Abstract
All vertically transmitted bacterial symbionts undergo a process of genome reduction over time, resulting in tiny, gene-dense genomes. Comparison of genomes of ancient bacterial symbionts gives only limited information about the early stages in the transition from a free-living to symbiotic lifestyle because many changes become obscured over time. Here, we present the genome sequence for the recently evolved aphid symbiont Serratia symbiotica. The S. symbiotica genome exhibits several of the hallmarks of genome evolution observed in more ancient symbionts, including elevated rates of evolution and reduction in genome size. The genome also shows evidence for massive genomic decay compared with free-living relatives in the same genus of bacteria, including large deletions, many pseudogenes, and a slew of rearrangements, perhaps promoted by mobile DNA. Annotation of pseudogenes allowed examination of the past and current metabolic capabilities of S. symbiotica and revealed a somewhat random process of gene inactivation with respect to function. Analysis of mutational patterns showed that deletions are more common in neutral DNA. The S. symbiotica genome provides a rare opportunity to study genome evolution in a recently derived heritable symbiont.
Keywords: Serratia symbiotica, genome reduction, pseudogene, recent symbiont
Introduction
Maternally transmitted symbionts exhibit some of the smallest bacterial genomes known, with the smallest currently described a mere 143 kb in size (organellar genomes excluded, McCutcheon et al. 2009a). Many of these bacteria formed symbiotic associations millions of years ago (Moran et al. 1993; Baumann 2005) and are dependably inherited through generations due to their essential function of providing nutrients to hosts (Buchner 1965; Douglas 1989; Nakabachi and Ishikawa 1999; Moran et al. 2008; Moya et al. 2008). Compared with free-living bacteria, symbionts that undergo strict vertical transmission experience some important differences in lifestyle, including population bottlenecks every generation from vertical transmission, further restriction by the population size of hosts, and asexuality (Moran 1996; Mira and Moran 2002). These lifestyle differences lead to greater genetic drift, allowing fixation of slightly deleterious mutations such as the deletions that result in tiny genomes and other changes characteristic of many symbiont genomes (Ohta 1992, reviewed in Moran et al. 2008). Symbionts that experience occasional horizontal transfer resulting in coinfections within hosts may be able to purge deleterious mutations through homologous recombination (Muller 1964).
Reconstruction of the evolutionary history of genomes from ancient symbiotic associations suggests the occurrence of major architectural change early after establishment of symbiosis, including large deletions and numerous rearrangements (reviewed in Moran 2003; Moran and Plague 2004). However, the nature of early genomic changes becomes obscured over time due to the gradual deletion of all unnecessary DNA in long-established symbionts.
Some symbionts, particularly some that are facultative for hosts, are more recently derived compared with ancient nutritional symbionts (Buchner 1965), and despite being related to pathogens, have conditionally beneficial effects upon their hosts (reviewed in Oliver et al. 2010). Maternally transmitted symbionts that are facultative for hosts are often referred to as “secondary” symbionts and include several species of Enterobacteriaceae that infect aphids (Oliver et al. 2010), Arsenophonus species in many insects (Wilkes et al. 2009), Wolbachia species (Wu et al. 2004; Klasson et al. 2009), and others. Occasional horizontal transfer of secondary symbionts between host species has been observed (Sandström et al. 2001; Russell et al. 2003; Russell and Moran 2005; Oliver et al. 2010), but unlike pathogens, these symbionts are primarily transmitted vertically (Sandström et al. 2001; Russell and Moran 2005). Secondary symbionts can be subject to a population genetic structure similar to that of more ancient symbionts, although coinfections of hosts may occur, allowing recombination among strains (e.g., Baldo et al. 2006; Degnan and Moran 2007). Thus, their genomes may display effects of host restriction but are expected to undergo occasional recombination and to represent earlier stages in the process of genome reduction.
Serratia symbiotica is a secondary symbiont of pea aphids (Acyrthosiphon pisum), providing defense against environmental heat stress in the form of improved survival, fecundity, and developmental time compared with uninfected aphids (Chen et al. 2000; Montllor et al. 2002; Russell and Moran 2006; Burke et al. 2010). This symbiont coinhabits pea aphids with the ancient symbiont Buchnera, whose association with aphids is more than 100 My old and is involved in biosynthesis of essential amino acids unavailable in the aphids’ diet (Moran et al. 1993; Sandström and Moran 1999; Shigenobu et al. 2000; Akman Gündüz and Douglas 2009). Phylogenetic studies based on both protein-coding and rRNA genes show that S. symbiotica is nested within the genus Serratia (Burke et al. 2009; Lamelas et al. 2008). This genus generally contains free-living bacteria that can be pathogenic to animals (Grimont F and Grimont PAD 2004) and for which several genomes have been fully sequenced. Although the absolute age of this symbiotic lineage has not been estimated, we note that the 16S rRNA of S. symbiotica is only 2–3% divergent from 16S rRNA of Serratia species with a free-living stage, indicating relatively recent origin of the symbiotic habit in this lineage (Russell et al. 2003). In contrast, 16S rRNA of Buchnera is >10% divergent from the closest free-living species. Thus, our analysis of the genome of S. symbiotica allows an evaluation of genome evolution in a beneficial symbiont in comparison with closely related free-living bacteria within the same genus.
Materials and Methods
DNA Preparation
Preparation of S. symbiotica genomic DNA for sequencing was challenging due to the low density of the symbionts in pea aphids. Genomic DNA was prepared for sequencing through several steps: size selection of cells using filtration techniques, selective whole-genome amplification (WGA), and finally standard randomly primed WGA.
“Tucson” genotype pea aphids (established from a single female collected in Tucson, AZ in 1999) were grown as a continuous line of parthenogenetic females in a growth chamber at 20 °C with a long day cycle of 16L:8D. Aphids of 2–3 g were crushed in 30 ml cold phosphate buffered saline (PBS) with a mortar and pestle. The filtrate was passed through a 100 μm filter twice and centrifuged at 1,000 rpm for 25 min at 4 °C to pellet heavier aphid cells (including bacteriocytes that house Buchnera). The supernatant was filtered through 20 and 11 μm filters twice, respectively, and centrifuged at 4,000 rpm at 4 °C for 15 min. The pellet, comprised of smaller cells, was resuspended in 300 μl of PBS and 50 μl of 10× DNAse buffer and 5 μl of DNAse I were added and incubated at room temperature for 30 min to digest any DNA not contained in whole cells. 61 μl of 0.5 M ethylenediaminetetraacetic acid inactivated the DNAse treatment. The cells were washed in PBS three times and pelleted by centrifugation at 4,000 × g. The cells were lysed and DNA extracted using the Gentra Puregene Tissue Kit (QIAGEN) following the standard kit protocol, including RNAse A treatment.
The genomic composition of DNA was estimated using quantitative polymerase chain reaction (PCR) using a Lightcycler (Roche Molecular Biochemicals) at each stage of preparation. The number of copies of a single-copy gene was measured for each genome present (S. symbiotica, Buchnera, and A. pisum). The percentage composition of each genome in the total DNA was calculated by weighting genome copy numbers by the estimated genome size. Multiple single-copy genes were measured for S. symbiotica and A. pisum in an attempt to check for major biases in coverage due to selective WGA. The single-copy genes dnaK and gyrB were used for S. symbiotica, dnaK for Buchnera, and EF-1α and hsp70 for A. pisum.
The primers used for quantitative PCR are in supplementary table 1, Supplementary Material online. Quantitative PCR conditions were used as described in Oliver et al. (2006), and DNA copies were quantified using an absolute standard curve specific for each gene. The regression lines for the standard curves had mean squared error <0.1, and fidelity of the amplication was checked using diagnostic melting curves for each amplicon.
The DNA was enriched for the S. symbiotica genome via WGA using G + C biased primers. The S10 primer, 5′-SSSSSSSSSS-3′, was designed with ten randomly chosen guanine or cytosine nucleotides, and two phosphorothioate modified nucleotides at the 3′ end. Given the G + C content of the respective genomes present, S10 was expected to anneal every 550 bp for S. symbiotica (52% G + C), 600,000 bp for Buchnera (26% G + C), and 200,000 bp for A. pisum (30% G + C) on average. This primer was used in WGA following the protocol of Pan et al. (2008), which utilizes trehalose to minimize template-independent products (TIPs) and amplification bias. The general procedure of Pan et al. (2008) was followed, with the following modifications: Each 100 μl reaction contained 4.8 nmol of S10 primer, 0.2 M trehalose, 1 μl of H2O (for negative controls) or DNA (∼100 ng), and 1 μl of Enzyme Mix from the Illustra GenomiPhi V2 DNA amplification kit (GE Lifesciences). The reaction was incubated at 30 °C for 12.5 h, then 70 °C for 20 min. The DNA was cleaned using the QIAGEN Cleanup of Genomic DNA protocol and DNAeasy kit reagents and eluted in 50 μl of H2O.
The selectively amplified DNA was amplified with a second round of WGA, this time using 1 μl of 1:10 diluted preamplified DNA or negative control reaction, 1 nmol random hexamers, and 0.66 M Trehalose per reaction. This was done a total of three times to prepare DNA for each of the three types of sequencing used. Between five and eight, 100 μl reactions were cleaned using the QIAGEN DNA clean-up protocol, resuspended in 30 μl of H2O each, and pooled.
Quantitative PCR was used again to confirm that the genomic composition of the DNA had not changed due to the second round of amplification.
DNA Sequencing
Three types of sequencing run were performed in an effort to obtain sequence data that would enable more complete assembly of genomic regions containing repetitive sequences; these consisted of a 454 single reads, 454 paired reads with an insert size of 2.5 kb, and Illumina paired reads with an insert size of 3.5–4 kb. DNA of 5 μg was used to construct two libraries for 454 single read and “mate pair” sequencing. Libraries were prepared as described in Margulies et al. (2005) using kits supplied by Roche Applied Science and sequenced on a Roche GS-FLX Titanium machine using 454 technology at the University of Arizona Genetics Core facility. For Illumina sequencing, 20 μg of DNA was prepared as directed in a Mate Pair Library Prep kit supplied by Illumina. 8 pmol of the library was sequenced on one lane as directed by the manufacturer in a 2× 36 bp run at the Genome Sequencing Center at the Washington University School of Medicine.
Genome Assembly
The single read 454 run generated a total of 222 Mb in 686,450 reads, with an average read size of 324 bp. The paired read run generated 238 Mb of sequence in 1,141,997 read fragments after trimming out linker sequences, which were 208 bp on average. Buchnera and A. pisum reads were identified and removed using nucleotide Blast to reference genomes (NC_002528, NZ_ABLF00000000) with an e-value cutoff of 7 × 1009 and requiring 92% identity. The remaining reads (94.4% of total) from both 454 runs were assembled with Newbler (v 2.3) using an overlap minimum match identity of 94%, and the “–large” algorithm; 63.1% of reads were assembled into 341 scaffolds and 4,627 contigs, of which 31 scaffolds and 470 contigs had greater than 20× coverage and were at least 200 bp in length (supplementary fig. 1, Supplementary Material online). A total of 106 of 318 gaps in these scaffolds were filled with contigs that overlapped both sides of the gap from an assembly of single 454 reads only. Contigs overlapping with scaffold sequences were removed, giving 358 final contigs. Ribosomal RNA operons were not assembled, as they are repeated several times in the genome and are longer than the span of the paired-end reads. Five operons were sequenced with Sanger technology and were assembled to the ends of scaffolds.
The paired-read Illumina run generated 1.8 Gb of sequence in 50,724,700 36 bp reads. These reads were mapped to the reference scaffolds and contigs using the CLC Genomics Workbench (CLC bio) and used to correct the errors inherent in 454 sequencing, such as the incorrect resolution of homopolymer lengths.
Genome Annotation
Gene prediction was performed by PRODIGAL (http://compbio.ornl.gov/), and predicted genes were annotated by combining the results of BlastX searches against Escherichia coli K12 MG1655 and the National Center for Biotechnology Information nr database, and hmmpfam searches with HMMER v2.3.2 (http://hmmer.janelia.org) against the Pfam 24.0 ls database (Finn et al. 2010), and the TIGRFAM v 9.0 database (Selengut et al. 2007). tRNAs were identified using ARAGORN (Laslett and Canback 2004), tmRNAs by BRUCE v1.0 (Laslett et al. 2002), and other functional RNAs by INFERNAL v1.0.2 (Nawrocki et al. 2009) and the RFAM 10.0 database (Gardner et al. 2008). Clusters of orthologous group (COG) categories were assigned using a BlastP search to the COG database (Tatusov et al. 2003). Metabolic pathways were built by hand, using Ecocyc (Keseler et al. 2009) and Metacyc (Caspi et al. 2010) as guides. Average coding sequence (CDS) size was calculated from complete genes on scaffolds that were not spanning gaps between contigs.
Phylogenetic Analyses
Core Gene Tree
Orthologous genes that do not show signatures of horizontal transfer were used for reconstructing phylogenetic relationships among S. symbiotica and related bacteria. We started with a set of 203 orthologous genes previously identified as present as exactly one copy in every genome of a set of 13 Gammaproteobacteria and as showing no evidence for horizontal transfer between lineages (Lerat et al. 2003). In our analysis, 26 Gammaproteobacterial genomes were searched for these 203 genes using a cutoff of >30% BlastP score relative to the alignment score of the sequence with itself as described in Lerat et al. (2003). One hundred and thirteen genes were identified with a single copy per genome, representing 3,632 amino acid sites after protein alignments were trimmed for all invariant and gap-containing sites (a protein alignment with only gap-containing sites removed resulted in an equivalent tree). Maximum likelihood searches for the best tree were performed by RAxML v7.0.4 (Stamatakis et al. 2005) with 100 rapid bootstrap replicates and a full Maximum Likelihood search with the PROTGAMMA algorithm (estimation of rate heterogeneity with four rate categories) and the WAG substitution matrix.
Rates of Evolution
To explore the rate of evolution of S. symbiotica compared with free-living bacteria, S. proteamaculans and Yersinia pseudotuberculosis were chosen to serve as a free-living relative and an outgroup, respectively. The evolutionary relationships of lineages in the genus Serratia could not be resolved with complete confidence (fig. 2), so Serratia species were not suitable as outgroup taxa for these relative rate tests.
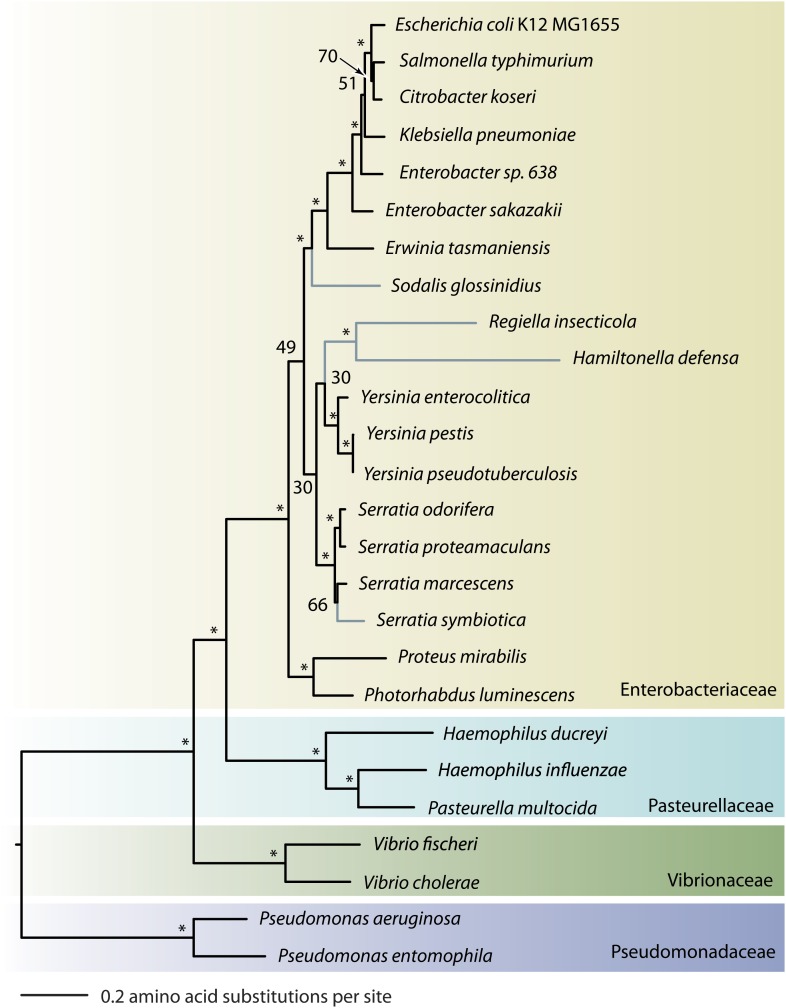
FIG. 2.
The relatedness of Serratia symbiotica to other Gammaproteobacterial lineages. Asterisks represent bootstrap support values >80, and symbiotic lineages are highlighted in blue. S. symbiotica belongs within the genus Serratia. S. symbiotica has diverged less than other facultative symbionts from the closest free-living bacterial lineages, suggesting a more recent lifestyle transition.
Orthologous protein-coding genes from S. symbiotica and Y. pseudotuberculosis were identified using reciprocal Blast hits (RBH) and by imposing an 80% length cut-off to ensure quality of alignments. Orthologous protein sequences were aligned with MUSCLE (Edgar 2004) and back-translated into codon-based nucleotide alignments using PAL2NAL (Suyama et al. 2006). Codon-based alignments revealed saturation at synonymous sites (average dS = 2.27) using codeml from PAML (Yang 2007) implementing the model of Nei and Gojobori (1986). To avoid synonymous site saturation, relative rates of evolution were tested using amino acid alignments in codeml and calculating branch lengths with the WAG substitution matrix for each gene tree constrained to the same tree topology. All statistical analyses were performed in JMP8 (SAS Institute).
Pseudogene Analyses
S. symbiotica pseudogene orthologs were identified in S. proteamaculans using BlastX of S. symbiotica DNA queries to a S. proteamaculans protein database and TBlastN for the reciprocal search. The Blast scores for disrupted gene fragments were added to give an overall score. Of the 525 scaffold pseudogenes, 296 had best reciprocal Blast hits (BRH) and a Blast score ratio >30 to orthologs in S. proteamaculans (Lerat et al. 2003), and an additional 176 had BRHs to proteins in the nr database (90% of scaffold pseudogenes had a good ortholog). S. symbiotica DNA sequences were aligned to orthologous protein sequences using “estwise” of Wise2 v2.2.0 (Birney et al. 2004), and mutations were counted using a custom perl script. Pseudogenes were considered to have large deletions if the S. symbiotica pseudogene was <80% of the length of its ortholog.
Synteny Analyses
Orthologous genes between S. proteamaculans and S. symbiotica scaffolds or S. marcescens were identified using BlastP to identify BRHs. The start coordinate and orientation of each gene was compared with those of its orthologs using custom perl scripts. S. symbiotica scaffolds and S. marcescens gene coordinates were arranged or shifted, respectively, to best showcase syntenous blocks of genes. Average dS was calculated as described in the “phylogenetic comparisons” section above.
GC Content Analysis
Orthologous genes between S. proteamaculans, S. marcescens, and S. symbiotica were identified and aligned using the method described above under “phylogenetic analyses and rates of evolution.” The GC content for third positions and across all sites was calculated using codeml from PAML (Yang 2007).
Results
Selective WGA
S. symbiotica infects pea aphids at low density, so in a DNA extraction of whole aphid bodies, S. symbiotica genomes make up only 0.8% of the total DNA in the sample as estimated by quantitative PCR (table 1). A combination of filtration to remove large aphid cells from a tissue sample followed by WGA selective for G + C rich DNA, and nonselective WGA to increase yield improved the percentage of S. symbiotica DNA, yielding approximately 5 μg of DNA, of which 91–96% was S. symbiotica DNA (table 1).
Table 1.
Selective WGA Enriched for Serratia symbiotica DNA from a Mixed Sample
| Absolute Number of Genic Copies in Sample |
DNA Composition from qPCR |
|||||||
| S. symbiotica dnaK | S. symbiotica gyrB | Buchnera dnaK | A. pisum hsp70 | A. pisum EF-1α | S. symbiotica Average (%) | Buchnera (%) | A. pisum Average (%) | |
| Tucson genotype aphid DNA | 136,800 | NA | 4,655,000 | 91,780 | NA | 0.8 | 6 | 93 |
| Filtered Tucson aphid DNA | 205,800 | NA | 2,485,000 | 1,338 | NA | 22 | 53 | 25 |
| Selectively amplified Tucson DNA | 807,500 | NA | 500,500 | 445.6 | NA | 82 | 10 | 8 |
| Single read 454 library | 788,500 | 7,499,000 | 474,000 | 5,91.9 | 613.9 | 95 | 2 | 2 |
| Paired read 454 library | 618,100 | 7,847,000 | 449,200 | 329.7 | 806.7 | 96 | 2 | 2 |
| Paired read Illumina library | 698,300 | 5,743,000 | 787,500 | 809.3 | 1,185 | 91 | 4 | 5 |
Note.—DNA composition was estimated using single gene copy numbers and an estimated genome size of 3 Mb for S. symbiotica. The Buchnera and Acyrthosiphon pisum genomes are 0.6 and 517 Mb in size, respectively (Shigenobu et al. 2000; International Aphid Genomics Consortium 2010).
The S. symbiotica Genome Sequence
The genomic sequence assigned to S. symbiotica is 2,789,218 bp in size and comprised 30 large scaffolds and 358 contigs <2 kb in size. This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number AENX00000000. The version described in this paper is the first version, AENX01000000. The assembly of sequence reads into a complete circular genome was prevented by repetitive sequences that were longer than the insert size used for paired reads, such as ribosomal RNA operons. Despite lack of complete assembly, essentially all the S. symbiotica genome is represented in these sequences. This is evident because the average coverage in 454 and Ilumina sequence reads was 62.6× and 459×, respectively. Furthermore, all the 203 single copy genes conserved among Gammaproteobacterial genomes (Lerat et al. 2003) are present in the genome sequence, although nine are pseudogenes. Thus, the sequences represent the complete content of unique genes in the S. symbiotica genome, but resolution of numbers of repetitive sequences and their assembly into contigs was not possible.
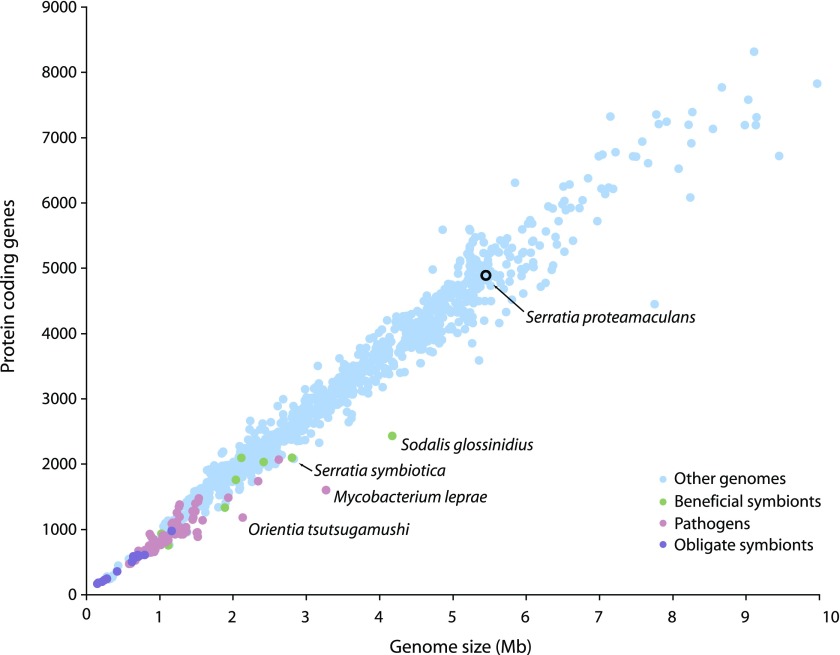
The S. symbiotica genome encodes 2,098 intact protein-coding sequences and 550 pseudogenes, resulting in a coding density of 60.9% (table 2). Bacterial species typically feature a tight correlation between genome size and the number of protein-coding genes (fig. 1). Compared with other bacterial genomes, S. symbiotica has a larger genome than expected given the number of intact coding sequences due to the large number of inactivated genes in the genome (fig. 1). The average gene length, 845 bp, is less than that of Buchnera, and free-living S. proteamaculans. Potentially, this lower average size reflects the inclusion of some inactivated genes, under our criterion that genes were considered intact if the CDS was >80% of the length of the ortholog.
Table 2.
Comparison of Serratia symbiotica Genome Features to Those of an Obligate and a Free-Living Bacterial Genome
| Buchnera aphidicola APS | S. symbiotica | S. proteamaculans | |
| Chromosome (bp) | 640,681 | 2,789,218 | 5,448,853 |
| Extrachromosomal elements | 2 | Unknown | 1 |
| Total G + C (%) | 26.2 | 52.0 | 55.0 |
| Total predicted CDS | 571 | 2,098 | 4,942 |
| Coding density (%) | 86.7 | 60.9 | 87.1 |
| Average CDS size (bp) | 984 | 845 | 968 |
| Pseudogenes | 13 | 550 | 12 |
| rRNA operons | 2 | 5 | 7 |
| tRNAs | 32 | 44 | 85 |
| Lifestyle | Obligate | Facultative | Free-living |
FIG. 1.
Plot of size and number of protein-coding genes for 1,190 bacterial genomes. Serratia symbiotica has a larger genome than expected given the number of intact genes.
On a tree constructed using 113 protein-coding genes, S. symbiotica falls within the genus Serratia, consistent with previous studies of the evolution of this symbiont based on a few genes only (fig. 2, Moran et al. 2005; Russell and Moran 2005; Lamelas et al. 2008; Burke et al. 2009). Compared with the secondary insect symbionts Sodalis glossinidius (of tsetse flies), and Hamiltonella defensa and Regiella insecticola (of aphids), which are considerably diverged from their closest free-living relatives, the short branch length of S. symbiotica indicates a more recent divergence from its free-living relatives.
Metabolic Reconstruction of the S. symbiotica Genome
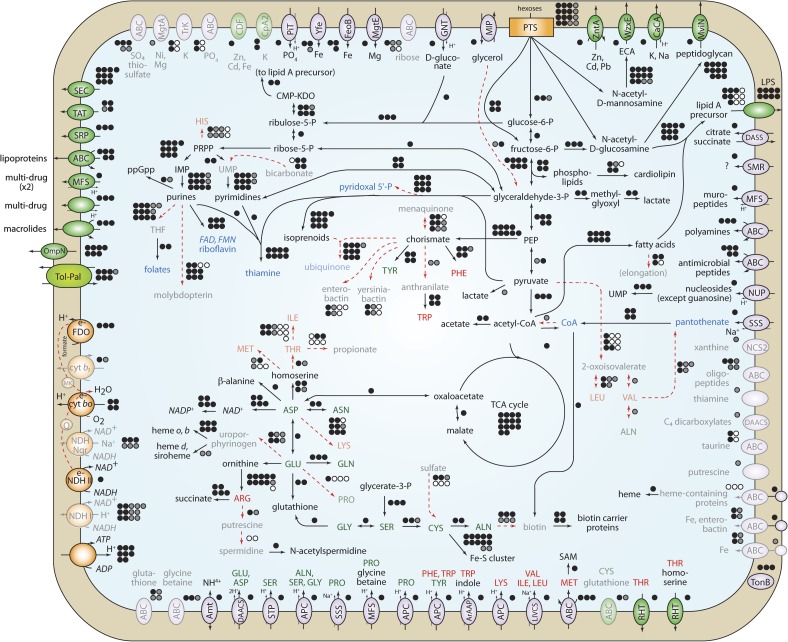
Figure 3 shows the reconstruction of S. symbiotica metabolism, using gene homology and biochemical studies in other bacteria to assign function to S. symbiotica genes.
FIG. 3.
Reconstruction of Serratia symbiotica metabolism. Intact pathways are shown with black lines, and inactive pathways in red dashed lines or opaque symbols and lettering. The number of genes involved in each pathway is shown as circles next to each pathway, with black representing intact genes, gray as pseudogenes, and white as genes absent from the genome. Vitamins and cofactors are in blue lettering, and essential and nonessential amino acids for aphids are shown in red and green lettering, respectively.
Similar to other Serratia lineages, S. symbiotica is an aerobic heterotroph, sharing pathways involved in converting organic carbon into energy with most members of the Enterobacteriaceae. It can also convert acetyl-coA to acetate and energy under oxygen-limiting conditions. S. symbiotica can grow on a variety of carbon sources, including glucose, fructose, mannitol, mannose, N-acetylglucosamine, and trehalose. S. symbiotica is able to synthesize four vitamins (thiamine, flavins, pyridoxal 5′-phosphate, and coenzyme A) and three essential and eight nonessential amino acids. Many transporters are present to import into the cell most amino acids that cannot be synthesized, except histidine and threonine.
Depsite these similarities to its relatives, S. symbiotica has developed metabolic dependencies that are not typical of free-living bacteria. It is almost certainly reliant upon the presence of Buchnera for full functionality. The essential amino acids imported from outside the cell are not available in large quantities in the aphid diet, nor produced by the aphid host, but are synthesized by Buchnera. In the pathway to make tryptophan, the two genes required to convert chorismate to anthranilate are inactivated in S. symbiotica, implying a requirement for exogenous anthranilate. These genes (trpE and trpG) are present on a Buchnera plasmid and may provide a source of anthranilate for S. symbiotica. S. symbiotica also appears to be reliant upon import of nucleosides from outside of the cell with a NUP family transporter in order to make one of the precursors for pyrimidine nucleotides.
In many cases, intact pathways contain pseudogenes for functions that are encoded by another gene in the genome. For example, in arginine biosynthesis, ornithine is converted into arginine through several intermediates requiring products of 11 genes in E. coli. Ten of these genes are present in S. symbiotica, but one gene, argD, has been inactivated. In E. coli, argD encodes acetylornithine transaminase, which converts N-acetyl-L-glutamate 5-semialdehyde into N-acetyl-L-ornithine. The enzyme AstC can perform the same function, but in E. coli is specialized to function in arginine degradation. In S. symbiotica, argD is inactivated, but astC is intact. In E. coli, both argF and argI, encoding ornithine carbamoyltransferase, are present, but in S. symbiotica, only argI is present. Another example is NADH dehydrogenases: 13 genes are required for NDH I, and 7 are inactivated, whereas NDH II is encoded by a single intact gene.
Many pathways are inactive due to the presence of pseudogenes or gene deletions (fig. 3). In addition to gene inactivation and apparent streamlining of functional pathways, S. symbiotica contains remnants of pathways that are no longer functional. Many amino acid biosynthesis pathways have been inactivated, including those for production of the essential amino acids histidine, isoleucine, leucine, valine, threonine, methionine, and lysine, and the nonessential amino acids alanine and proline. Other vitamins and cofactors usually essential for bacterial growth such as pantothenate, folates, ubiquinone, menaquinone, molybdopterin, biotin, and heme all have inactivated biosynthesis pathways, and the only transporter for specific import of a missing metabolite is that for pantothenate transport.
Many genes usually involved in bacterial pathogenesis, such as iron acquisition genes, have become pseudogenized. The pathways to make siderophores enterobactin and yersiniabactin, and outer membrane proteins (fhuA, fepA, fecA) and ABC transporters (fep and fec genes) for uptake of siderophores and iron are inactivated. Also, many inactivated genes for Type IV pili involved in bacterial attachment to host cells are present in the genome.
Pseudogene Decay
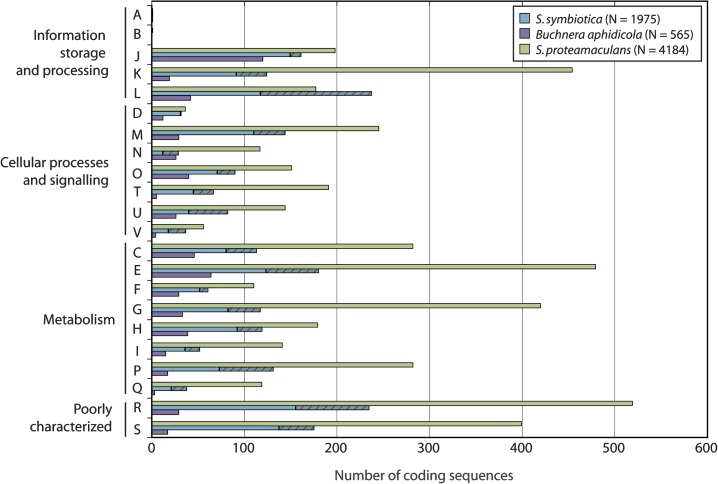
Five hundred and fifty pseudogenes were identified in the S. symbiotica genome, with 520 located on scaffolds. Figure 4 shows the functional categorization of intact genes and pseudogenes in the S. symbiotica genome into COG categories, compared with genes in the Buchnera and S. proteamaculans genomes. Every major category contains a substantial number of pseudogenes, except category D, for genes involved in cell cycle control, cell division, and chromosome partitioning. In general, S. symbiotica has more intact genes in each category compared with Buchnera but fewer than S. proteamaculans.
FIG. 4.
Serratia symbiotica genes represented as COG functional categories. For each category, three genomes, Buchnera, S. symbiotica, and S. proteamaculans are shown. Hashed segments of bars represent pseudogenes.
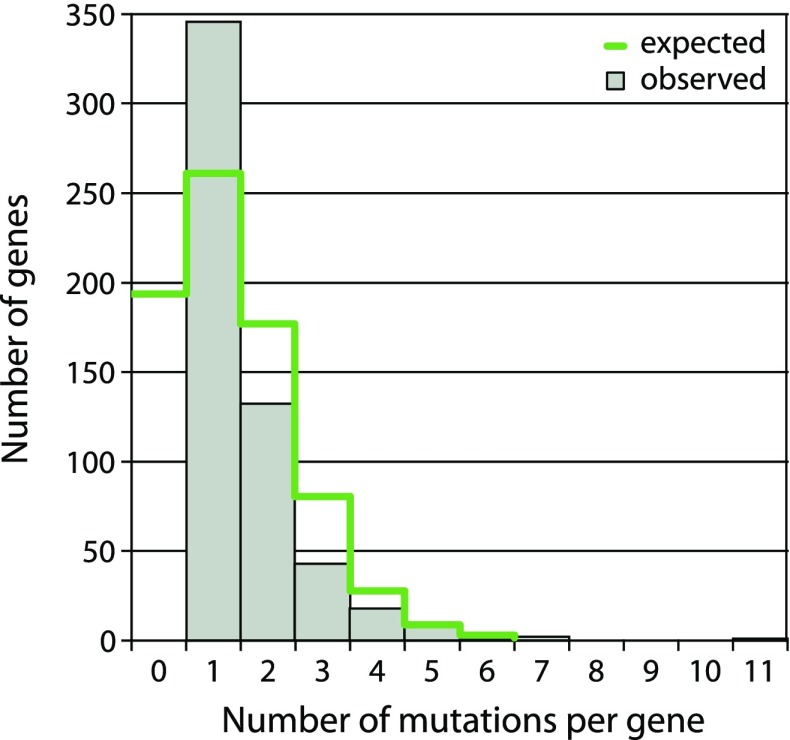
A total of 472 of 520 scaffold pseudogenes had good orthologs, and mutations were counted compared with the intact sequence. The most common inactivating mutations were large deletions resulting in loss of >20% of protein length (249), deletions causing frameshifts (221), then frameshift-causing insertions (187), and stop codons (102). A histogram of the number of inactivating mutations per pseudogene is shown in figure 5.
FIG. 5.
The distribution of mutations in neutralized genes in the Serratia symbiotica genome. One hundred and ninety three genes are expected to be neutral but to lack evident inactivating mutations. Genes with exactly one mutation are overrepresented possibly due to large deletions masking older mutations in a gene.
Genome Rearrangements and Mobile DNA
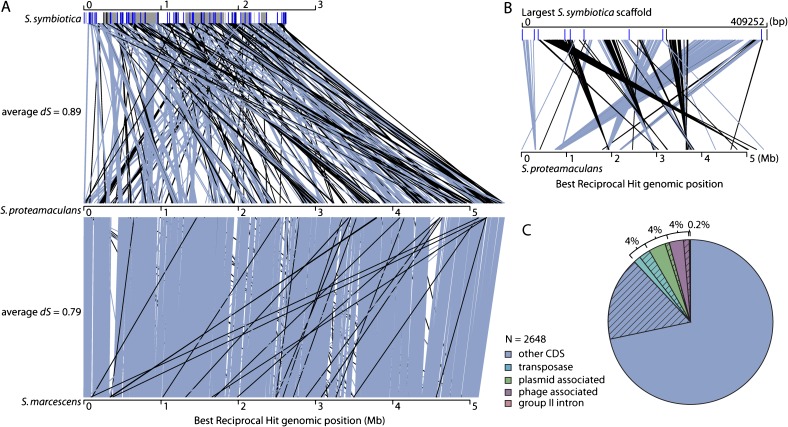
Divergence (given by dS, the number of synonymous substitutions per synonymous site) is similar between the free-living S. proteamaculans and S. symbiotica (dS = 0.89) and between S. proteamaculans and S. marcescens (dS = 0.79). However, comparison of levels of synteny between S. proteamaculans/S. symbiotica and S. proteamaculans/S. marcescens shows that much more rearrangement has occurred in the lineage leading to S. symbiotica (figure 6A). Gene order is highly conserved between the two free-living lineages, but only small blocks of synteny are observed between the symbiotic and free-living Serratia genomes, with the largest such block containing 124 genes. Many rearrangements are observed within scaffolds (fig. 6B), indicating that rearrangements depicted in figure 6A are not due to suboptimal ordering of scaffolds to form the S. symbiotica genome. Sometimes, syntenic blocks are flanked by intact or pseudogenized transposases that may have been part of insertion sequence (IS) elements (fig 6A and B). The massive amount of rearrangement in the S. symbiotica genome may have been facilitated by mobile DNA. Transposases, plasmid-associated genes, and phage-associated genes each make up 4% of the total number of genes present in the genome, although many are pseudogenes (fig. 6C). The presence of plasmid and phage genes likely reflects the insertion of plasmid or phage DNA into the genome and subsequent degradation. There were 22 families of transposases present in the S. symbiotica genome. The method of genome assembly resulted in compression of identical transposable element copies into a single representative sequence, so their copy numbers could not be evaluated. However, divergent copies of transposases in the genome could be enumerated, and members of the IS256 family were most common, with 16/106 (15%) of distinct transposase sequences belonging to this family.
FIG. 6.
(A) Rearrangements in three Serratia genomes. Lines indicate positions of orthologous genes in each genome, with grey-blue indicating genes the same orientation on chromosomes, and black indicating the opposite orientation. The Serratia symbiotica genome is shown as alternating white and gray blocks that represent different scaffolds. Black and blue lines on these blocks indicate positions of intact and inactive transposases, respectively. (B) Rearrangements between the largest scaffold of the S. symbiotica genome and the S. proteamaculans genome. Lines are colored as for (A). Transposases often flank blocks of conserved synteny. (C) Distribution of intact genes and pseudogenes in three categories of mobile DNA in the S. symbiotica genome. Percentages indicate the proportion of genes in each category, and hashing indicates pseudogenes.
The Rate of Evolution of S. symbiotica
Due to the constraints of a symbiotic lifestyle, it is expected that genes will accumulate slightly deleterious substitutions faster in symbiotic bacteria than in their free-living relatives. One test for this difference in rates of evolution is to compare dN/dS between a symbiotic lineage and a free-living lineage with respect to an outgroup. Unfortunately, the most suitable outgroup was divergent enough to result in saturation at synonymous sites. Another method that does not rely on synonymous sites is comparison of rates of protein evolution. Averaged over 1,224 orthologous gene sets, the amino acid substitution rate for the lineage leading to S. symbiotica was two times faster than for the lineage leading to S. proteamaculans (two-sided t-test, t2364 = 19.3, P < 0.0001, fig. 7). Because the transition to symbiosis occurred after the split between S. symbiotica and S. proteamaculans, the acceleration may be greater in the symbiotic lineage.
FIG. 7.
Tree representing relationships and rates of evolution between Serratia symbiotica, S. proteamaculans, and Yersinia pseudotuberculosis. Branch lengths represent average divergence in amino acid substitutions per site for 1,224 genes, and numbers in parentheses give the 95% confidence interval for the means.
Discussion
The divergence of S. symbiotica from related free-living lineages is small compared with the divergence of other studied symbiotic lineages from free-living lineages, indicating a relatively recent transition of S. symbiotica to a symbiotic lifestyle (fig. 2). Several members of the genus Serratia are known to infect insects and plants, but the direct free-living ancestor of S. symbiotica remains unresolved. Assuming the rate of evolution of these symbiotic lineages is comparable, S. symbiotica seems to be a younger symbiont than So. glossinidius, H. defensa, or R. insecticola. The maximum age of S. symbiotica as an aphid symbiont can be estimated at approximately 90 My by comparing the divergence between S. symbiotica and free-living Serratia lineages to expected divergences based on two different approximate rates for bacterial evolution (supplementary table 2, Supplementary Material online). However, the transition to symbiosis could have occurred long after this divergence, so the age could be much less.
When bacteria become host-restricted symbionts, they experience a reduction in effective population size and a change in environment, which has consequences for their genome evolution. High levels of genetic drift and reduced selection result in genomic changes, the severity of which relates to the duration of the symbiotic association.
Gene Decay and Loss
Genome reduction occurs in symbiotic bacterial genomes as a result of the loss of genes that are superfluous or beneficial but not essential. In ancient symbionts such as Buchnera and some secondary symbionts such as H. defensa and R. insecticola, most lost genes are entirely absent (deleted) and recognizable pseudogenes are rare. More recently derived secondary symbionts such as So. glossinidius and S. symbiotica have many pseudogenes, resulting in fewer protein-coding genes than expected given their genome size (fig. 1). This pattern likely reflects a period of massive gene inactivation following the initiation of the host-restricted lifestyle, with the result that gene inactivation outpaces DNA deletion. This pattern was first observed in Mycobacterium leprae, a recently evolved host-restricted pathogen (Cole et al. 2001).
S. symbiotica provides the rare opportunity to examine the process of pseudogene formation. This genome has approximately half the number of genes found in free-living Serratia lineages, and losses affect all major categories of genes (fig. 4). Many intact pathways contain pseudogenes for which the previous function can be performed by another gene. Other pathways have lost genes essential for activity, and are undergoing degradation. Many genes involved in pathogenesis in free-living relatives have been inactivated. In contrast to the ancient symbiont Buchnera, more genes functioning in biosynthesis of nonessential amino acids and cell surface components, regulation, DNA repair, and recombination are present. S. symbiotica has likely developed dependence upon compounds produced by Buchnera, including essential amino acids, and anthranilate, a metabolite intermediate in the pathway to produce tryptophan. Other secondary symbionts H. defensa and R. insecticola are also reliant on Buchnera for some essential amino acids but show no obvious need for the exchange of metabolic intermediates. The tsetse fly symbiont So. glossinidius may cooperate with the anciently derived symbiont Wigglesworthia to synthesize thiamine (Belda et al. 2010). These examples could represent early stages in the evolution of the metabolic complementation that has evolved repeatedly among long-established symbiont pairs (McCutcheon and Moran 2007, 2010; McCutcheon et al. 2009b).
Comparison of the nucleotide sequences of S. symbiotica pseudogenes and intact genes from closely related bacteria allows categorization and numeration of mutations becoming fixed within pseudogenes. The most common mutations were deletions, a bias observed in bacterial genomes generally (Parkhill et al. 2003; Kuo and Ochman 2010) that are an important force in the reduction in genome size. We hypothesize that many genes became effectively neutral (i.e., no longer subject to purifying selection preserving functionality) immediately or almost immediately after S. symbiotica became a symbiont, due to a change in environment and/or to reduced efficacy of selection resulting from reduced population size and asexuality imposed by the symbiotic lifestyle. In this case, the decay in pseudogenes should be random in terms of the number of inactivating mutations per newly neutralized gene (adjusted for gene length). Thus, the number of such mutations per neutralized gene should fit a Poisson distribution (fig. 5). Given the observed frequency of inactivating mutations per bp of 759/461,743 = 0.0016, the average expected number of mutations is 1.35 per pseudogene of average length 845 bp. From the Poisson distribution, the probability that a pseudogene has one or more inactivating mutations is 0.74, which corresponds to the 550 sequences categorized as pseudogenes. Extrapolation using these numbers leads to the estimate that the total number of neutralized genes in the S. symbiotica genome is 743, including 193 genes that are not preserved by purifying selection but that have not yet acquired a mutation (fig. 5). Compared with the numbers of inactivating mutations expected per gene given random accumulation of mutations (fig. 5), there is an overrepresentation of genes with exactly one inactivating mutation, which may reflect large deletions masking mutations that occurred in deleted regions. Another explanation for the paucity of pseudogenes with more than one mutation could be negative selection upon inactivated genes, as proposed for Salmonella pseudogenes (Kuo and Ochman 2010). This scenario is unlikely due to the underlying processes in action within these different bacterial populations: Salmonella have a large free-living population in which genes are gradually being inactivated, whereas S. symbiotica experienced a transitory event (free-living to symbiotic) in which many genes were likely neutralized within a short period of time. Within many enzymatic pathways, some genes are inactivated and others are intact (fig. 3). The latter are good candidates for neutralized genes that have not acquired an inactivating mutation, as predicted above. Possibly some of these genes are effectively inactivated due to loss of promoter features or to amino acid replacements that eliminate function of the protein. Furthermore, the observation of shorter than average gene length (845 bp) in the S. symbiotica genome strongly suggests that some sequences annotated as intact genes, because they did not meet our criteria for pseudogenes, are in fact truncated pseudogenes. Thus, some number of CDS scored as intact genes may in fact be already neutralized sequences that would be expected to be eliminated over evolutionary time.
AT Bias
Symbiotic genomes generally have low GC content likely due to mutational biases present in replication or DNA repair that create neutral mutations fixed through genetic drift (Moran 1996; Wernegreen and Funk 2004). S. symbiotica has an overall GC content of 52%; in other members of the genus, GC contents range from 52% to 60% (Grimont F and Grimont PAD 2004). However, for 633 genes orthologous between S. marcescens, S. proteamaculans, and S. symbiotica, S. symbiotica has the lowest GC content at third positions in codons, and across all sites, suggestive of a small decrease in GC content in this symbiont lineage compared with free-living relatives (table 3). The GC content of S. symbiotica is higher than GC contents of any other fully sequenced beneficial secondary symbiont genome (excluding So. glossinidius), which likely further reflects its recent derivation as a symbiotic lineage.
Table 3.
GC Content for 633 Orthologous Genes in the Genus Serratia
| Organism | GC Percentage, Third Position in Codon | GC Percentage, Overall |
| Serratia symbiotica | 0.63 | 0.55 |
| S. proteamaculans | 0.68 | 0.57 |
| S. marcescens | 0.79 | 0.61 |
Genome Dynamics
The genome dynamics of intracellular bacteria range from major change soon after the establishment of symbiosis to extreme genome stasis on an evolutionary timescale. Reconstruction of the deletion history of Buchnera suggests that large deletions occurred early in the establishment of symbiosis possibly because selection was reduced across a large proportion of genes due to a new environment and population structure (Moran and Mira 2001). S. symbiotica, a recently derived symbiont, appears to be undergoing rapid changes in genomic architecture. Many large deletions have already occurred resulting in a genome that is half the size of its closest relatives, consistent with the hypothesis of large early deletions in Buchnera. In addition to large deletions, which are the main basis for DNA loss and genome reduction, individual genes are also undergoing inactivation and shortening. The early stages of this process (gene inactivation, before deletion) are in progress in S. symbiotica (see above) as well as in the genomes of So. glossinidius and the pathogens M. leprae and Orientia tsutsugamushi (Cole et al. 2001; Toh et al. 2006; Cho et al. 2007).
Many rearrangements have occurred in the S. symbiotica genome compared with free-living Serratia lineages of similar relatedness (fig. 6A). Rearrangements may be facilitated by transposases, which are sometimes flanking syntenous blocks of genes in S. symbiotica and some pathogen genomes (Parkhill et al. 2003). The genomes of recently derived symbionts and of ancient symbionts experiencing occasional horizontal transfer have relatively large amounts of mobile DNA, such as transposases, plasmid-related genes, and phage-related genes, compared with both free-living bacteria or to strictly vertically transmitted ancient symbionts such as Buchnera (Toh et al. 2006; Gil et al. 2008; Plague et al. 2008; Degnan et al. 2009; Newton and Bordenstein 2011). S. symbiotica is no exception with approximately 12% of its genes in these categories. Proliferation of mobile DNA most likely reflects ineffectual purifying selection for insertion events, and occasional horizontal transfer between symbionts could prevent extinction of mobile elements (Moran and Plague 2004; Bordenstein and Reznikoff 2005; Newton and Bordenstein 2011). High frequencies of mobile DNA are observed in more anciently derived symbiont genomes such as H. defensa, R. insecticola, and Wolbachia (Wu et al. 2004; Degnan et al. 2009, Degnan et al. 2010; Klasson et al. 2009). These symbionts may be more sexual; paternal transmission has been observed for H. defensa and R. insecticola but not S. symbiotica, providing more opportunity for genetic exchange and mobile element transfer (Moran and Dunbar 2006).
Extreme genome stasis (no gene acquisition or rearrangement) has been observed for distantly related genomes of Buchnera and other long-term obligate symbionts, suggesting its establishment fairly early in symbiosis (e.g., Tamas et al. 2002, van Ham et al. 2003; Sabree et al. 2010). Genome stasis is not observed in the sister taxa H. defensa and R. insecticola, which show very little conservation of gene order, possibly due to occasional horizontal transfer and recombination (Degnan et al. 2010). Comparison of S. symbiotica Tucson to other S. symbiotica lineages will reveal the extent of genome rearrangements in this symbiont group; it is expected to be large given the abundance of mobile DNA and functional recombination machinery in S. symbiotica.
Rate of Evolution
The rate of nonsynonymous substitution in the lineage leading to S. symbiotica is twice as high as for the lineage leading to S. proteamaculans, its free-living relative (fig. 7). The test used could not distinguish between an increase in mutation rate and an increase in fixation of slightly deleterious nonsynonymous mutations due to genetic drift. In Buchnera, both factors play a role in its accelerated evolution (Moran 1996; Clark et al. 1999).
Concluding Remarks
S. symbiotica is a recently acquired symbiont undergoing the early stages of genomic change affecting bacteria that transition to a host-restricted lifestyle. Comparison with free-living Serratia species revealed a decaying genome dramatically reduced in size, with many pseudogenes and rearrangements, and increased levels of mobile DNA. Study of the formation of pseudogenes allowed examination of the past and current metabolic capabilities of S. symbiotica and suggested a somewhat random process of gene inactivation, with all functional categories of genes affected fairly equally. Analysis of mutational patterns suggested a bias toward deletion, and a prediction that a substantial number of genes remain to be inactivated and removed by deletion. The S. symbiotica genome is a rare example of the process of genome reduction in a recently derived symbiont and contributes to our knowledge of the early stages of symbiotic bacterial genome evolution.
Supplementary Material
Supplementary tables 1 and 2 and figure 1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was funded by National Science Foundation grants 0723472 and 0313737 to N.M.
References
- Akman Gündüz E, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Belda E, Moya A, Bentley S, Silva FJ. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics. 2010;11:449. doi: 10.1186/1471-2164-11-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Reznikoff WS. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- Buchner P. Endosymbionts of animals with plant microorganisms. New York: Interscience; 1965. [Google Scholar]
- Burke G, Fiehn O, Moran N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 2010;4:242–252. doi: 10.1038/ismej.2009.114. [DOI] [PubMed] [Google Scholar]
- Burke GR, Normark BB, Favret C, Moran NA. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 2009;75:5328–5335. doi: 10.1128/AEM.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38(Database issue):D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DQ, Montllor CB, Purcell AH. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl. 2000;95:315–323. [Google Scholar]
- Cho N-K, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci U S A. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- Cole ST, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Moran NA. Evolutionary genetics of a defensive facultative symbiont of aphids: exchange of toxin-encoding bacteriophage. Mol Ecol. 2007;17:916–929. doi: 10.1111/j.1365-294X.2007.03616.x. [DOI] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, et al. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2010;12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Mycetocyte symbiosis in insects. Biol Rev. 1989;69:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database Issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2008;37(Database issue):D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, et al. Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int Microbiol. 2008;11:41–48. [PubMed] [Google Scholar]
- Grimont F, Grimont PAD. Genus XXXIV Serratia. In: Brenner DJ, Krieg NR, Staley JR, Garrity G, editors. Bergey’s manual of systematic bacteriology. The Proteobacteria, Part B: The Gammaproteobacteria. 2nd ed. Vol. 2. New York: Springer; 2004. pp. 799–811. [Google Scholar]
- International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6:e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, et al. Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in aphid species of the subfamily lachninae. Appl Environ Microbiol. 2008;74:4236–4240. doi: 10.1128/AEM.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B, Andersson S. BRUCE: a program for the detection of transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 2002;30:3449–3453. doi: 10.1093/nar/gkf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009a;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009b;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Moran NA. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 2002;44:137–143. doi: 10.1007/s00248-002-0012-9. [DOI] [PubMed] [Google Scholar]
- Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. [Google Scholar]
- Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Moran NA, Dunbar HE. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci USA. 2006;103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:54. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiont bacteria is calibrated using the insect host. Proc R Soc Lond B Biol. 1993;253:167–171. [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Moran NA, Russell JA, Koga R, Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005;71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Newton IL, Bordenstein SR. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 2011;62:198–208. doi: 10.1007/s00284-010-9693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol. Syst. 1992;23:263–286. [Google Scholar]
- Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts of aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Moran NA, Hunter MS. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc Biol Sci. 2006;273:1273–1280. doi: 10.1098/rspb.2005.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, et al. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc Natl Acad Sci USA. 2008;105:15499–15504. doi: 10.1073/pnas.0808028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Plague GR, Dunbar HE, Tran PL, Moran NA. Extensive proliferation of transposable elements in heritable bacterial symbionts. J Bacteriol. 2008;190:777–779. doi: 10.1128/JB.01082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol. 2003;12:1061–1075. doi: 10.1046/j.1365-294x.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- Russell JA, Moran NA. Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol. 2005;71:7987–7994. doi: 10.1128/AEM.71.12.7987-7994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc Biol Sci. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree Z, Degnan PH, Moran NA. Chromosome stability and gene loss in cockroach endosymbionts. Appl Environ Microbiol. 2010;76:4076–4079. doi: 10.1128/AEM.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström JP, Moran N. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 1999;91:203–210. [Google Scholar]
- Sandström JP, Russell JA, White JP, Moran NA. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol. 2001;10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- Selengut JD, et al. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35(Database issue):D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. Raxml-iii: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RC, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Funk DJ. Mutation exposed: a neutral explanation for extreme base composition of an endosymbiont genome. J Mol Evol. 2004;59:849–858. doi: 10.1007/s00239-003-0192-z. [DOI] [PubMed] [Google Scholar]
- Wilkes TE, Darby AC, Choi J-H, Colbourne JK, Hurst GDD. The draft genome sequence of Arsenophonus nasoniae, son-killer bacterium of Nasonia vitripennis, reveals genes associated with virulence and symbosis. Insect Mol Biol. 2009;19:59–73. doi: 10.1111/j.1365-2583.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]