(1):49–53. doi: 10.1016/s0047-6374(02)00169-0. [DOI] [PubMed] [Google Scholar]
- 69.Gavrilov LA, Gavrilova NS. Reliability theory of aging and longevity. In: Austad S, Kirkwood T, editors. Handbook of the Biology of Aging. New York, NY, USA: Academic Press; 2006. pp. 3–47. [Google Scholar]
- 70.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genetics. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 71.Bua E, Johnson J, Herbst A, et al. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. American Journal of Human Genetics. 2006;79(3):469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meissner C, Bruse P, Mohamed SA, et al. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Experimental Gerontology. 2008;43(7):645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 73.Busuttil RA, Rubio M, Dollé MET, Campisi J, Vijg J. Oxidative damage, somatic mutations and cellular aging. In: Von Zglinicki T, editor. Aging at Molecular Level. Norwell, Mass, USA: Kluwer Academic Publisher; 2003. pp. 79–90. [Google Scholar]
- 74.Hamilton ML, Van Remmen H, Drake JA, et al. Does oxidative damage to DNA increase with age? Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoffmann S, Spitkovsky D, Radicella JP, Epe B, Wiesner RJ. Reactive oxygen species derived from the mitochondrial respiratory chain are not responsible for the basal levels of oxidative base modifications observed in nuclear DNA of mammalian cells. Free Radical Biology and Medicine. 2004;36(6):765–773. doi: 10.1016/j.freeradbiomed.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 76.Barja G, Cadenas S, Rojas C, Lopez-Torres M, Perez-Campo R. A decrease of free radical production near critical targets as a cause of maximum longevity in animals. Comparative Biochemistry and Physiology B. 1994;108(4):501–512. doi: 10.1016/0305-0491(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 77.Abring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation within Hella cell mitochondrial DNA near its origin of replication. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nass MMK, Nass S, Afzelius BA. The general occurrence of mitochondrial DNA. Experimental Cell Research. 1965;37(3):516–539. doi: 10.1016/0014-4827(65)90204-1. [DOI] [PubMed] [Google Scholar]
- 79.Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mechanisms of Ageing and Development. 2005;126(9):929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 80.Gredilla R, Barja G. Caloric restriction, aging and oxidative stress. Endocrinology. 2005;146(9):3713–3717. doi: 10.1210/en.2005-0378. [DOI] [PubMed] [Google Scholar]
- 81.Colman RJ, Anderson RM, Johnson SC, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325(5937):201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gredilla R, Sanz A, Lopez-Torres M, Barja G. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. The FASEB Journal. 2001;15(9):1589–1591. doi: 10.1096/fj.00-0764fje. [DOI] [PubMed] [Google Scholar]
- 83.Gómez J, Caro P, Naudí A, Portero-Otin M, Pamplona R, Barja G. Effect of 8.5% and 25% caloric restriction on mitochondrial free radical production and oxidative stress in rat liver. Biogerontology. 2007;8(5):555–566. doi: 10.1007/s10522-007-9099-1. [DOI] [PubMed] [Google Scholar]
- 84.Sanz A, Caro P, Ibañez J, Gómez J, Gredilla R, Barja G. Dietary restriction at old age lowers mitochondrial oxygen radical production and leak at Complex I and oxidative DNA damage in rat brain. Journal of Bioenergetics and Biomembranes. 2005;37(2):83–90. doi: 10.1007/s10863-005-4131-0. [DOI] [PubMed] [Google Scholar]
- 85.McCarter R, Masoro EJ, Yu BP. Does food restriction retard aging by reducing the metabolic rate? The American Journal of Physiology. 1985;248(4):E488–490. doi: 10.1152/ajpendo.1985.248.4.E488. [DOI] [PubMed] [Google Scholar]
- 86.Yen K, Mastitis JW, Mobbs CV. Lifespan is not determined by metabolic rate: evidence from fishes and C. elegans. Experimental Gerontology. 2004;39(6):947–949. doi: 10.1016/j.exger.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 87.Drew B, Dirks PA, Selman C, et al. Effects of aging and caloric restriction on mitochondrial energy production in gastrocnemius muscle and heart. American Journal of Physiology. 2003;284(2):R474–R480. doi: 10.1152/ajpregu.00455.2002. [DOI] [PubMed] [Google Scholar]
- 88.Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. The FASEB Journal. 2004;18(3):595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- 89.Lee YS, Choi JY, Park MK, Choi EM, Kasai H, Chung MH. Induction of oh8Gua glycosylase in rat kidneys by potassium bromate (KBrO3), a renal oxidative carcinogen. Mutation Research. 1996;364(3):227–233. doi: 10.1016/s0921-8777(96)00038-9. [DOI] [PubMed] [Google Scholar]