Abstract
Translocase mediates preprotein translocation across the Escherichia coli inner membrane. It consists of the SecYEG integral membrane protein complex and the peripheral ATPase SecA. Here we show by functional assays, negative-stain electron microscopy and mass measurements with the scanning transmission microscope that SecA recruits SecYEG complexes to form the active translocation channel. The active assembly of SecYEG has a side length of 10.5 nm and exhibits an ∼5 nm central cavity. The mass and structure of this SecYEG as well as the subunit stoichiometry of SecA and SecY in a soluble translocase–precursor complex reveal that translocase consists of the SecA homodimer and four SecYEG complexes.
Keywords: preprotein translocation/SecA/SecY/translocase
Introduction
Escherichia coli translocase consists in its minimal form of three integral membrane proteins termed SecY, SecE and SecG, and a peripheral ATPase SecA (Brundage et al., 1990; Hanada et al., 1994). SecY and SecE form a stable stoichiometric complex in the cytoplasmic membrane that does not dissociate in vivo (Matsuyama et al., 1990; Joly et al., 1994). SecYE is purified from the cytoplasmic membrane as a soluble complex together with the SecG protein. SecG strongly stimulates the in vitro translocase activity (Nishiyama et al., 1993) and, therefore, the SecYEG complex is used for the reconstitution of the translocation reaction in proteoliposomes (Hanada et al., 1994; Douville et al., 1995; van der Does et al., 1998). SecA is a homodimeric protein that serves both as a receptor for precursor proteins and as an ATP-driven molecular motor during the translocation reaction (Driessen et al., 1998; Economou, 1998). SecA binds to the cytoplasmic membrane with a low affinity for phospholipids and a high affinity for the SecYEG complex (Hartl et al., 1990; Hendrick and Wickner, 1990). It binds to SecYEG, at least partially, via a direct interaction with SecY (Manting et al., 1997; Snyders et al., 1997). Upon binding to SecYEG, SecA is activated for precursor-stimulated cycles of ATP binding and hydrolysis (Lill et al., 1990). This process permits the stepwise movement of a translocating polypeptide chain across the membrane (Schiebel et al., 1991) by a two-stroke reaction (van der Wolk et al., 1997). Translocation is stimulated further by the presence of a proton-motive force, which ensures unidirectionality of the translocation reaction and facilitates the SecA reaction cycle (Driessen, 1992; Nishiyama et al., 1999).
SecA is a highly dynamic protein. Calorimetric studies of the nucleotide-modulated structural changes of the soluble SecA molecule demonstrate that it exists in a compact, ADP-bound state and an extended, ATP-bound state (den Blaauwen et al., 1996). During translocation, SecA may insert into the cytoplasmic membrane with both a 30 kDa C–terminal domain and a 65 kDa N–terminal domain (Economou et al., 1994; Price et al., 1996; Eichler and Wickner, 1997). SecYEG appears to form a sufficiently large transmembrane structure, which accommodates at least a part of SecA together with a translocating precursor protein, shielding both from phospholipids (Joly and Wickner, 1993; Eichler et al., 1997; van Voorst et al., 1998). The Bacillus subtilis SecYE complex and the homologous eukaryotic Sec61p complex have been visualized by electron microscopy as quasi-pentagonal structures with a diameter of 8.5 nm and a central cavity with a diameter of 1.5–2 nm (Hanein et al., 1996; Meyer et al., 1999). Beckmann et al. (1997) have reported the three-dimensional structure of the ribosome-bound Sec61p at 2.6 nm resolution. It is a 4 nm thick disk with a diameter of 9.5 nm, containing a 1.5–3.5 nm wide central pore.
In this study, we determined the oligomeric nature of the E.coli SecYEG complex. The majority of purified E.coli SecYEG forms dimers that resemble the previously reported B.subtilis SecYE structure (Meyer et al., 1999). However, membrane insertion of SecA triggers the assembly of SecYEG into a larger complex, consisting of four SecYEG subunits. The subunit stoichiometry of a soluble translocase–preprotein complex revealed that translocase contains four SecY molecules per SecA dimer. We propose that SecA recruits and assembles four SecYEG complexes to form the translocase.
Results
SecYEG is solubilized as dimers and monomers
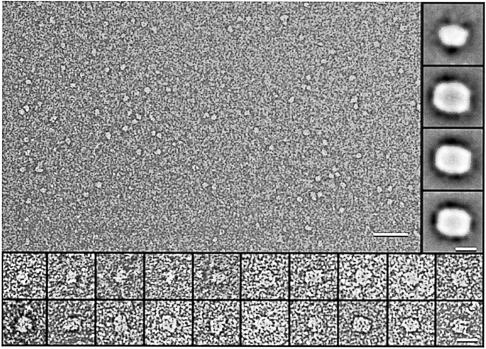
Translocase was studied in a purified system using proteoliposomes reconstituted with SecYEG (van der Does et al., 1998). First, SecYEG was re-solubilized in dodecyl maltoside without prior incubation with the other components of the translocation reaction. Excess phospholipids were removed from the soluble SecYEG complex by anion-exchange chromatography, without a detectable loss of SecYEG activity (data not shown; see Materials and methods). The complex was then visualized using negative-stain electron microscopy (EM). In uranyl formate-stained preparations, the SecYEG complex was visible as an elongated structure with a length of 8–9 nm and a central stain-filled indentation (Figure 1). Single particle analysis and averaging of the 8 nm structures resulted in classes of structures that resembled each other in shape and size (Figure 1, right inserts). Smaller particles were discerned as well, but their structural features were less distinct. The analysis of the small particles yielded an average of 6.8 nm length and 4.5 nm width (top right inset in Figure 1). The remarkable similarity of the three major class averages (Figure 1, inserts) suggests that SecYEG binds to the supporting carbon EM grids in a specific orientation, probably representing a top or bottom view of the complex. Its width and length estimated from the class averages are 6.7 and 8.7 nm, respectively. From the 2-fold symmetry in the particles and the particle size, we conclude that the structure of soluble SecYEG represents an assembly of two SecYEG subunits (see also Table I). The smaller particles that were observed indicate that part of the SecYEG dimers dissociated into monomers due to the solubilization and purification procedure.
Fig. 1. Structural analysis of soluble SecYEG. Electron microscopy of negatively stained SecYEG complexes solubilized in dodecyl maltoside reveals particles of two distinct sizes (overview, scale bar = 50 nm). While particles with a size of ∼5 nm lack distinct structural features, the larger particles of 8–9 nm are elongated, sometimes having a central stain-filled pit (bottom inserts, scale bar = 10 nm). Single particle analysis and class averaging yield projection maps with a width of 6.7 nm, a length of 8.7 nm and a more or less pronounced 2 nm wide stain-filled indentation with all three major classes, containing ∼450 particles each (right inserts, scale bar = 5 nm). The major class average of the smaller particles is shown in the top right insert and has a width of 4.5 nm and a length of 6.8 nm. It is close to half an elongated particle.
Table I. Size and interpretation of the recurring peaks in STEM mass analysis.
| Peak positiona | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| SecYEG | (SecYEG)2 | (SecYEG)4 | (SecYEG)2 SecA proOmpA | (SecYEG)4 SecA proOmpA | |
| Proteinb | 75 | 150 | 300 | 385 | 535 |
| Micellec | 75 | 100 | 150 | 150 | 150 |
| SecYEG | 176 ± 32 | 268 ± 32 | – | – | – |
| SecYEG after pre-incubation with SecA + AMP–PNP | 183 ± 57 | 289 ± 57 | 421 ± 57 | – | – |
| SecYEG, SecA + ATP, proOmpA | ND | 276 ± 48 | 416 ± 48 | 531 ± 48 | 686 ± 48 |
| Calculatedd | 151 | 287 | 477 | ND | ND |
ND, not determined.
aPeak positions relate to the STEM analysis shown in Figure 7.
bCalculated from the molecular masses in kilodaltons of the indicated proteins.
cThe amount of dodecyl maltoside micelle bound to the purified SecYEG complex was determined to be 100 kDa as described in Materials and methods, and found to be 100 kDa for the SecYEG dimer. The other values are estimates.
dCalculated masses of the SecYEG dimer and tetramer were determined by their surface observed in negative-stain EM after single particle analysis (349 nm2 for the dimer and 582 nm2 for the tetramer), a height of 6 nm, and a density of 1.35 g/cm3, with no correction for the putative pore.
Membrane-inserted SecA triggers tetramerization of SecYEG
In the next experiment, SecYEG proteoliposomes were incubated with SecA in the presence of AMP–PNP. This non-hydrolysable ATP analogue triggers membrane insertion of the SecA protein even in the absence of preprotein (Economou et al., 1994). Proteoliposomes were then collected by centrifugation, solubilized in dodecyl maltoside solution and subjected to anion-exchange chromatography. The His-tagged SecYEG complex elutes at the very beginning of the salt gradient (van der Does et al., 1998) and was separated easily from SecA and excess phospholipids. After incubation with membrane-inserted SecA, a significant fraction of SecYEG was visible in negative-stain EM as large, 10–12 nm sized structures (Figure 2). Incubation of SecYEG proteoliposomes with SecA in the absence of AMP–PNP did not result in any change in shape or size of SecYEG, as observed with B.subtilis SecYE (Meyer et al., 1999) (see also Figure 3A). Owing to the bi-directional reconstitution of SecYEG into proteoliposomes (van der Does et al., 1998), ∼50% of the total SecYEG was inactive. Therefore, an inhomogeneous particle distribution emerged, with a major fraction of dimeric SecYEG and the larger particles comprising ∼30% of the number of particles (Figure 2, see also Figure 7 and Table I). The large SecYEG particles triggered by membrane-inserted SecA often exhibited a square shape and a stain-filled central cavity. Class averages of these square-shaped structures had a side length of 10.5–12 nm and a 5 nm wide central stain-filled depression (Figure 2, right inserts). The size of these structures suggests that SecYEG had assembled into a tetrameric structure upon interaction with membrane-inserted SecA. As with the dimeric SecYEG, the major class averages of the SecYEG tetramer resembled each other in size and shape and appeared to represent top or bottom views. The size difference in the class averages, however, suggests a slightly asymmetrical particle, with a 10.5 nm projection and a 12 nm projection (Figure 2, right inserts).
Fig. 2. Formation of large SecYEG structures after incubation with membrane-inserted SecA. SecYEG proteoliposomes were incubated with SecA and AMP–PNP, solubilized and subjected to anion-exchange chromatography. The fraction containing SecYEG was visualized by negative-stain EM, revealing a heterogeneous size distribution (overview, scale bar = 50 nm). A significant fraction of particles had a size of 10–12 nm, a 4–6 nm central stain-filled cavity and a squarish shape (bottom inserts, scale bar = 10 nm). Major class averages (200–400 particles) were generated by single particle analysis (right inserts, scale bar = 5 nm).
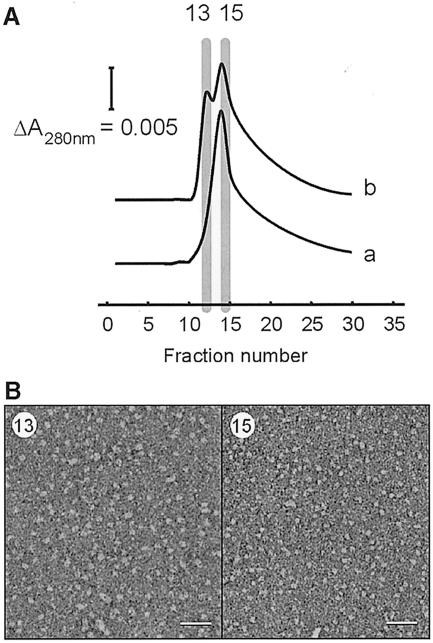
Fig. 3. Separation of small and large SecYEG complexes by Ni2+ affinity chromatography. (A) SechisYEG proteoliposomes were incubated with SecA and AMP–PNP, and solubilized with dodecyl maltoside. The material was loaded onto an Ni2+–NTA column and eluted with a sharp imidazole gradient. SecYEG complex that was solubilized directly eluted as a single peak, while the SecYEG incubated with SecA and AMP–PNP contained a second peak at lower imidazole strength. SDS–PAGE and silver staining revealed that these fraction contained SecYEG protein only. SecA protein did not bind to the Ni2+–NTA column, and was found in the flow-through fraction. (B) Peak fractions of the SecYEG proteoliposomes incubated with SecA and AMP–PNP were visualized by negative-stain EM. The early eluting SecYEG fraction (13) contained particles with a size range of 10–12 nm (left panel), while the other fraction (15) contained the smaller particles of 5–9 nm (right panel) (scale bar = 50 nm).
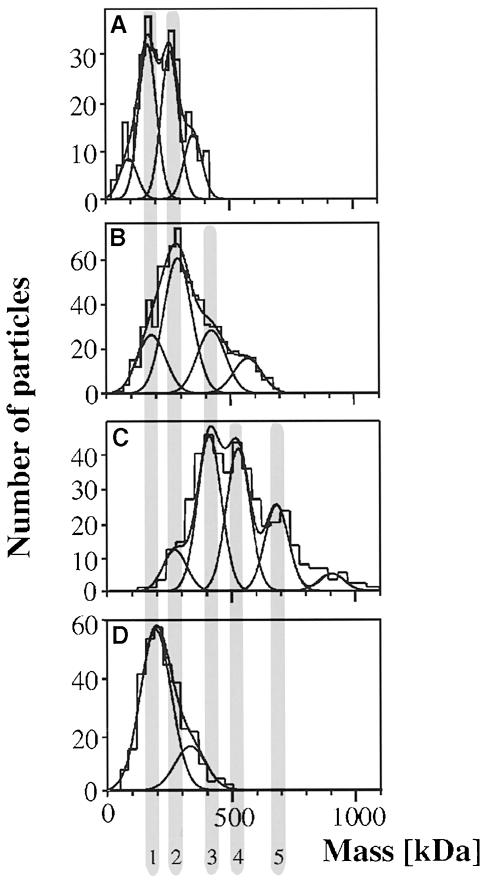
Fig. 7. Mass analysis. The scanning transmission electron microscope (STEM) allows mass analysis of heterogeneous particle mixtures. Unstained preparations prepared by freeze–drying were recorded and evaluated as described (Müller and Engel, 1998). (A) Solubilized SecYEG reveals two major peaks [at 176 (peak 1) and 268 kDa (peak 2)] and two minor peaks at the left and right sides of the histogram. (B) SecYEG after incubation with SecA and AMP–PNP demonstrates a striking shift of the particle mass. The three major peaks are centred at 183 kDa (peak 1), 289 kDa (peak 2) and 421 kDa (peak 3). The peak at 580 kDa includes ∼10% of all particles measured. (C) Sucrose gradient fractionation of solubilized proteoliposomes after the accumulation of the I31 translocation intermediate reveals a further shift towards larger complexes. The histogram includes particles larger than 10 nm. The three major peaks are centred at 416 kDa (peak 3), 531 kDa (peak 4) and 686 kDa (peak 5), with a minor peak at 276 kDa (peak 2). (D) Mass analysis of the same preparation, but of particles <10 nm. The major Gauss peak is located at 200 kDa (peak 1), the minor peak at 330 kDa (peak 2).
To obtain more insight into the distribution of the various SecYEG complexes, solubilized proteoliposomes pre-treated with SecA and AMP–PNP were subjected to alternative chromatography. Ni2+–NTA affinity purification resulted in the elution of two distinct fractions containing SecYEG protein (Figure 3A). The SecYEG-containing fractions were examined further by negative-stain EM (Figure 3B). The early fraction contained the largest oligomer, while the more tightly bound fraction contained the dimeric and monomeric SecYEG species. SecA protein was not retained on the Ni2+–NTA, and was found to elute in the non-bound fraction.
Formation of a soluble translocase–precursor complex
To determine the number of SecY molecules in translocase, a stable translocation intermediate was established in SecYEG proteoliposomes complemented with SecA and ATP. The precursor of outer membrane protein A, proOmpA, translocates efficiently in this system (Brundage et al., 1990; Figure 4A, lanes 1–4). Under oxidizing conditions, however, proOmpA translocates into the SecYEG proteoliposomes as an intermediate with an apparent molecular mass on SDS–PAGE of 31 kDa (Figure 4A, lanes 6–9). This translocation intermediate, termed I31, resembles the processed I29 intermediate formed in inner membrane vesicles due to a disulfide bond between the unique cysteine residues C290 and C302 of proOmpA (Tani et al., 1990; Schiebel et al., 1991). The I29 intermediate is associated with SecA and is unstable at 37°C upon depletion of ATP, reduction of the disulfide bond or the addition of antibodies against SecA, but is completely stable at low temperature (Schiebel et al., 1991).
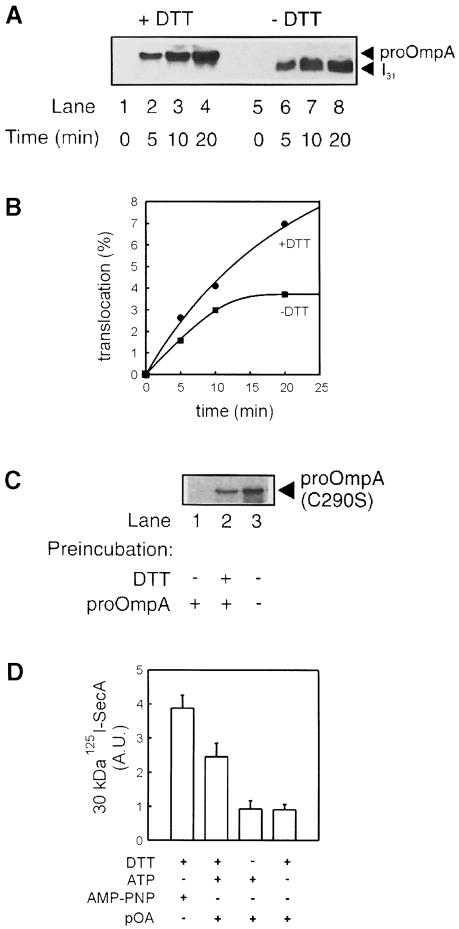
Fig. 4. Characterization of the I31 intermediate. (A) Translocation of proOmpA (lanes 1–4) or translocation intermediate I31 (lanes 5–8). Reconstituted SecYEG (10 μg/ml) was mixed with SecA (60 μg/ml) and proOmpA (20 μg/ml) in the presence or absence of 10 mM DTT. Reactions were energized with 2 mM ATP, incubated at 37°C for the time indicated, chilled on ice and digested with protease K (0.1 mg/ml) for 15 min. Accumulation of translocated preprotein was analysed by Western blotting using monoclonal antibodies against proOmpA. (B) Densitometric quantitation of the data in (A). (C) I31 blocks active translocation sites. Proteoliposomes that had undergone translocation of I31 (lane 1), proOmpA (lane 2) or no translocation (lane 3) were tested for their ability to support a second round of translocation. Reactions were performed for 20 min in the absence of DTT similarly to those in (A), except that proOmpA was replaced by 35S-labelled single-cysteine proOmpA (C290S). (D) I31 does not support formation of the 30 kDa SecA fragment. Translocations were performed with 125I-labelled SecA and in the presence or absence of 10 mM DTT, proOmpA, 2 mM AMP–PNP or 2 mM ATP, as indicated. After 20 min incubation at 37°C, or with AMP–PNP at 4°C, reactions were digested with protease K. The 30 kDa SecA fragment was visualized using autoradiography and quantitated by densitometry. The averages and error margins of two experiments are shown as arbitrary densitometric units.
I31 reached its maximum level within 15 min of incubation at 37°C, while the translocation reaction in the presence of dithiothreitol (DTT) continued nearly linearly up to at least 20 min (Figure 4B). To confirm that I31 occupied all active translocation sites, proteoliposomes bearing I31 and proteoliposomes that had undergone complete translocation cycles were harvested and tested for a second round of translocation. SecYEG proteoliposomes that had not been pre-incubated with proOmpA were used as controls. For this round of translocation, we used [35S]methionine-labelled (C290S)proOmpA, a site-directed single cysteine mutant that is unable to form an intramolecular disulfide bond and translocates completely under both reducing and oxidizing conditions. Proteoliposomes that had accumulated I31 were inactive for proOmpA translocation (Figure 4C, lane 1), whereas those that underwent full translocation were nearly as active as control proteoliposomes not incubated with preprotein (lanes 2 and 3). Since I31 quantitatively occupies the translocation sites and the accumulation level of fully translocated proOmpA exceeds that of I31 (Figure 4B), SecYEG proteoliposomes must allow multiple rounds of translocation. This is supported by their activity after their re-isolation from a previous translocation reaction. Membrane insertion of SecA in the presence of proteoliposomes was assayed using 125I-labelled protein. As expected, a proteolytically stable 30 kDa fragment was either formed during translocation or with AMP–PNP. In contrast, I31-bound SecA yielded only background levels of the 30 kDa fragment, similar to a control experiment where ATP was omitted. This indicates that I31 retains SecA in a deinserted state (Figure 4D).
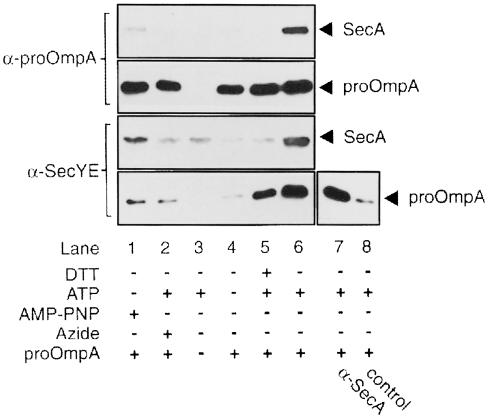
The requirements for a solubilized translocase–precursor complex were established using immunoprecipitation. SecYEG proteoliposomes were incubated with SecA and proOmpA under various conditions, as described below. They were then collected and, after solubilization, the interactions between the SecYEG complex, SecA and proOmpA were determined by co-immunoprecipitation (Figure 5). AMP–PNP (Economou and Wickner, 1994) or the ATPase inhibitor sodium azide (van der Wolk et al., 1997) enforce SecA membrane insertion. However, neither condition gave rise to a translocase–precursor complex that is stable in micellar solution, as only minute amounts of SecA were immunoprecipitated with anti-SecYE or anti-OmpA serum (lanes 1 and 2). After incubation under conditions that resulted in the completion of translocation, some proOmpA but no SecA remained associated with the SecYEG complex upon solubilization (lane 5). This may represent an incomplete release of proOmpA after translocation, due to the absence of the accessory translocase subunit SecDF (Matsuyama et al., 1993) or the proton-motive force (Geller, 1990). A marked increase in the amount of immunoprecipitable proOmpA was observed when the translocation reaction was performed in the absence of DTT to yield I31 (lane 6). In addition, SecA co-immunoprecipitated with SecYE and proOmpA, whereas in control samples lacking proOmpA or ATP no immunoprecipitation of SecA was observed (lanes 3 and 4). In the reciprocal experiment, proOmpA was immunoprecipitated by anti-SecA (lane 7). These data establish that the translocation intermediate I31 is sufficiently stable to allow its solubilization.
Fig. 5. Immunoprecipitation of soluble translocase–precursor complexes. Translocation reactions were incubated for 20 min in the presence or absence of DTT, 2 mM ATP, 2 mM AMP–PNP, or 2 mM ATP together with 10 mM NaN3, as indicated. Solubilized proteoliposomes were incubated with protein A–Sepharose beads coated with antiserum against OmpA (top panels), SecYE (bottom panels), SecA (lane 7) or no antiserum (control, lane 8), and were precipitated. Precipitates were eluted and analysed by Western blotting using monoclonal antibodies against SecA or proOmpA as indicated.
Translocase contains four SecY molecules per SecA dimer
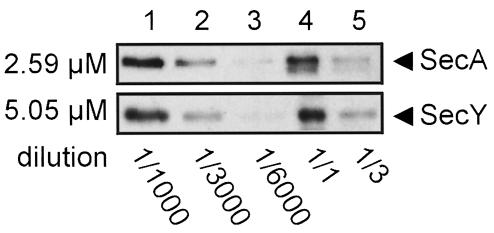
The purification of a translocation intermediate via antibodies against the precursor offered the possibility to determine the amount of SecA and SecY in translocase by quantitative immunoblotting. First, SecY and SecA concentration standards were prepared. His6-tagged SecY was purified from the soluble SecYEG complex via Ni2+–NTA chromatography (Manting et al., 1997). Amino acid analysis of this SecY sample determined its concentration as 5.05 ± 0.05 μM, on the basis of 10 amino acids in a duplo analysis. A SecA standard with a concentration of 2.58 ± 0.03 μM was used as a comparison. To obtain purified translocase, we immunoprecipitated the translocase–I31 complex with antibodies against OmpA. The amounts of SecA and SecY associated with the translocation intermediate were then analysed by quantitative Western blotting, using dilution series of SecA and SecY as standards (Figure 6). The SecY:SecA molar ratio in the precipitated samples was determined to be 2.0 ± 0.3 (n = 6) by densitometry on films from chemoluminescent blots. As SecA is a dimeric molecule (Akita et al., 1991; Driessen, 1993), the SecY:SecA ratio implies a tetrameric organization of SecY in translocase.
Fig. 6. The SecY:SecA ratio in I31-bound translocase. Translocase was immunoprecipitated via I31 using NHS–Sepharose coated with IgG against OmpA. Samples incubated without proOmpA were used as negative controls to verify the specificity of the immunoprecipitation. The amounts of SecA and SecY in the immunoprecipitate were determined by densitometry, using purified SecA (2.6 μM) and SecY (5.1 μM) as standards. Dilutions of the immunoprecipitate (lanes 4 and 5) were compared with dilutions of the protein standards (lanes 1 and 2) that gave similar intensities after immunostaining, taking the average as the outcome of one experiment. Three immunoprecipitates were compared in this manner with two independent dilution series of the protein standards, yielding a total of six experiments in which the SecY:SecA ratio was determined as 2.0 ± 0.3.
Mass analysis of solubilized SecYEG and translocase–I31
To substantiate the structural and biochemical data on the different oligomeric states of the SecYEG complex, scanning transmission electron microscopy (STEM; Müller et al., 1992) was employed to determine the particle masses in all three cases described above. Proteoliposomes were solubilized directly, or pre-incubated with SecA and AMP–PNP, then solubilized and subjected to anion-exchange chromatography that removed SecA. Proteoliposomes that had accumulated I31 were solubilized and subjected to sucrose gradient density centrifugation to remove excess phospholipids.
Samples were freeze-dried after adsorption to thin carbon film and extensive washing by double-distilled water. Particle masses were evaluated from dark-field micrographs recorded at a dose of typically 300 electrons/nm2. Figure 7 shows a striking upshift of the mass values observed with solubilized SecYEG (Figure 7A), SecA- and AMP–PNP-treated SecYEG (Figure 7B) and the translocase–I31 complexes (Figure 7C). The histogram in Figure 7D shows the mass histogram of all small particles present in the translocase–I31 preparation. Distinct peaks emerged reproducibly in the mass histograms, as summarized in and interpreted in Table I. Although the corresponding Gaussian profiles in Figure 7A–C had the same position within the error limits, their standard deviation varied between 32 and 57 kDa, because the background signal was different in different preparations. To interpret these masses, they were compared with the particle masses estimated from the average particle projections, and masses calculated from the assumed stoichiometry of the protein complexes. Soluble SecYEG that had not been incubated with translocation ligands was present in equal amounts as monomers and dimers, demonstrating that the majority (∼65%) of SecYEG is organized as a dimer (Figure 7A). The minor shoulders on both sides of this histogram indicated two further particle types, but the corresponding mass values cannot be related to distinct oligomers.
Importantly, the STEM analysis confirms the tetrameric assembly of the SecYEG that was pre-incubated with SecA and AMP–PNP (Figure 7B). These particles, with a mass of ∼420 kDa, were also observed in sucrose gradient fractions prepared from proteoliposomes that had been incubated with proOmpA and ATP under oxidizing conditions (Figure 7C). Two other classes of large particles were observed in these preparations. Particles with a mass of 686 kDa are consistent with the presence of translocase containing the I31 translocation intermediate. Another particle mass of 531 kDa represents a complex that was not observed with the samples containing purified SecYEG (Figure 7C). As SecA binds efficiently to soluble SecYEG (van der Does et al., 1998), we propose that these complexes consist of SecA together with the precursor and unassembled, dimeric SecYEG. Minor peaks required to fit the histograms in Figure 7B–D represent ∼10% of the measurements and were not assigned.
The particle masses observed in STEM analysis corroborate the apparent increase in size of the SecYEG complex after incubation with SecA and AMP–PNP observed in negative-stain EM. The masses and structures of the soluble SecYEG complexes before and after such incubation suggest that SecYEG dimers assemble into a tetramer upon interaction with membrane-inserted SecA. The mass of the soluble translocase–I31 particle and the presence of subcomplexes of translocase in solubilized proteoliposomes containing I31 support the SecA-mediated assembly of four SecYEG complexes in translocase.
Discussion
This study demonstrates that E.coli translocase consists of the dimeric SecA ATPase and a channel structure composed of four SecYEG complexes. A remarkable enzymological event underlies formation of the SecYEG tetramer: SecYEG complexes are recruited by the SecA protein and assembled upon SecA membrane insertion. The tetrameric organization of active SecYEG elucidates how the complex provides a structure that allows ATP-driven membrane insertion of SecA domains and the concomitant translocation of preproteins. The recruitment of additional subunits also explains the plasticity observed with the protein-conducting channel in the mammalian endoplasmic reticulum (ER) membrane, which increases its pore size during translocation (Hamman et al., 1997, 1998). It is conceivable that SecA covers the pore in SecYEG, thereby preventing the non-specific passage of macromolecules and ions through the translocation channel.
We have used proteoliposomes reconstituted with SecYEG as a minimal system to study SecA- and ATP-driven translocation. SecG is a specific subunit of the bacterial translocase, and its function appears to be related to that of SecA. The membrane insertion and deinsertion of SecA are accompanied by topology inversions of the highly hydrophobic SecG, which facilitates the SecA reaction cycle (Nishiyama et al., 1996). Our studies do not include SecD, SecF and the uncharacterized YajC protein, all of which form a complex that interacts with SecYE (Sagara et al., 1994; Duong and Wickner, 1997). SecG and SecDF(YajC) have apparent complementary functions, but SecYEG is the most abundant SecYE-containing complex present in the cytoplasmic membrane (Pogliano and Beckwith, 1994; Duong and Wickner, 1997). Purified SecYEG suffices to reconstitute translocation sites that allow multiple rounds of translocation mediated by SecA and ATP (Figure 3B; Bassilana and Wickner, 1993). Finally, Duong and Wickner (1998) have shown that this complement of translocase subunits also permits the integration of hydrophobic transmembrane segments into the lipid bilayer.
Solubilized SecYEG exists as monomers and dimers, the latter representing a majority having a mass of ∼280 kDa, including detergent. This value is the average of three independent STEM measurements and the mass estimated from the size of negatively stained particles (Table I). We suspect that part of the SecYEG dimers have dissociated during purification. Alternatively, SecYEG may be present in the membrane in equilibrium between its monomeric and dimeric form. Upon interaction with SecA and AMP–PNP, a substantial amount (30%) of SecYEG had assembled into tetramers, with a measured mass of 420 kDa (two independent experiments) and a calculated mass of 477 kDa (Table I). The latter value is an overestimate, because the central indentation in the SecYEG tetramers has not been accounted for. These particles were also observed in the STEM analysis of solubilized proteoliposomes that were pre-incubated with oxidized proOmpA and ATP. In addition, the proteoliposomes harboured translocase–precursor complexes with a mass of 686 kDa, and smaller 530 kDa complexes probably consisting of SecA and dimeric SecYEG. SecYEG tetramers and monomeric SecA may also account for the latter particle, but this is unlikely as SecA functions as a dimer and does not dissociate during translocation (Driessen, 1993). In our calculations of the protein masses (Table I), we presume a stoichiometric organization of SecY, SecE and SecG within each SecYEG subunit, making it a 75 kDa protein complex. SecY and SecE form a stable complex that does not dissociate in the cytoplasmic membrane (Joly et al., 1994). From their interdependent stability and similar expression levels in the cytoplasmic membrane (Matsuyama et al., 1990; Pogliano and Beckwith, 1994), as well as their defined regions of interaction (Baba et al., 1994; Flower et al., 1995), it is apparent that they interact stoichiometrically. SecG is associated somewhat loosely with the SecYE complex (Joly et al., 1994), and may therefore be present in sub- or super-stoichiometric amounts to SecYE.
The soluble E.coli SecYEG dimer was visible in negative-stain EM mainly as a 2-fold symmetrical, elongated particle, but smaller particles were present as well. Averages of both particle types were calculated and compared. The smaller particle (top right inset in Figure 1) had the dimensions (4.5 × 6.8 nm) of half the larger particle (6.7 × 8.7 nm; right inset in Figure 1), consistent with the mass measurements. The central position of their stain-filled depression in the larger particle indicates that the structures represent a top or bottom view of the SecYEG complex. The resolution obtained by single particle analysis and class averaging approximates 2 nm, and is insufficient to allow a more detailed analysis of the structural organization and subunit assembly within the SecYEG dimer. The reported structure of B.subtilis SecYE resembles that of the SecYEG dimer, but has a semi-pentagonal appearance (Meyer et al., 1999). The pronounced shape of the B.subtilis complex may be the result of the biased particle selection that was used to obtain the averaged images, different fixation of the protein samples or the absence of the SecG subunit. Although translocation in B.subtilis is also enhanced by SecG (Swaving et al., 1999; van Wely et al., 1999), the subunit is apparently not required for formation of the SecYE dimer. This is consistent with data obtained in E.coli, which indicate that SecYE is sufficient to support translocation, albeit inefficiently (Duong and Wickner, 1997).
The active conformation of the preprotein-conducting channel is not the SecYEG dimer, but is represented by a larger structure consisting of a SecYEG tetramer. This square-shaped particle had a side length of 10.5–12 nm (Figure 2). First, it was formed upon interaction with membrane-inserted SecA, and not with SecA without activation with AMP–PNP. Secondly, it was present in proteoliposomes that had been incubated with all the components to produce complete translocation reactions. Thirdly, biochemical analysis of the SecY:SecA ratio in a soluble translocase–precursor complex demonstrated that SecY is organized as a tetramer. We propose a two-step model for the formation of translocase in which the SecA dimer first binds two SecYEG dimers, thereby bringing them together, and subsequently enforces their stable assembly into a functional channel upon membrane insertion. Speculatively, SecG and SecDF may influence the formation of this active channel by promoting the stability of the tetramer. The assembly of translocase via a complex between SecA and dimeric SecYEG is supported by the putative of such protein complexes formation under translocation conditions. The ability of SecA to bind unassembled SecYEG is also apparent from results obtained in detergent solution (van der Does et al., 1998). We have also employed negative-stain EM for the sucrose gradient fractions containing translocase–I31. However, due to a lack of preferred orientation of the protein particles on the EM grid and the large heterogeneity of the samples, we were unable to obtain averaged images of these complexes.
The eukaryotic Sec61p complex of the ER membrane is homologous to SecYEG (Hartmann et al., 1994; Matlack et al., 1998). Purified Sec61p has a closed ring structure and a central pore or indentation with a diameter of 1.5–2 nm, and these structures are also observed in the ER membrane (Hanein et al., 1996). When bound to the ribosome, the central cavity of Sec61p aligns the putative nascent chain-conducting channel in the large ribosomal subunit (Beckmann et al., 1998). The size of the protein-conducting channel in the mammalian ER membrane is dramatically larger (4–6 nm) during translocation than the putative pore observed in structural analysis of Sec61p in the absence of nascent chains (Hamman et al., 1997). It was therefore suggested that the Sec61p structure does not represent the active protein-conducting channel. Indeed, biophysical studies with microsomal membranes confirm the presence of two distinct populations of Sec61p complexes, with a pore size of either 0.9–1.5 or 4–6 nm. The latter is formed only upon incubation with ribosome–nascent chain complexes and collapses after release of the nascent polypeptide (Hamman et al., 1998). Our results with the purified and reconstituted SecYEG channel support a model in which the large ‘active’ channels are formed via recruitment and assembly of small subcomplexes.
Both the dimeric and tetrameric SecYEG complexes will be deleterious to the integrity of the cytoplasmic membrane as a chemo-osmotic barrier. In the ER membrane, the lumenal side of the Sec61p channel is closed by the BiP ATPase. BiP remains bound to Sec61p after ribosomal targeting and channel opening, but is released when translocating nascent chains have reached a length of 70 amino acids or more (Hamman et al., 1998). In analogy, the SecA molecule may be involved in closing the pore of the SecYEG complex, either by binding at the cytosolic side or through the insertion of specific domains, or both. The presence of SecA during the translocation reaction may limit the effective pore size of the protein-conducting channel, as the small disulfide-bonded loop of amino acids in oxidized proOmpA is apparently sufficiently large to block the translocase. In addition to SecA, the SecDF complex may contribute to the maintenance of the proton-motive force during translocation (Arkowitz and Wickner, 1994). The SecA membrane topology appears complex and its integral membrane state is not dependent on translocation (Kim et al., 1994; Chen et al., 1996; van der Does et al., 1996; Ramamurthy and Oliver, 1997). Topology studies on SecA may be obscured because the SecA protein is part of a large transmembrane structure. It may be accessible for chemical probes or proteases added from the periplasmic side of the membrane via the proteinaceous channel constituted by SecYEG. To understand this complex nanomachine, the crystal structure of the complex formed by SecYEG and SecA is required. Although our experiments did not yield immediate structural information, the stabilization of translocase via a translocation intermediate is an important advance towards the purification, characterization and crystallization of this complex. In addition, it will be important to establish the thermodynamics of the translocation reaction and the surface topography of the SecYEG-bound SecA during the various stages of the translocation reaction. A major question also lies in the structural and mechanistic features of the protein-conducting channel during the integration of membrane proteins.
Materials and methods
Materials
ProOmpA (Crooke et al., 1988), SecB (Weiss et al., 1988) and SecA (Cabelli et al., 1988) were purified as previously described. A plasmid encoding (C290S)proOmpA was constructed by PCR starting from plasmid pET149 (van der Wolk et al., 1997) using the mutagenesis primer 5′-GGCAACACCTCTGACAACGTG-3′. The resulting plasmid pET503 was used for the synthesis of 35S-labelled (C290S)proOmpA with an in vitro transcription–translation reaction and immunopurified as described (van der Wolk et al., 1997). His6-tagged SecYEG was purified and reconstituted into proteoliposomes as described by van der Does et al. (1998). Rabbit polyclonal antisera against SecY, SecE, OmpA, SecA and SecB were raised against the purified proteins at the animal facility of the Department of Chemistry, University of Groningen, The Netherlands. The antisera against SecY or SecE were raised against the His6-tagged proteins, and contained specific cross-reactivity with His6 tags on other proteins. Monoclonal antibodies against OmpA were raised and selected by Professor Dr L.de Leij, Academic Hospital Groningen. SecA was detected with a mixture of monoclonal antibodies (oligoclonal) (den Blaauwen et al., 1997). Protein samples were analysed by SDS–PAGE using 12–15% acrylamide gels, followed by Western blotting or silver staining (Bio-Rad, Hercules, CA). Western blots were developed as films using chemoluminescence (Tropix, Bedford, MA). For densitometry, a Dextra DF-2400T scanner (Dextra Technology Corp., Taipei, Taiwan) and SigmaScan/Image Software (Jandel Corp., San Rafael, CA) were used. Protein A– or N-hydroxy-succinimide(NHS)– Sepharose beads were from Pharmacia (Uppsala, Sweden), dodecyl maltoside and octyl glucoside were from Sigma (St Louis, MO), and E.coli phospholipids were from Avanti polar lipids (Alabaster, AL).
Translocation reactions
Translocation reactions were performed with 6.0 μg of SecA, 2.0 μg of urea-denatured proOmpA, 1.0 μg of reconstituted SecYEG (8–10 μl of proteoliposomes) and 2 mM ATP in 100 μl of translocation buffer consisting of 50 mM HEPES–KOH pH 7.6, 100 mM KCl and 5 mM MgAc2. For oxidation or reduction of proOmpA, complete reaction mixtures were pre-incubated for 10 min on ice in the absence or presence of 10 mM DTT. Reactions were started by placing them at 37°C and stopped by chilling on ice, followed by digestion with protease K, trichloroacetic acid (TCA) precipitation and a washing step with ice-cold acetone (van der Wolk et al., 1997). For densitometric quantitation of translocation, bands representing accumulated proOmpA or I31 were compared with 5 and 10% of a reaction mixture that was not digested with protease K, as a standard. For a second round of translocation, proteoliposomes were collected by centrifugation (Beckman Airfuge, 30 p.s.i., 10 min) and resuspended in ice-cold translocation buffer. Translocation reactions were then performed as described above, except for the proOmpA, which was replaced by 2 μl of urea-denatured 35S-labelled (C290S)proOmpA.
Immunoprecipitations
Antibodies (20 μl) were mixed with 10 μl of protein A–agarose suspension in a total volume of 200 μl with buffer S, consisting of 50 mM Tris–HCl pH 8.0, 50 mM KCl, 20% (w/v) glycerol and 0.05% (w/v) dodecyl maltoside. Mixtures were incubated for 1.5 h at 4°C and the beads were then washed with 0.5 ml of buffer S (Eppendorf centrifuge, 14 000 r.p.m., 3 min). Isolated proteoliposomes from two translocation reactions were solubilized with 2% (w/v) dodecyl maltoside in buffer S (200 μl) and mixed with the protein A-bound antibodies for 1 h at 4°C. Beads were then washed extensively with buffer S and bound proteins were eluted with SDS sample buffer (10 min, 50°C). Samples were analysed on Western blots using mouse monoclonals as primary antibodies to avoid cross-reaction of the secondary antibody with the rabbit IgG used for the precipitation. Alternatively, purified IgG was covalently coupled to NHS–Sepharose, according to the manufacturer's recommendations. Immunoprecipitations were carried out as described above with 10 μl of NHS-coupled IgG–Sepharose. Bound proteins were eluted with SDS sample buffer in the absence of reducing agents, to avoid dissociation of the IgG molecules.
For quantitative immunoblotting, purified SecA in 0.1% (w/v) SDS was prepared by buffer exchange using a PD10 gel filtration column (Pharmacia). His-tagged SecY was purified from 0.5 mg of soluble SecYEG complex by Ni2+ affinity chromatography (Manting et al., 1997) and dialysed against water. Precipitated protein was collected by centrifugation (5 min, 14 000 r.p.m., Eppendorf) and solubilized in 0.1% (w/v) SDS (10 min, 50°C). Non-solubilized material was removed by centrifugation (10 min, 14 000 r.p.m., Eppendorf). Amino acid concentrations of the SecY and SecA standards were determined in duplicate, using RNase as a standard, by Eurosequence, Groningen. Using a dilution range of 1–1/20 000 of the SecA and SecY standards, the protein contents in immunoprecipitates of translocase–I31 complexes were estimated by immunoblotting. Next, the appropriate dilutions were used to determine densitometrically the amounts of SecA and SecY in the immunoprecipitates from films of chemoluminescent blots.
Isolation of SecYEG complexes
Either proteoliposomes containing purified SecYEG (50 μl, 0.4 mg/ml in 50 mM Tris–HCl, 50 mM KCl), or reaction mixtures (500 μl) containing SecYEG proteoliposomes (20 μg), SecA (120 μg) and 2 mM AMP–PNP in translocation buffer, were solubilized on ice in a total volume of 2 ml of 2% (w/v) dodecyl maltoside, 10 mM Tris–HCl pH 8.0, 10 mM KCl and 10% (w/v) glycerol. The sample was injected on a miniQ anion-exchanger mounted in a Smartsystem (Pharmacia) equilibrated at 6°C with a buffer containing 10 mM Tris–HCl pH 8.0, 10% glycerol, 10 mM KCl and 0.03% (w/v) dodecyl maltoside. The column was eluted with a linear gradient of 0.01–0.3 M KCl in the same buffer and 75 μl fractions were collected. To separate the tetrameric and dimeric SecYEG complexes, proteins were solubilized on ice in a total volume of 2 ml of 2% (w/v) dodecyl maltoside, 10 mM imidazole pH 8.0, 100 mM KCl and 10% glycerol, and injected on a 1 ml Pharmacia high-Trap chelating column equilibrated with 0.05% (w/v) dodecyl maltoside, 10 mM imidazole pH 8.0, 100 mM KCl and 10% glycerol. The protein was eluted in the same buffer containing 200 mM imidazole pH 7.0. The protein content of these fractions was assayed by SDS–PAGE and appropriate fractions were used for electron microscopy. Activity of the purified SecYEG complex was confirmed by reconstitution in E.coli phospholipids via rapid dilution (van der Does et al., 1998) and measuring the proOmpA-stimulated SecA ATPase activity.
To isolate translocase–precursor complexes, proteoliposomes yielding the I31 translocation intermediate were harvested from a total of 2 ml of translocation reactions performed in the absence of DTT. Proteoliposomes were collected by ultracentrifugation (120 000 g, 30 min), solubilized in 250 μl of buffer S and loaded onto 10 ml linear sucrose gradients ranging from 10 to 45% (w/v) in 50 mM Tris–HCl pH 8.0, 50 mM KCl and 0.03% (w/v) dodecyl maltoside. After equilibration centrifugation (16 h, 120 000 g, 4°C), gradients were fractionated as 1 ml samples by pipetting from the top. All translocase subunits (SecA, SecY, SecE and SecG) co-fractionationed with I31 and were separated from phospholipids, which were contained at the top of the gradient.
Determination of dodecyl maltoside concentration
The final dodecyl maltoside concentration was determined by a modified protocol for α-naphthol staining of glycolipids. In brief, 40 μl of sample were mixed with 40 μl of 0.5% α-naphthol and 400 μl of 95% sulfuric acid and incubated for 10 min at 95°C. The absorbance of the samples was determined at 555 nm and compared with a standard of known dodecyl maltoside concentrations in column buffer.
Electron microscopy
Fractions from anion-exchange chromatography or sucrose gradients were absorbed to carbon films rendered hydrophilic by glow discharge. Grids were washed briefly with distilled water and stained with saturated uranyl formate. Images were taken in a Hitachi H-7000 transmission electron microscope at 100 kV and a magnification of 50 000×. Suitable films were scanned at 50 lines/mm (corresponding to a pixel size of 0.4 nm) using a Leafscan 45. Particles were selected automatically based on their size by calculating the cross-correlation function with a ring-shaped reference of either 7 or 10 nm diameter, or were selected interactively, using the SEMPER image processing system (Saxton et al., 1979). Multivariate statistical classification (Frank et al., 1988) was achieved on data sets containing >3000 particles prior to angular alignment, and cluster averages were calculated to allow for picking an unbiased reference. Aligning ∼3000 particles from each set of experimental conditions using the respective reference laterally and angularly, and subsequent classification yielded a final set of cluster averages from which class averages containing closely related structures were calculated. Typically, the class averages shown included 50–60% of all selected particles.
STEM mass analysis of detergent-solubilized particles was carried out as described previously (Müller and Engel, 1998). Elastic dark images of unstained complexes were recorded using a VG-HB5 STEM at electron doses of typically 300 electrons/nm2 and 80 kV acceleration voltage. The particle mass was evaluated by measuring the number of electrons elastically scattered by a circular region enclosing the particle and subtracting the background contribution due to the thin carbon film using the IMPSYS software (Müller et al., 1992). Gaussian curves were fitted to the mass histogram peaks by a Marquart algorithm (Bevington, 1962).
Acknowledgments
Acknowledgements
The expert assistance of Shirley Müller, Sabine Wirtz and Bettina Wolpensinger with STEM analysis and electron microscopy is highly appreciated. This work was supported by a PIONIER grant of the Netherlands Organisation for Scientific Research (N.W.O.), and by the Earth and Life Sciences Foundation (A.L.W.), the Swiss National Foundation for Scientific Research (grants 31-42435.94 and 4036-44062.96 to A.E.) and the Maurice E.Müller Foundation of Switzerland. C.van der Does was supported by a short–term EMBO fellowship.
References
- Akita M., Shinkai, A., Matsuyama, S. and Mizushima, S. (1991) SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem. Biophys. Res. Commun., 174, 211–216. [DOI] [PubMed] [Google Scholar]
- Arkowitz R.A. and Wickner, W. (1994) SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J., 13, 954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Taura, T., Shimioke, T., Akiyama, Y., Yoshihisa, T. and Ito, K. (1994) A cytoplasmic domain is important for the formation of a SecY–SecE translocator complex. Proc. Natl Acad. Sci. USA, 91, 4539–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M. and Wickner, W. (1993) Purified Escherichia coli preprotein translocase catalyses multiple cycles of precursor protein translocation. Biochemistry, 32, 2626–2630. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck, D., Grassucci, R., Penczek, P., Verschoor, A., Blobel, G. and Frank, J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Bevington P.R. (1969) Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill, New York, NY. [Google Scholar]
- Brundage L.A., Hendrick, J.P., Schiebel, E., Driessen, A.J.M. and Wickner, W. (1990) The purified Escherichia coli integral membrane protein SecY/E is sufficient for the reconstitution of SecA-dependent precursor protein translocation. Cell, 62, 649–657. [DOI] [PubMed] [Google Scholar]
- Cabelli R.J., Chen, L., Tai, P.C. and Oliver, D.B. (1988) SecA protein is required for secretory protein translocation into E.coli membrane vesicles. Cell, 55, 683–692. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu, H. and Tai, P. (1996) A significant fraction of SecA is permanently embedded in the membrane. J. Biol. Chem., 271, 29698–29706. [DOI] [PubMed] [Google Scholar]
- Crooke E., Guthrie, B., Lecker, S., Lill, R. and Wickner, W. (1988) ProOmpA is stabilized for membrane translocation by either purified E.coli trigger factor or canine signal recognition particle. Cell, 54, 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T., Fekkes, P., de Wit, J.G., Kuiper, W. and Driessen, A.J.M. (1996) Domain interactions of the peripheral preprotein translocase subunit SecA. Biochemistry, 35, 11994–12004. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T., de Wit, J.G., Gosker, H., van der Does, C., Breukink, E.–J., de Leij, L. and Driessen, A.J.M. (1997) Inhibition of preprotein translocation and reversion of the membrane inserted state of SecA by a carboxyl terminus binding mAb. Biochemistry, 36, 9159–9168. [DOI] [PubMed] [Google Scholar]
- Douville K., Price, A., Eichler, J., Economou, T. and Wickner, W. (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem., 270, 20106–20111. [DOI] [PubMed] [Google Scholar]
- Duong F. and Wickner, W. (1997) Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J., 16, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner, W. (1998) Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J., 17, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A.J.M. (1992) Precursor protein translocation by the Escherichia coli translocase is directed by the proton motive force. EMBO J., 11, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A.J.M. (1993) SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry, 32, 13190–13197. [DOI] [PubMed] [Google Scholar]
- Driessen A.J.M., Fekkes, P. and van der Wolk, J.P.W. (1998) The Sec system. Curr. Opin. Microbiol., 1, 216–222. [DOI] [PubMed] [Google Scholar]
- Economou A. (1998) Bacterial preprotein translocase: mechanism and conformational dynamics of a processive enzyme. Mol. Microbiol., 27, 511–518. [DOI] [PubMed] [Google Scholar]
- Economou A. and Wickner, W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell, 78, 835–843. [DOI] [PubMed] [Google Scholar]
- Eichler J. and Wickner, W. (1997) Both an N–terminal 65-kDa domain and a C–terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc. Natl Acad. Sci. USA, 94, 5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J., Brunner, J. and Wickner, W. (1997) The protease-protected 30 kDa domain of SecA is largely inaccessible to the membrane lipid phase. EMBO J., 16, 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A.M., Osborne, R.S. and Silhavy, T.J. (1995) The allele-specific synthetic lethality of prlA–prlG double mutants predicts interactive domains of SecY and SecE. EMBO J., 14, 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Bretaudiere, J.-P., Carazo, J.-M., Verschoor, A. and Wagenknecht, T. (1988) Classification of images of biomolecular assemblies. A study of ribosomes and ribosomal subunits of Escherichia coli. J. Microsc., 150, 99–115. [DOI] [PubMed] [Google Scholar]
- Geller B.L. (1990) Electrochemical potential releases a membrane-bound secretion intermediate of maltose-binding protein in Escherichia coli. J. Bacteriol., 172, 4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B.D., Chen, J.-C., Johnson, E.E. and Johnson, A.E. (1997) The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell, 89, 535–544. [DOI] [PubMed] [Google Scholar]
- Hamman B.D., Hendershot, L.M. and Johnson, A.E. (1998) BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell, 92, 747–758. [DOI] [PubMed] [Google Scholar]
- Hanada M., Nishiyama, K., Mizushima, S. and Tokuda, H. (1994) Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE and SecG (p12). J. Biol. Chem., 269, 23625–23631. [PubMed] [Google Scholar]
- Hanein D., Matlack, K.E.S., Jungnickel, B., Plath, K., Kalies, K.-U., Miller, K.R., Rapoport, T.A. and Akey, C.W. (1996) Oligomeric rings of the Sec61 complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Hartl F.-U., Lecker, S., Schiebel, E., Hendrick, J.P. and Wickner, W. (1990) The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E.coli plasma membrane. Cell, 63, 269–279. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Sommer, T., Prehn, S., Görlich, D., Jentsch, S. and Rapoport, T.A. (1994) Evolutionary conservation of the components of the protein translocation complex. Nature, 367, 654–657. [DOI] [PubMed] [Google Scholar]
- Hendrick J.P. and Wickner, W. (1991) SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J. Biol. Chem., 266, 24596–24600. [PubMed] [Google Scholar]
- Joly J.C. and Wickner, W. (1993) The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J., 12, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly J.C., Leonard, M. and Wickner, W. (1994) Subunit dynamics in Escherichia coli preprotein translocase. Proc. Natl Acad. Sci. USA, 91, 4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Rajapandi, T. and Oliver, D.B. (1994) SecA protein is exposed to the periplasmic surface of the E.coli inner membrane in its active state. Cell, 78, 845–853. [DOI] [PubMed] [Google Scholar]
- Lill R., Dowham, W. and Wickner, W. (1990) The ATPase activity of SecA is regulated by acidic phospholipids, SecY and the leader and mature domains of precursor proteins. Cell, 60, 271–280. [DOI] [PubMed] [Google Scholar]
- Manting E.H., van der Does, C. and Driessen, A.J.M. (1997) In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J. Bacteriol., 179, 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Akimura, J. and Mizushima, S. (1990) SecE-dependent overproduction of SecY in Escherichia coli. FEBS Lett., 269, 96–100. [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Fujita, Y. and Mizushima, S. (1993) SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J., 12, 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T.H., Ménétret, J.-F., Breitling, R., Miller, K.R., Akey, C.W. and Rapoport, T.A. (1999) The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J. Mol. Biol., 285, 1789–1800. [DOI] [PubMed] [Google Scholar]
- Müller S.A. and Engel, A. (1998) Mass measurement in the scanning transmission electron microscope: a powerful tool for studying membrane proteins. J. Struct. Biol., 121, 219–230. [DOI] [PubMed] [Google Scholar]
- Müller S.A., Goldie, K.N., Bürki, R., Häring, R. and Engel, A. (1992) Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy, 46, 317–334. [Google Scholar]
- Nishiyama K.-i., Mizushima, S. and Tokuda, H. (1993) A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J., 12, 3409–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K.-i., Suzuki, T. and Tokuda, H. (1996) Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell, 85, 71–81. [DOI] [PubMed] [Google Scholar]
- Nishiyama K.-i., Fukuda, A., Morita, K. and Tokuda, H. (1999) Membrane-deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. EMBO J., 18, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K.J. and Beckwith, J. (1994) Genetic and molecular characterization of the Escherichia coli secD operon and its products. J. Bacteriol., 176, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A., Economou, A., Duong, F. and Wickner, W. (1996) Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J. Biol. Chem., 271, 31580–31584. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V. and Oliver, D.B. (1997) Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J. Biol. Chem., 272, 23239–23246. [DOI] [PubMed] [Google Scholar]
- Sagara K., Matsuyama, S. and Mizushima, S. (1994) SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J. Bacteriol., 176, 4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.O., Pitt,T.J. and Horner,M. (1979) Digital image processing: the Semper system. Ultramicroscopy, 4, 343–354. [Google Scholar]
- Schiebel E., Driessen, A.J.M., Hartl, F.-U. and Wickner, W. (1991) ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell, 64, 927–939. [DOI] [PubMed] [Google Scholar]
- Snyders S., Ramamurhty, V. and Oliver, D.B. (1997) Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J. Biol. Chem., 272, 11302–11306. [DOI] [PubMed] [Google Scholar]
- Swaving J., van Wely, K.H.M. and Driessen, A.J.M. (1999) Host specificity is determined by the core subunits of preprotein translocase. J. Bacteriol., 181, 7021–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani K., Tokuda, H. and Mizushima, S. (1990) Translocation of proOmpA possessing an intramolecular disulphide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J. Biol. Chem., 265, 17341–17347. [PubMed] [Google Scholar]
- van der Does C., den Blaauwen, T., de Wit, J.G., Manting, E.H., Groot, N., Fekkes, P. and Driessen, A.J.M. (1996) SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxy–terminus to the periplasm. Mol. Microbiol., 22, 619–629. [DOI] [PubMed] [Google Scholar]
- van der Does C., Manting, E.H., Kaufmann, A., Lutz, M. and Driessen, A.J.M. (1998) Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry, 37, 201–210. [DOI] [PubMed] [Google Scholar]
- van der Wolk J.P.W., de Wit, J.G. and Driessen, A.J.M. (1997) The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J., 16, 7297–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Voorst F., van der Does, C., Brunner, J., Driessen, A.J.M. and de Kruijff, B. (1998) Translocase-bound SecA is largely shielded from the phospholipid acyl chains. Biochemistry, 37, 12261–12268. [DOI] [PubMed] [Google Scholar]
- van Wely K.H.M., Swaving, J., Broekhuizen, C.P., Rose, M., Quax, W.J. and Driessen, A.J.M. (1999) Functional identification of the product of the Bacillus subtilis yvalL gene as a SecG homologue. J. Bacteriol., 181, 1786–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.B., Ray, P.H. and Bassford, P.J.,Jr (1988) Purified secB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc. Natl Acad. Sci. USA, 85, 8978–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]