Abstract
The non-enveloped fusogenic avian and Nelson Bay reoviruses encode homologous 10 kDa non-structural transmembrane proteins. The p10 proteins localize to the cell surface of transfected cells in a type I orientation and induce efficient cell–cell fusion. Mutagenic studies revealed the importance of conserved sequence-predicted structural motifs in the membrane association and fusogenic properties of p10. These motifs included a centrally located transmembrane domain, a conserved cytoplasmic basic region, a small hydrophobic motif in the N-terminal domain and four conserved cysteine residues. Functional analysis indicated that the extreme C-terminus of p10 functions in a sequence-independent manner to effect p10 membrane localization, while the N-terminal domain displays a sequence-dependent effect on the fusogenic property of p10. The small size, unusual arrangement of structural motifs and lack of any homologues in previously described membrane fusion proteins suggest that the fusion-associated small transmembrane (FAST) proteins of reovirus represent a new class of membrane fusion proteins.
Keywords: fusion protein/membrane fusion/reovirus/syncytium formation
Introduction
Membrane fusion is an essential cellular process involved in the regulated interaction between intracellular membrane compartments, as occurs during constitutive vesicular transport and regulated exocytosis (Ferro-Novick and Jahn, 1994; Rothman, 1994; Bock and Scheller, 1997). Exoplasmic fusion events are also known to occur and represent essential steps, for example during sperm–egg fusion and myoblast differentiation, and as part of the entry pathway of enveloped viruses (Kielian and Jungerwith, 1990; White, 1990; Bentz, 1993; Lanzrein et al., 1994). It is widely accepted that all of these biological fusion events are mediated by specific fusion proteins that function to overcome the energy barrier to spontaneous membrane fusion (Hernandez et al., 1986; Stegmann et al., 1989; Hoekstra, 1990; Zimmerberg et al., 1993). In spite of considerable study, the nature of the minimal fusion machinery and the precise sequence of events that regulate and mediate protein-mediated membrane fusion have not been discerned.
Structural and functional studies of enveloped virus fusion proteins, primarily the influenza virus haem– agglutinin (HA) fusion protein, have been instrumental in the development of a model for protein-mediated membrane fusion (Gaudin et al., 1995; Ramalho-Santos and de Lima, 1998). Similarities in the structural arrangement of the transmembrane-anchored polypeptides of several enveloped virus fusion proteins suggest that the working model for HA-mediated fusion may extend to many enveloped virus fusion proteins (Gaudin et al., 1995; Hughson, 1997; Weissenhorn et al., 1997; Joshi et al., 1998; Ben-Efraim et al., 1999). Furthermore, the studies of enveloped virus fusion proteins have been complemented by recent investigations of SNARE-mediated intracellular vesicular fusion events (Sollner et al., 1993; Sutton et al., 1998; Weber et al., 1998). The convergence of these two lines of investigation has suggested that the mechanism of action of both SNAREs and viral fusion proteins may be similar. In both cases, the energy required to overcome the thermodynamically unfavourable process of lipid leaflet mixing may be contributed by rearrangements of extended heptad repeats to generate coiled-coil structures in membrane-anchored proteins (Skehel and Wiley, 1998; Weber et al., 1998). Although the generation of coiled-coils is clearly an essential step in membrane fusion mediated by these viral and cellular proteins, the precise function of this interaction in the actual fusion reaction remains unclear (Ungermann et al., 1998; Otter-Nilsson et al., 1999). Furthermore, structural analysis of certain enveloped virus fusion proteins indicates that the paradigm of extensive coiled-coil rearrangements is not universal (Kielian, 1995; Rey et al., 1995).
We have been investigating an unusual example of exoplasmic fusion, i.e. the induction of syncytium formation by a group of non-enveloped viruses, the fusogenic orthoreoviruses (Duncan et al., 1995, and references therein). The orthoreoviruses are one of nine genera in the family Reoviridae, a large diverse family of non-enveloped viruses with segmented double-stranded RNA genomes (Nibert et al., 1996). The majority of the members of the Reoviridae do not induce cell fusion, a typical phenotype for non-enveloped viruses that do not require fusion proteins to facilitate virus entry or egress from cells. However, within the genus Orthoreovirus, all of the avian reovirus (ARV) isolates induce syncytium formation in cell culture (Kawamura et al., 1965). There are also two atypical mammalian reoviruses that share the syncytium-inducing properties of ARV: Nelson Bay virus (NBV) and baboon reovirus (BRV) (Gard and Compans, 1970; Duncan et al., 1995). The nature of the viral protein responsible for reovirus-induced cell fusion, and its mechanism of promoting membrane fusion, have not been determined.
We previously have shown that, unlike enveloped virus-induced membrane fusion, the mechanism responsible for ARV-induced cell fusion is not related directly to either the viral entry or exit pathways (Duncan, 1996; Duncan et al., 1996). The primary purpose of the ARV fusion protein may be to direct cell–cell fusion, a process that contributes to a rapid lytic response and enhanced rate of virus release (Duncan et al., 1996). Since the ARV fusion protein is not required for virus entry or egress, it is conceivable that this accessory viral protein may not be subject to the mechanisms (i.e. ligand binding and/or low pH) that regulate the fusion activity of enveloped virus fusion proteins. Such a fusion protein might offer a simplified system for investigating the minimal determinants required for protein-mediated membrane fusion.
Using transfection studies, we have now identified the homologous fusion proteins of ARV and NBV. These 10 kDa non-enveloped virus fusion proteins are the smallest known viral or cellular fusion proteins. Moreover, the p10 proteins lack any extended heptad repeat structures or obvious fusion peptide motif typical of many enveloped virus fusion proteins. These simple fusion proteins appear to represent a new class of membrane fusion proteins whose structural features indicate that they mediate membrane fusion through a coiled-coil-independent pathway.
Results
ARV and NBV encode 10 kDa non-structural proteins that are responsible for cell fusion
Previous genetic studies implicated the S1 genome segment in the fusogenic activity of ARV (Duncan and Sullivan, 1998). The genetic implication of the ARV S1 genome segment in reovirus-induced syncytium formation was confirmed by expressing the full-length cloned S1 cDNA in transfected cells (Figure 1a). Similar results were obtained by expression of the S1 cDNA of the related NBV (Figure 1b). None of the other cloned S–class genome segment cDNAs of ARV or NBV were capable of inducing cell fusion in transfected cells (data not shown), indicating that an S1–specific gene product was responsible for syncytium formation.
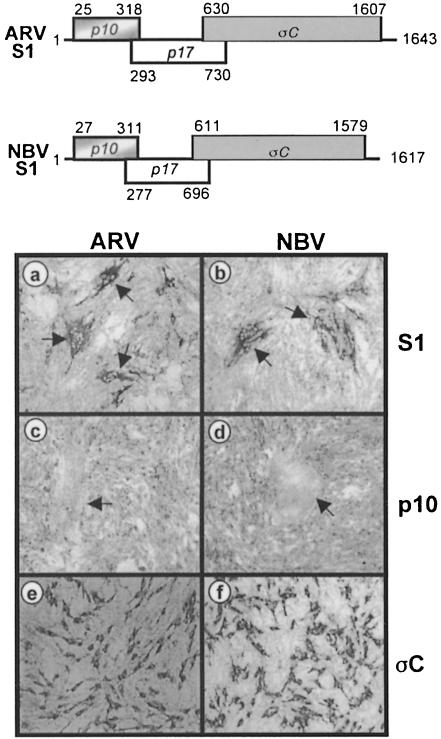
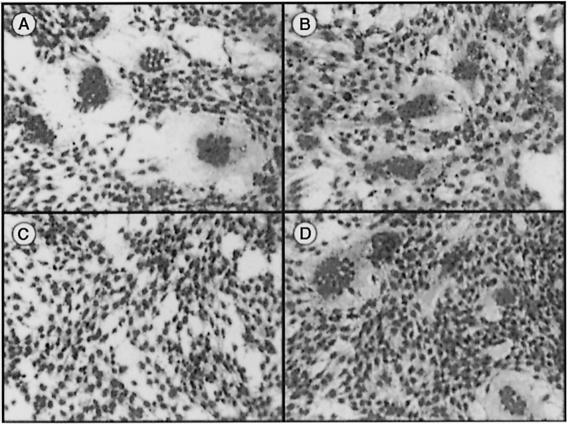
Fig. 1. The p10 ORF of the S1 genome segment is sufficient for fusion. A schematic representation of the S1 genome segments of ARV and NBV is presented at the top of the figure. The three sequential overlapping ORFs encoding the 10 kDa fusion protein, a putative 17 kDa protein and the cell attachment protein, σC, are indicated, along with the nucleotide positions corresponding to the first position of the start codon and the last position of the ORF. The lower part of the figure represents transfected QM5 cells immunostained using antibodies raised against the structural proteins of ARV (a, c and e) or NBV (b, d and f). The monolayers were transfected with expression plasmids encoding the ARV or NBV S1 segment (a and b, respectively), the ARV or NBV p10 protein (c and d, respectively) or the ARV or NBV σC protein (e and f, respectively). Arrows in (a) and (b) indicate syncytial foci that stained antigen-positive due to the presence of the σC protein expressed from the full-length S1 cDNA, while those in (c) and (d) indicate the location of syncytia induced by transfection of the p10 ORF alone that failed to react with the polyclonal antisera against virus structural proteins. Cells were photographed at 100× magnification.
The S1 genome segment of ARV and NBV contains three sequential, overlapping open reading frames (ORFs) (Kool and Holmes, 1995). Only the 3′-terminal ORF has been shown previously to be functional. This ORF encodes the receptor-binding protein of ARV, σC (Varela and Benavente, 1994; Shapouri et al., 1996; Martinez-Costas et al., 1997), which was previously implicated in syncytium formation (Theophilos et al., 1995). However, expression of the σC ORF of either ARV or NBV in transfected quail cells, as revealed by immunostaining (Figure 1e and f), failed to induce syncytium formation. Identical results were obtained in σC–transfected COS-7 and Vero cells (data not shown). Conversely, expression of the 5′-terminal S1 ORF alone (which encodes a predicted 10 kDa protein) resulted in cell–cell fusion (Figure 1c and d), implying that a previously unidentified reovirus protein (p10) was responsible for the fusogenic property of the virus. Interestingly, the polyclonal antisera raised against purified virus particles failed to stain syncytial foci induced by transfection of the p10 ORF alone (Figure 1c and d), suggesting that the predicted p10 protein might be a non-structural protein of the virus.
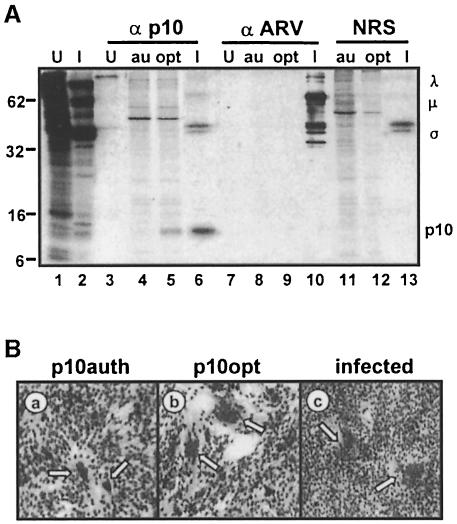
We confirmed that the first ORF of the S1 genome segment encodes a 10 kDa protein responsible for cell–cell fusion using a specific antiserum. Polyclonal antiserum was raised against the C-terminal half of the predicted ARV p10 protein by expression in Escherichia coli as a chimeric maltose-binding protein (MBP)–p10 construct. The p10 antiserum precipitated a 10 kDa protein from radiolabelled transfected and infected cell lysates (Figure 2A). The specificity of the p10 antiserum was evident from the lack of significant cross-reaction with other ARV structural or non-structural proteins (Figure 2A, lane 6). The low level of ARV structural proteins precipitated by the anti-p10 antiserum reflects non-specific trapping of radiolabelled virus particles, as shown by a similar degree of trapping when using normal rabbit serum (Figure 2A, lane 6 versus lane 13). These results confirmed that the 5′-terminal ORF of the reovirus S1 genome segment is indeed functional, and encodes a 10 kDa protein that is responsible for virus-induced cell fusion. The ARV p10 antiserum did not cross-react with the NBV protein (data not shown); therefore, all subsequent experiments were performed with ARV alone.
Fig. 2. Increased p10 expression corresponds to enhanced fusion. (A) Cells were transfected with the authentic p10 gene (au) or with p10 containing an optimized translation initiation sequence (opt), or were infected with ARV (I) or mock infected (U). Radiolabelled cell lysates were immunoprecipitated using anti-p10 (α p10), polyclonal anti-ARV serum (α ARV) or normal rabbit serum (NRS), and the cell lysates (lanes 1 and 2) or immune complexes were resolved on a 15% acrylamide gel and detected by fluorography. Numbers on the left indicate the location of molecular weight markers. The locations of the major λ-, μ- and σ-class viral proteins, and of p10, are indicated on the right. (B) QM5 cells were transfected with an expression plasmid encoding p10 containing the authentic translation start site (a), with p10 containing an optimized translation initiation sequence (b) or infected with ARV (c), and syncytium formation was detected by Wright–Giemsa staining. Arrows indicate multinucleated syncytia. Cells were photographed at 100× magnification.
As added proof that the ARV p10 protein is responsible for cell fusion, the suboptimal translation start site for the p10 ORF was modified to an optimal context (from CGUCAUGC to CCACCAUGG), which resulted in both enhanced syncytium formation (Figure 2B) and increased p10 expression (Figure 2A, lane 4 versus lane 5). The level of p10 expression from the optimized construct was still less than that observed in cell lysates infected with limiting virus dilutions that generated approximately equivalent numbers of syncytial foci as observed in transfected cells (Figure 2A, lane 5 versus lane 6, and B, panels b and c), indicating that cell fusion mediated by the p10 ORF alone was not an artefact of protein overexpression. These results conclusively demonstrated that ARV and, by inference, NBV encode the smallest known viral fusion-associated proteins.
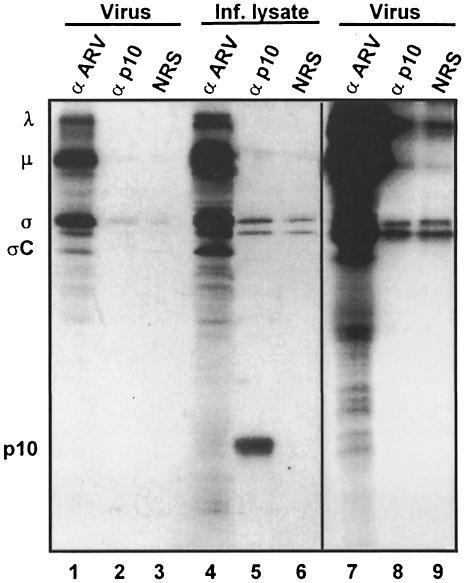
The inability of a polyclonal antiserum specific for ARV structural proteins to immunostain p10-transfected cells (Figure 1c and d) or to precipitate radiolabelled p10 from infected or transfected cell lysates (Figure 2A, lanes 8–10) suggested that, unlike all enveloped virus fusion proteins (Bentz, 1993), the reovirus p10 protein might be a non-structural protein of the virus. This speculation was confirmed by the inability of the p10 antiserum to detect p10 in radiolabelled virus particles. Virus particles were disrupted with SDS and heat (to expose inner, as well as outer, capsid proteins), and the solubilized proteins were immunoprecipitated using the p10-specific antiserum. Contrary to the ability of the polyclonal anti-ARV serum to recognize the known λ-, μ- and σ-class virus structural proteins, the p10 antiserum failed to precipitate any protein present in the virus pellets (Figure 3). The absence of p10 in the virus pellets was apparent even after extended autoradiographic exposure of the gels (Figure 3, lanes 7–9), whereas the minor σC receptor-binding protein of the virus, present at only 36 copies per virus particle (Strong et al., 1991; Shapouri et al., 1996), was clearly detected. In addition, the ability of the p10 antiserum to precipitate SDS-denatured p10 obtained from whole-cell lysates (Figure 3, lane 5) indicated that the inability of this serum to precipitate p10 from solubilized virus pellets was not the result of p10 epitope destruction due to SDS denaturation. The cumulative evidence strongly implies that p10 is not only the first non-enveloped virus protein capable of promoting fusion from within, it is also the first non-structural virus protein capable of inducing cell–cell fusion.
Fig. 3. The p10 protein is a non-structural viral protein. Detergent-solubilized, radiolabelled virus pellets (Virus) or detergent-solubilized virus-infected cell lysates (Inf. lysate) were immunoprecipitated with polyclonal anti-ARV serum (α ARV), anti-p10 (α p10) or normal rabbit serum (NRS), and the immune complexes were resolved on 15% acrylamide gels and detected by fluorography. The locations of the major λ-, μ- and σ-class viral proteins, and of p10 are indicated on the left. Lanes 7–9 are an extended exposure of lanes 1–3.
Sequence-predicted structural motifs in the p10 fusion proteins
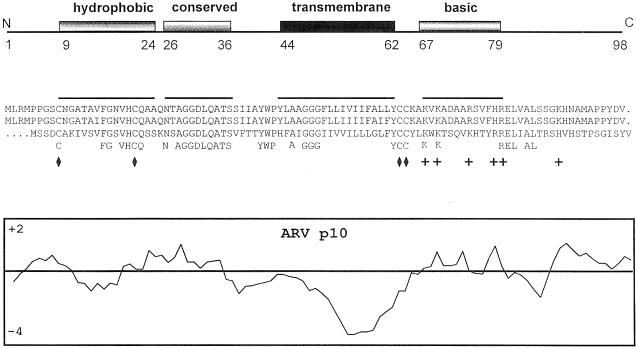
Assuming that ARV p10 initiates from the first in-phase methionine codon (there are two methionine codons at residues 1 and 4 in the predicted p10 ORF of ARV; both exist in a suboptimal initiation consensus sequence), then the aligned ARV and NBV p10 proteins possess an overall sequence identity of only 33%, with an obvious clustering of conserved residues in the N–proximal domain of p10. Both proteins are small (98 or 95 amino acids for ARV and NBV, respectively), hydrophobic and basic (pI = 8.8). A gapped BLAST search failed to identify any known homologues of p10. The p10 proteins possess no identifiable N-terminal signal peptide but they do have a predicted 19 residue transmembrane (TM) domain in the centre of each sequence (Figure 4) that could serve as a signal/anchor sequence (Zheng and Gierasch, 1996; Martoglio and Dobberstein, 1998). This highly hydrophobic 19 amino acid sequence was identified as a TM domain using the TMAP algorithm of Persson and Argos (1994). The majority of the basic residues reside in a conserved basic region on the C–proximal side of the TM domain, suggesting that the p10 proteins assume a type I orientation (N-terminus out) based on the positive-inside rule (Matlack et al., 1998).
Fig. 4. Sequence and structural conservation in the p10 proteins of ARV and NBV. The top panel indicates the locations of conserved structural motifs, and the first and last amino acid of each motif in the ARV sequences. The centre panel shows the aligned p10 amino acid sequences of ARV strains 176 (first line) and 138 (second line), and of NBV (third line). The locations of conserved identical amino acids are indicated (fourth line), along with the locations of the four conserved cysteine residues (diamonds) and the conserved basic residues (+). The overlining corresponds to the locations of the conserved structural motifs identified in the top panel. The bottom panel represents a hydropathy profile of the ARV p10 protein according to the algorithm of Kyte and Doolittle, averaged over a window of seven residues (hydrophobic below the line).
The ARV and NBV p10 proteins contain four cysteine residues in conserved locations, two immediately adjacent to the C-terminus of the TM domain and the other two located in the N–proximal domain of p10 (Figure 4). The two conserved cysteine residues in the N–proximal domain reside adjacent to the most conserved region of the ARV and NBV p10 proteins. These cysteine residues lie near the ends of a 16 amino acid region (residues 9–24 in ARV) that can be modelled as a short, moderately hydrophobic helix. This small hydrophobic region is the only portion of the p10 proteins that bears any resemblance to a fusion peptide motif. However, the overall hydrophobicity of this region is considerably less than that of the N-terminal fusion peptides of enveloped viral fusion proteins as determined using the normalized consensus scale of Eisenberg (1984) (0.29 for ARV and 0.41 for NBV, versus an average of 0.61 for enveloped virus fusion peptides) (White, 1990). Moreover, the p10 hydrophobic helix does not display an obviously amphipathic nature or preference for bulkier amino acids on one side of the helix, common features of enveloped virus fusion peptides (reviewed in White, 1990). If this region does function as a fusion peptide by interacting directly with the phospholipid bilayer of target membranes, then it represents an unusual fusion peptide motif.
The reovirus p10 protein is a surface-localized type I transmembrane protein
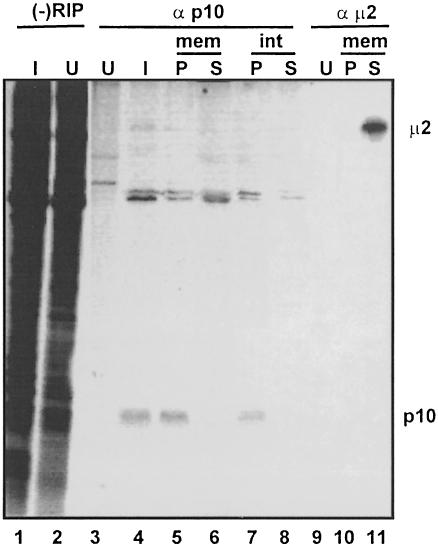
If the reovirus p10 fusion-associated proteins contribute directly to membrane fusion, then one would expect that these proteins should be surface-localized TM proteins. Immunoprecipitation of the membrane fraction from ARV-infected cells clearly indicated that p10 exists exclusively in the membrane pellet (Figure 5). As a control, antiserum specific for a major outer capsid protein of the virus, μ2, was used to demonstrate that this soluble viral protein resided entirely in the supernatant fraction (Figure 5, lanes 10 and 11), indicating that the membrane pellet was devoid of detectable contamination with the soluble fraction of the cell lysate. Moreover, the removal of proteins peripherally associated with the membrane fraction by extraction with high salt and pH did not remove p10 (Figure 5, lane 7), indicating that p10 is an integral membrane protein, consistent with the presence of a predicted central TM domain (Figure 4).
Fig. 5. The p10 protein is an integral membrane protein. Uninfected (U) or ARV-infected (I) cell lysates (–RIP) were immunoprecipitated using anti-p10 (α p10) or anti-μ2C (α μ2C). The infected cell lysates were also fractionated into the membrane pellet (P) or membrane supernatant (S) fractions before immunoprecipitation, either without (mem) or with (int) prior extraction of peripheral membrane proteins to reveal the integral nature of p10 membrane association. Samples were resolved by SDS–PAGE using a 15% acrylamide gel, and detected by fluorography.
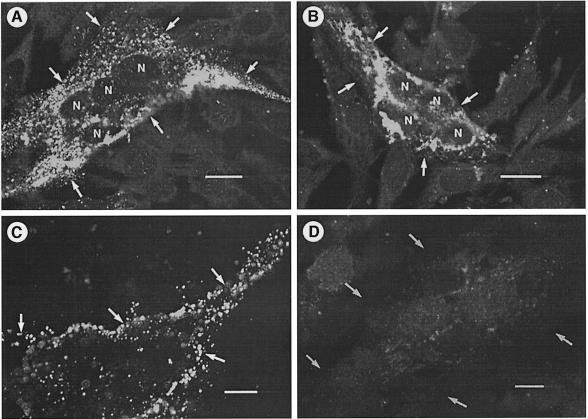
To assess the membrane orientation and surface localization of p10, the N– and C–termini of the ARV p10 protein were tagged using the influenza virus HA epitope, and an anti-HA monoclonal antibody was used for immunofluorescent staining of permeabilized and non-permeabilized transfected cells (Figure 6). The N– and C-terminal epitope tags had no significant effect on p10-induced syncytium formation (see Figure 7). Immunofluorescent staining of permeabilized transfected cells revealed a diffuse punctate staining pattern in the cytoplasm of syncytial cells generated by transfection with either modified p10 construct (Figure 6A and B), indicating that both proteins were expressed in transfected cells. Staining of non-permeabilized cells transfected with the N–tagged p10 construct revealed fluorescent staining of the periphery of syncytial foci (Figure 6C), clearly indicating the presence of cell surface-localized p10. Conversely, no specific fluorescence was detected in non-permeabilized cells expressing the C-terminal tagged p10 construct (Figure 6D), confirming the surface specificity of the fluorescence observed with the N-terminal tagged p10 construct. These results indicated that p10 localizes to the cell surface in a type I (N–out) orientation.
Fig. 6. p10 is a surface-localized type I transmembrane protein. Cells were transfected with the N-terminal (A and C) or C-terminal (B and D) HA-tagged p10 constructs. The transfected cells were stained using anti-HA monoclonal antibody and FITC–conjugated secondary antibody. The cells in (A) and (B) were permeabilized prior to incubation with the antibody to reveal intracellular expression of the tagged p10 constructs. Cells in (C) and (D) were stained without permeabilization of the cells to reveal surface-localized p10. The arrows indicate the membrane boundaries of a single syncytial focus. The nuclei present within a syncytium in the permeabilized cells are indicated (N). The bars represent 10 μm.
Fig. 7. Anti-HA monoclonal antibody inhibits syncytium formation by the N-terminal tagged p10 construct. Quail cells were transfected with plasmids expressing the N-terminal (A and C) or C-terminal (B and D) HA-tagged p10 constructs, and incubated in the absence (A and B) or presence (C and D) of anti-HA monoclonal antibody. Monolayers were fixed and Wright–Giemsa stained to reveal the presence of multinucleated syncytial foci.
As further evidence of the surface localization and membrane orientation of p10, we used the anti-HA monoclonal antibodies in a syncytial inhibition assay. Both the N– and C-terminally tagged p10 constructs induced syncytium formation in transfected cells (Figure 7A and B). Addition of the anti-HA monoclonal antibody to the medium on transfected cells abrogated syncytium formation induced by the N-terminally modified p10 construct (Figure 7C), but had no effect on syncytium formation induced by the C-terminally tagged p10 construct (Figure 7D). These results confirmed the type I surface orientation of p10, and provided indirect evidence that p10 might be involved directly in the fusion reaction.
Mutational analysis of the reovirus p10 fusion proteins
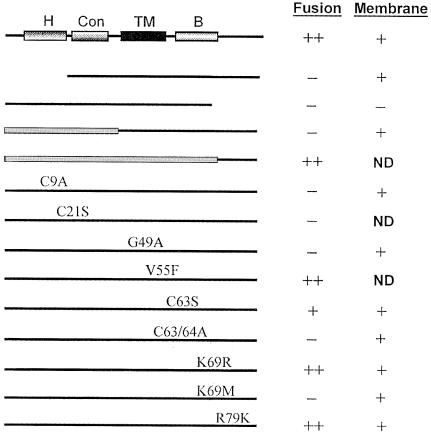
In order to assess the significance of the sequence-predicted structural motifs we identified in p10, a series of mutations were engineered into the ARV p10 protein, and the fusogenic activity and membrane association of the altered proteins were determined. The results obtained from HA tagging indicated that alteration of the termini of p10 had no effect on the fusogenic activity of the protein (Figure 7). However, deletion of the extreme N– or C-terminus of p10 abrogated the fusion-inducing ability of the protein (Figure 8). Deletion of the N-terminal domain did not affect p10 membrane association, indicating that the N-terminal domain influences the functional structure of the protein independently of its membrane association. Interestingly, while deletion of the ARV C-terminus eliminated both the membrane association and fusogenic capability of p10, substitution of the ARV C-terminus with the non-conserved C-terminus of NBV (Figure 8), or with the HA tag (Figure 7), restored both properties. The C-terminal domain of p10 apparently functions in a sequence-independent manner to effect p10 membrane association.
Fig. 8. Deletion and substitution analysis of p10. Various deletions or site-specific substitutions were constructed in the ARV p10 protein, the modified proteins were expressed in transfected cells, and the ability of the proteins to induce cell fusion or to localize to the membrane fraction was assessed. Similar approaches were used to assess the effects of chimeric constructs of the ARV and NBV p10 proteins (filled rectangles indicate NBV sequences). The identities and approximate locations of the site-specific substitutions are indicated, using the single letter amino acid code to indicate the identity of the authentic amino acid, its position and the identity of the substituted residue. The C21S and V55F substitutions were constructed in the NBV p10 protein. ND, not determined.
Since the N– and C-terminal domains of p10 are physically separated in distinct subcellular environments by the intervening TM domain and are likely to fold independently, it might be expected that alterations in the N-terminal domain should not affect the folding of the C-terminal domain. In conjunction with the extensive sequence conservation between the ARV and NBV p10 N-terminal domains, and the presence of conserved structural motifs in this region, we anticipated that the N-terminal domains of ARV and NBV should be interchangeable. However, this was found not to be the case; substitution of the ARV N-terminal domain with that of NBV eliminated the fusogenic property of p10 but did not influence membrane association (Figure 8). This somewhat surprising result suggested that the N-terminal domain of p10 functions in a sequence-dependent manner, and in concert with the TM and/or C-terminal domains of p10, to influence p10 structure or function.
To evaluate the role of the conserved cysteine residues in p10 membrane localization and fusion, site-specific substitutions were engineered into the p10 protein. Alteration of the N-terminal cysteine residues (C9A and C21S constructs) ablated the fusogenic property of p10 (Figure 8). The substitution of Cys21 by serine, an alteration that conserves both hydrophobicity and mass, suggested an essential requirement for a cysteine residue in this location. These cysteine residues are unlikely to mediate disulfide-stabilized dimer formation of p10 since the electrophoretic mobility of p10 was not altered under non-reducing conditions (data not shown). Similarly, the importance of the conserved di-cysteine motif adjacent to the TM domain was confirmed by substitution analysis. A single substitution (C63S) of the di-cysteine motif reduced, but did not abrogate, p10-induced cell fusion, while alanine substitution of both cysteine residues (C63/64A) eliminated the fusogenic properties of p10 (Figure 8). Alteration of these cysteine residues did not affect p10 membrane association.
Site-directed substitutions were also engineered into the conserved basic region and TM domain of p10 (Figure 8). Conservative substitution of basic residues in the C-terminal basic region (K69R and R79K) had no effect on p10 membrane fusion, while a non-conservative substitution (K69M) eliminated p10-induced fusion (Figure 8). A conservative substitution in the predicted TM domain (V55F) had no effect on p10 function, while a conservative substitution in the conserved polyglycine region of the TM domain (G49A) eliminated p10-induced syncytium formation. Interestingly, the substitution in the polyglycine region and the non-conservative substitution in the basic region, all of which eliminated the fusogenic activity of p10, did not affect p10 membrane association. These results indicated that minor alterations to the TM domain and basic region in p10 alter protein structure or function and affect the fusogenic property of p10 independently of the influence of these regions on p10 membrane association.
Discussion
The analysis of the influenza virus HA and of the cellular SNARE proteins involved in constitutive vesicle transport and regulated exocytosis has contributed to the development of a model for protein-mediated membrane fusion (Carr and Kim, 1993; Rothman, 1994; Pfeffer, 1996; Weimbs et al., 1997; Skehel and Wiley, 1998; Weber et al., 1998). Structural and functional studies suggest that the rearrangement of extended heptad repeat structures in membrane-anchored fusion proteins may function to supply the energy required to overcome the thermodynamic barriers that prevent spontaneous membrane fusion. This current model is unlikely to be the complete story, however, since certain viral fusion proteins do not conform to the current paradigm of membrane fusion induced by enveloped virus fusion proteins. For example, although the 14 kDa fusion protein of vaccinia virus contains a coiled-coil motif, this small atypical fusion protein lacks an identifiable fusion peptide and is anchored in membranes not through a TM, but via interactions with another vaccinia-encoded protein (Vazquez et al., 1998). Furthermore, structural analysis of the fusion proteins of various alphaviruses and flaviviruses indicates that a requirement for extensive rearrangements mediated by heptad repeats is not universal (Kielian, 1995; Rey et al., 1995). Consequently, alternative models of proteinmediated membrane fusion need to be developed.
The unusual properties of the ARV and NBV fusion proteins described in this report are without precedent amongst the viral and cellular proteins implicated in membrane fusion. In conjunction with the absence of any identifiable homologues, the unique structural features of the reovirus p10 proteins suggest that these fusion-associated small transmembrane (FAST) proteins represent a new class of membrane fusion-inducing proteins, the first example of non-structural proteins encoded by a non-enveloped virus that are capable of inducing fusion from within. The FAST proteins contain only a small 39–43 amino acid ectodomain that lacks an extended heptad repeat; therefore, the extensive conformational changes that accompany membrane fusion induced by certain enveloped virus fusion proteins are unlikely to be possible in these simple fusion-inducing proteins. How such a simple protein could overcome the thermodynamic barriers to membrane fusion presently is unknown, although it seems clear that the FAST proteins are likely to use a novel mechanism to promote membrane fusion.
Our results indicate that the FAST proteins are the only reovirus proteins required to promote syncytium formation. It is not possible, however, to state that the FAST proteins function independently to induce membrane fusion and, hence, are true fusion proteins per se. It is conceivable, for example, that the FAST proteins might function indirectly to effect cell–cell fusion, possibly through the auspices of an unidentified host factor. However, the ability of a viral protein to trigger a cellular fusion response indirectly has never been reported. Furthermore, actinomycin D inhibits host cell transcription, but has no effect on reovirus transcription or on virus-induced syncytium formation (Ni and Ramig, 1993; R.Duncan, unpublished). Consequently, the FAST proteins would need to modulate the activity of a pre-existing host factor that never functions independently to promote exoplasmic fusion, but is capable of doing so only in the presence of p10. Such a scenario seems unlikely. It seems more probable that the FAST proteins are, in fact, fusion proteins that contribute directly to lipid bilayer mixing. This contention is supported by the cell surface localization of p10, and by the ability of HA monoclonal antibodies to abrogate syncytium formation induced by the N-terminally tagged p10 construct. Direct evidence that p10 alone is sufficient to induce membrane fusion will require the demonstration that purified p10 promotes fusion of pure phospholipid bilayers. Such studies currently are under way.
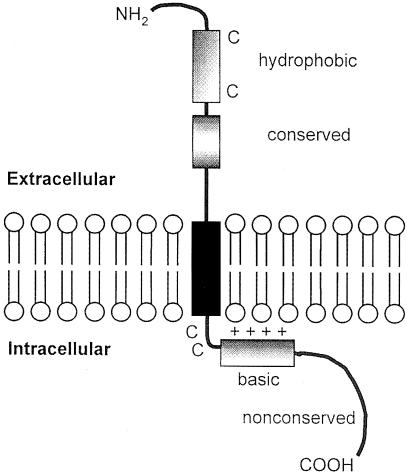
Our preliminary sequence and functional analyses of the reovirus fusion-associated proteins provide the basis for a working model of p10 structure and function (Figure 9). The p10 proteins are surface-localized type I transmembrane proteins. Our observation that brefeldin A, an inhibitor of vesicular transport, abrogates ARV-induced cell fusion (Duncan et al., 1996) is consistent with p10 transport through the endoplasmic reticulum (ER)–Golgi pathway (Einfeld and Hunter, 1991). The deletion and substitution analysis of the C-terminus of p10 suggests that p10 localization to the ER most probably occurs via a signal recognition particle (SRP)-dependent targeting mechanism, mediated by the TM domain serving as a signal/anchor sequence (Pugsley, 1990; Zheng and Gierasch, 1996; Wilkinson et al., 1997; Matlack et al., 1998). This conclusion is based on the absence of a cleavable N-terminal signal peptide in p10 (Martoglio and Dobberstein, 1998), and on the fact that deletion of the non-conserved C-terminus eliminates p10 membrane association while substitution of this region with the NBV C-terminus or with an HA epitope restores both p10 membrane association and fusion. The fact that substitution of the C-terminal portion of p10 with heterologous sequences restores membrane association indicates that this region functions in a sequence-independent manner to effect targeting of p10 to the membrane fraction of cells. Since the SRP only recognizes nascent signal peptides, ∼30–40 amino acids (the length of polypeptide protected by a translating ribosome and the approximate length of the p10 C-terminal domain) must lie on the C–proximal side of the signal/anchor peptide to allow it to be exposed on the surface of the ribosome for interaction with the SRP particle (Pugsley, 1990). Therefore, we suggest that the C-terminal tail of p10 may serve as a ‘stuffer’ to permit SRP-dependent ER insertion.
Fig. 9. Structural model of the reovirus p10 fusion protein. The membrane orientation of the p10 protein is diagrammed, along with the locations of the conserved cysteine residues, hydrophobic heptad, basic region, transmembrane domain, a short stretch of completely conserved residues and the non-conserved C-terminal region.
Additional mutagenic analyses demonstrated the importance of several conserved motifs present in the ARV and NBV p10 proteins. A conservative substitution in the predicted TM domain (V55F) had no effect on syncytium formation, while a single alteration to the polyglycine region in the TM domain (G49A) eliminated cell fusion but did not affect p10 membrane association. The ability to disrupt the fusogenic property of p10 without altering p10 membrane association suggests that the TM domain may serve as more than just a signal/anchor, either by destabilizing the donor membrane, as suggested by studies with GPI-anchored HA, which promotes hemifusion but not complete fusion (Kemble et al., 1994), or by promoting functional p10 folding or multimer formation as occurs with several integral membrane proteins (McGinnes et al., 1993; Lemmon et al., 1994; Shai, 1995; Mingarro et al., 1996; Burke et al., 1997).
The majority of the basic residues present in p10 reside in the cytoplasmic domain, immediately adjacent to the predicted TM domain (Figure 4). These basic residues probably contribute to the type I orientation of the protein. However, the presence of a basic domain adjacent to a TM domain is a hallmark feature of a large group of small membrane proteins, the viroporins (also referred to as holins in bacteriophages), encoded by numerous enveloped and non-enveloped viruses (Young, 1992; Carrasco, 1995). Viroporins appear to contribute to cellular membrane destabilization, possibly as a means to promote virus exit from cells (Tollefson et al., 1996; Tiganos et al., 1998). Our preliminary mutagenic analysis implicates the basic region in p10 function independently of any role it might have in p10 membrane association. Conservative changes in the p10 cytoplasmic basic domain had no effect on p10 function, while a single non-conservative substitution (K69M) eliminated the fusogenic activity, but not p10 membrane association. Since the cytoplasmic, TM and extracellular domains of transmembrane proteins generally fold independently (Doms et al., 1993), it is likely that a single substitution in the basic domain of p10 would have only local effects on p10 structure. It is conceivable, therefore, that the p10 basic domain may not only influence the membrane orientation of the protein, but may also contribute to destabilization of the donor lipid bilayer, analogously to the viroporins. A concerted mutagenic analysis of the basic region in the context of the N–terminal HA-tagged construct should reveal the influence of this region on the relationship between p10 membrane localization and membrane fusion.
Alteration of the conserved cysteine residues in p10 reduced, or eliminated, the fusion-inducing property of p10 but did not affect p10 membrane association. The two conserved cysteine residues in the predicted cytoplasmic domain of p10, immediately adjacent to the TM domain (Figure 9), may be palmitylated, similarly to the situation with the adenovirus death protein (Hausmann et al., 1998). Although several enveloped virus fusion proteins are also palmitylated on membrane-proximal cysteine residues, the role for palmitylation in the fusion activity of enveloped virus fusion proteins is variable (Yang et al., 1995; Veit et al., 1996; Fisher et al., 1998; Ryan et al., 1998). Similarly, alteration of either of the two cysteine residues flanking the small hydrophobic region in the N-terminal domain (Figure 4) abrogated p10-induced cell fusion. This is the only region of p10 that bears any resemblance to a fusion peptide motif, containing a moderately hydrophobic short heptad repeat structure that might exist in a membrane-seeking helical conformation. However, the biophysical properties of this region are quite distinct from those of any previously characterized fusion peptides from enveloped virus fusion proteins; hence, the identification of this region as a fusion peptide must be considered tentative.
The FAST proteins of the fusogenic reoviruses are clearly distinct from any previously identified fusion-inducing proteins, and may offer a minimalist model for investigating the mechanism of protein-mediated membrane fusion. The FAST proteins are not involved directly in virus entry into or exit from cells, and appear to be non-essential proteins of the virus whose sole, or primary, purpose is to promote membrane fusion (Duncan, 1996; Duncan et al., 1996). The accessory nature of the FAST proteins may have afforded these non-structural viral fusion-inducing proteins the ability to evolve a simplified structure with a specialized purpose. In addition, since they do not contribute directly to virus entry or exit, their fusion activity may not be subject to the triggering mechanisms that regulate the fusogenic activity of enveloped virus fusion proteins. The absence of a requirement for regulated fusion would further permit these novel fusion proteins to simplify their domain organization to include the minimal determinants required to direct membrane localization, destabilization and fusion.
Materials and methods
Plasmids, virus and cells
ARV strain 176 and NBV have been described previously (Duncan et al., 1995), and were grown and plaque-purified in a continuous quail cell line, QM5 (Duncan and Sullivan, 1998) or in Vero cells, respectively. All cells were maintained in growth medium consisting of medium 199 supplemented with 10% fetal bovine serum (FBS), 10% tryptose phosphate broth and penicillin/streptomycin (50 U/ml and 50 μg/ml, respectively). The QM5 cells were used for most of the transfection assays due to their high transfection efficiency.
The eukaryotic expression vector pcDNA3 (Invitrogen) was used for expression of viral genes in transfected cells. pMAL-c2 (New England Biolabs) was used for expression of the MBP–p10 chimeric protein in E.coli.
Cloning, site-directed PCR mutagenesis and epitope tagging
The full-length cDNAs corresponding to the S1 genome segments of ARV and NBV were cloned into pcDNA3, as previously described (Duncan, 1999). The sequences of the ARV-176 and NBV S1 genome segment cDNAs are deposited in DDBJ/EMBL/GenBank (accession Nos AF218358 and AF218360, respectively). These clones were used as templates for PCR subcloning, using Vent polymerase (New England Biolabs), to generate fragments corresponding to the p10 gene alone, the p10 gene with an optimized translational start sequence, p10 harbouring site-specific mutations and p10 containing the HA epitope at its N– or C-terminus. The sequence of all constructs was confirmed. To synthesize the p10 gene, and the p10 gene containing an optimized translation initiation sequence, forward primers 5′–TACTACTAAG– CTTGCTTTTTCAATCCCTTGTTCG–3′ and 5′–TACTACTAAGCTT– GCTTTTTCAATCCCTTGTTCCACCATGGTGCGTATGCC–3′ were used, respectively, along with the reverse primer 5′–TGAAGA– AGCGGCCGCGAAGTGATAGCGGAC–3′. Primers annealed to the non-coding sequences (underlined) flanking the 5′ and 3′ ends of the p10 ORF, and added HindIII and NotI sites to the 5′ and 3′ ends, respectively. Primers containing the HA nonapeptide sequence (5′–TACCCATACGATGTTCCTGACTATGCG–3′) and sequences complementary to the 5′ or 3′ ends of the p10 ORF were used to introduce the HA epitope as a nine residue N-terminal extension, or C-terminal replacement, of the p10 ORF. The final PCR consisted of 1× Vent polymerase buffer, 2 mM MgSO4, 0.2 mM each dNTP, 0.05 pmol of template, 40 pmol of each forward and reverse primer, and 0.5 μl of Vent polymerase in a final volume of 100 μl. Samples were heated at 94°C for 4 min and then cycled 30 times at 94°C for 1 min, 52°C for 30 s, then 72°C for 30 s. Products and vector (pcDNA3) were digested with HindIII and NotI, and gel purified using β–agarase (New England Biolabs) and Geneclean (BIO101) according to the manufacturer's instructions. The purified vector and insert were ligated, and transformed into Top-10 cells according to standard protocols.
All site-directed mutations were made using a rapid PCR-based technique. Internal primers were synthesized that incorporated the desired mutation flanked by extended sequence complementary to the template. In the first round of PCR, the forward primer containing the optimized translational start sequence (see above) was used along with the internal mutagenic primer to synthesize a fragment containing the mutation near the 3′ end. The original primers were removed using Qiaquick (Qiagen), and the first round PCR product was used as the primer for a second round of PCR, along with the reverse p10 primer (see above), producing a fragment corresponding to the entire p10 gene containing the mutation. Touch-down PCR was used for better product specificity and yield, which involved five cycles of 94°C for 1 min, 52°C for 40 s, 72°C for 40 s; five cycles of 94°C for 1 min, 50°C for 40 s, 72°C for 40 s; and 25 cycles of 94°C for 1 min, 48°C for 40 s, 72°C for 40 s. The products were then cloned into pcDNA3 as above. Using this method, a mutation can be made specifically and inserted into a vector in a single day using only one additional primer.
p10 antiserum production
Polyclonal antiserum was raised against the C-terminal domain of p10. To synthesize the MBP–p10 recombinant protein construct, the C-terminal portion of p10 (amino acids 63–98) was cloned in-frame with the MBP in the pMAL-c2 vector. PCR was performed as above using the forward primer 5′–TACTGTTGTAAGGCTAAGGTC–3′ and the reverse primer 5′–CGCGGATCCTCAGTTACGTCGTATGGCGGAG– C–3′ (underlined sequences are complementary to the p10 ORF), cloned into the XmnI and BamHI sights of pMAL-c2 (New England Biolabs), and transformed into E.coli Top-10 cells. The chimeric MBP–p10 protein was induced with isopropyl-β-d-thiogalactopyranoside (IPTG), purified from 1 l cultures on amylose affinity columns, and used to inject rabbits. The rabbits were immunized at three sites (two intramuscular and one subcutaneous) using 300–500 μg of the chimeric protein in Freund's complete adjuvant, then repeatedly boosted using a similar regime with Freund's incomplete adjuvant. Animals were exsanguinated when the antibody titre plateaued after seven injection series. A similar protocol failed to obtain an immune response against the N-terminal domain of p10.
Cell staining
Monolayers of QM5 cells were transfected with the p10-expressing pcDNA3 constructs using Lipofectamine (Life Technologies Inc.) and incubated for 24–36 h. Cell monolayers were also infected with ARV at a low multiplicity of infection (m.o.i.) to generate syncytial foci after 16 h. Transfected or infected cell monolayers were stained with Wright–Giemsa stain (Diff-Quik) according to the manufacturer's instructions (VWR Scientific) to visualize cell nuclei and polykaryon formation. Viral proteins were detected immunocytochemically using primary antiserum raised either against virus structural proteins (Duncan et al., 1996) or against p10. The p10 antiserum and the polyclonal anti-ARV serum were diluted 1:400 in antibody blocking buffer [2% bovine serum albumin (BSA), 10% normal goat serum, 20 mM Tris pH 7.5, 150 mM NaCl, 0.1% NP-40] and adsorbed to fixed monolayers for 60 min at room temperature. The monolayers were washed extensively before and after antibody additions with phosphate-buffered saline (PBS) containing 2% BSA. Foci were visualized using a secondary goat anti-rabbit IgG conjugated with alkaline phosphatase (Life Technologies; 1:600 dilution) according to standard protocols (Harlow and Lane, 1988). Cells stained with alkaline phosphatase were visualized and photographed on a Nikon Diaphot inverted microscope at 100× magnification.
Fluorescent staining and syncytial inhibition using HA monoclonal antibodies
The HA-tagged p10 constructs were expressed in transfected cells growing on multiwell chamber slides (Nunc) as described above. The medium was removed from the transfected cells 28 h post-transfection, the monolayers were washed twice with Hank's balanced salt solution and pre-blocked with antibody blocking buffer for 30 min at room temperature. The HA monoclonal antibody was prepared from 12CA5 hybridoma cell culture supernatants by ammonium sulfate precipitation (50% saturation) and dialysis against PBS. The antibody suspension (5 mg/ml protein) was diluted 1:100 in antibody blocking buffer and incubated with the unfixed cell monolayers for 1 h at room temperature to detect cell surface-localized p10. For visualization of intracellular p10 expression, cells were fixed and permeabilized using methanol prior to addition of a 1:200 dilution of the monoclonal antibody. Primary antibody was removed by four washes with Hank's balanced salt solution containing 2% FBS at room temperature for 30 min. A secondary rabbit anti-mouse fluorescein isothiocyanate (FITC)-conjugated antiserum (Life Technologies Inc.) was diluted 1:20 in antibody blocking buffer, and incubated with the monolayers for 1 h. Monolayers were washed extensively as above, fixed with 4% paraformaldehyde for 10 min at room temperature, and the slides mounted for examination by confocal microscopy. The cells were visualized and photographed on a Zeiss LSM510 scanning argon laser confocal microscope with appropriate filter sets using the 63× or 100× objectives.
For antibody inhibition of syncytium formation, the HA monoclonal antibody was diluted 1:400 in tissue culture medium and added to transfected cells 4 h post-transfection. At 36 h post-transfection, the cells were methanol fixed and Giemsa stained as described above, and examined for the presence of multinucleated syncytia.
Analysis of virus structural proteins
The analysis of virus structural proteins was essentially as previously described (Duncan, 1996). QM5 cells grown in 175 cm2 flasks (3.6 × 107 cells) were infected at an m.o.i. of 0.1, labelled at 14 and again at 17 h post-infection with [35S]methionine (50 μCi/ml), and the infection was allowed to proceed until cell lysis. Cell lysates were frozen and thawed three times to disrupt virus aggregates, centrifuged at 10 000 g for 20 min to remove cell debris, then centrifuged at 100 000 g for 1 h through a 30% (w/v) sucrose cushion to obtain the virus particles. The virus pellet was resuspended in 1% SDS, and the virus particles were disrupted by heating at 37°C for 30 min to liberate all of the structural proteins. The disrupted virions were diluted in RIPA to a final concentration of 0.1% SDS before proceeding to immunoprecipitation.
Immunoprecipitation
Immunoprecipitation was performed as previously described (Duncan and Sullivan, 1998). QM5 cells were transfected or infected with ARV at an m.o.i. of 0.1 and labelled with [35S]methionine (50 μCi/ml) for 1 h at 24 or 14 h post-transfection/infection, respectively. Cells were lysed on ice in RIPA buffer containing protease inhibitors, and cell lysates were centrifuged at 100 000 g for 25 min to remove virus particles. The supernatant was precipitated for 60 min on ice using rabbit antiserum raised against viral structural proteins, p10 or normal rabbit serum (all diluted 1:250). Immune complexes were recovered using IgGsorb (The Enzyme Center), washed extensively with RIPA and released by boiling in SDS protein sample buffer (Laemmli, 1970) before SDS–PAGE using 15% acrylamide gels.
Membrane fractionation of infected and transfected cells
QM5 cells, in 12-well cluster plates, were infected with ARV at an m.o.i. of 0.1, or cells were transfected using LipofectAMINE reagent (Life Technologies) according to product instructions, using 3 μl of LipofectAMINE and 0.75 μg of DNA on 70% confluent cell monolayers in 12-well cluster plates. Infected/transfected cells were labelled with [35S]methionine (50 μCi/ml) for 1 h when extensive syncytium was observed, washed twice with PBS, harvested by scraping into 1 ml of PBS, then passed through a 30 gauge needle 10 times. Nuclei and cell debris were removed by centrifugation at 600 g for 3 min, and the membrane fraction was recovered by centrifugation at 100 000 g for 25 min. The membrane pellet was either dissolved in electrophoresis sample buffer (Laemmli, 1970) for direct analysis by SDS–PAGE using 15% acrylamide gels, or was dissolved in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.02% NaN3) containing protease inhibitors (1 μg/ml each of aprotinin, leupeptin and pepstatin) for subsequent immunoprecipitation analysis. For removal of peripheral membrane-associated proteins, pellets consisting of membranous material were treated with 100 mM Na2CO3 pH 11.3, for 30 min on ice, followed by centrifugation at 100 000 g for 25 min to recover the membrane fraction and associated integral membrane proteins.
Acknowledgments
Acknowledgements
The authors thank Jingyun Shou for expert technical assistance and Jennifer Corcoran for assistance with the production of p10 antiserum. This research was funded by grants from the Medical Research Council of Canada and from the Natural Sciences and Engineering Research Council (NSERC) of Canada. M.S. was funded by scholarships from NSERC and from the Killam Foundation.
References
- Ben-Efraim I., Kliger, Y., Hermesh, C. and Shai, Y. (1999) Membrane-induced step in the activation of Sendai virus fusion protein. J. Mol. Biol., 285, 609–625. [DOI] [PubMed] [Google Scholar]
- Bentz J. (1993) Viral Fusion Mechanisms. CRC Press, Boca Raton, FL. [Google Scholar]
- Bock J.B. and Scheller, R.H. (1997) A fusion of new ideas. Nature, 387, 133–135. [DOI] [PubMed] [Google Scholar]
- Burke C.L., Lemmon, M.A., Coren, B.A., Engelman, D.M. and Stern, D.F. (1997) Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene, 14, 687–696. [DOI] [PubMed] [Google Scholar]
- Carr C.M. and Kim, P.S. (1993) A spring-loaded mechanism for the conformational change of influenza virus hemagglutinin. Cell, 73, 823–832. [DOI] [PubMed] [Google Scholar]
- Carrasco L. (1995) Modification of membrane permeability by animal viruses. Adv. Virus Res., 45, 61–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R.W., Lamb, R.A., Rose, J.K. and Helenius, A. (1993) Folding and assembly of viral membrane proteins. Virology, 193, 545–562. [DOI] [PubMed] [Google Scholar]
- Duncan R. (1996) The low pH-dependent entry of avian reovirus is accompanied by two specific cleavages of the major outer capsid protein μ2C. Virology, 219, 179–189. [DOI] [PubMed] [Google Scholar]
- Duncan R. (1999) Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology, 260, 316–328. [DOI] [PubMed] [Google Scholar]
- Duncan R. and Sullivan, K. (1998) Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities. Virology, 250, 263–272. [DOI] [PubMed] [Google Scholar]
- Duncan R., Murphy, F.A. and Mirkovic, R. (1995) Characterization of a novel syncytium-inducing baboon reovirus. Virology, 212, 752–756. [DOI] [PubMed] [Google Scholar]
- Duncan R., Chen, Z., Walsh, S. and Wu, S. (1996) Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology, 224, 453–464. [DOI] [PubMed] [Google Scholar]
- Einfeld D. and Hunter, E. (1991) Transport of membrane proteins to the cell surface. Curr. Top. Microbiol. Immunol., 170, 107–139. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. (1984) Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem., 53, 595–623. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S. and Jahn, R. (1994) Vesicle fusion from yeast to man. Nature, 370, 191–193. [DOI] [PubMed] [Google Scholar]
- Fischer C., Schroth-Diez, B., Herrmann, A., Garten, W. and Klenk, H. (1998) Acylation of the influenza hemagglutinin modulates fusion activity. Virology, 248, 284–294. [DOI] [PubMed] [Google Scholar]
- Gard G. and Compans, R.W. (1970) Structure and cytopathic effects of Nelson Bay virus. J. Virol., 6, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Ruigrok, R.W.H. and Brunner, J. (1995) Low-pH induced conformational changes in viral fusion proteins: implications for the fusion mechanism. J. Gen. Virol., 76, 1541–1556. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hausmann J., Ortmann, D., Witt, E., Veit, M. and Seidel, W. (1998) Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology, 244, 343–351. [DOI] [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman, L.R., Wolfsberg, T.G. and White, J.M. (1996) Virus–cell and cell–cell fusion. Annu. Rev. Cell Dev. Biol., 12, 627–661. [DOI] [PubMed] [Google Scholar]
- Hoekstra D. (1990) Membrane fusion of enveloped viruses: especially a matter of proteins. J. Bioenerg. Biomembr., 22, 121–154. [DOI] [PubMed] [Google Scholar]
- Hughson F.M. (1997) Enveloped viruses: a common mode of membrane fusion?Curr. Biol., 7, R565–R569. [DOI] [PubMed] [Google Scholar]
- Joshi S.B., Dutch, R.E. and Lamb, R.A. (1998) A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology, 248, 20–34. [DOI] [PubMed] [Google Scholar]
- Kawamura H., Shiinizu, F., Maeda, M. and Tsubahara, H. (1965) Avian reovirus: its properties and serological classification. Natl Inst. Anim. Health Q., 5, 115–124. [PubMed] [Google Scholar]
- Kemble G.W., Danieli, T. and White, J.M. (1994) Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell, 76, 383–391. [DOI] [PubMed] [Google Scholar]
- Kielian M. (1995) Membrane fusion and the alphavirus life cycle. Adv. Virus Res., 45, 113–152. [DOI] [PubMed] [Google Scholar]
- Kielian M. and Jungerwirth, S. (1990) Mechanisms of enveloped virus entry into cells. Mol. Biol. Med., 7, 17–31. [PubMed] [Google Scholar]
- Kool D.A. and Holmes,I.H. (1995) The S1 gene sequences of two Australian avian reoviruses. DDBJ/EMBL/GenBank direct submission.
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lanzrein M., Schlegel, A. and Kempf, C. (1994) Entry and uncoating of enveloped viruses. Biochem. J., 302, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. and Engelman, D.M. (1994) Specificity and promiscuity in membrane helix interactions. FEBS Lett., 346, 17–20. [DOI] [PubMed] [Google Scholar]
- Martinez-Costas J., Grande, A., Varela, R., Garcia-Martinez, C. and Benavente, J. (1997) Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J. Virol., 71, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B. and Dobberstein, B. (1998) Signal sequences: more than just greasy peptides. Cell Biol., 8, 410–415. [DOI] [PubMed] [Google Scholar]
- Matlack K.E.S., Mothes, W. and Rapoport, T.A. (1998) Protein translocation: tunnel vision. Cell, 92, 381–390. [DOI] [PubMed] [Google Scholar]
- McGinnes L., Sergel, T. and Morrison, T. (1993) Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology, 196, 101–110. [DOI] [PubMed] [Google Scholar]
- Mingarro I., Whitley, P., Lemmon, M.A. and von Heijne, G. (1996) Ala-insertion scanning mutagenesis of the glycophorin A transmembrane helix: a rapid way to map helix–helix interactions in integral membrane proteins. Protein Sci., 5, 1339–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y. and Ramig, R.F. (1993) Characterization of avian reovirus-induced cell fusion: the role of viral structural proteins. Virology, 194, 705–714. [DOI] [PubMed] [Google Scholar]
- Nibert M.L., Schiff,L.A. and Fields,B.N. (1996) Reoviruses and their replication. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fundamental Virology. 3rd edn. Lippincott-Raven Press, New York, NY, pp. 1557–1596. [Google Scholar]
- Otter-Nilsson M., Hendriks, R., Pecheur-Huet, E.-I., Hoekstra, D. and Nilsson, T. (1999) Cytosolic ATPases, p97 and NSF, are sufficient to mediate rapid membrane fusion. EMBO J., 18, 2074–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B. and Argos, P. (1994) Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol., 237, 182–192. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. (1996) Transport vesicle docking: SNAREs and associates. Annu. Rev. Cell Dev. Biol., 12, 441–461. [DOI] [PubMed] [Google Scholar]
- Pugsley A.P. (1990) Translocation of proteins with signal sequences across membranes. Curr. Opin. Cell Biol., 2, 609–616. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos J. and Pedroso de Lima, M.C. (1998) The influenza virus hemagglutinin: a model protein in the study of membrane fusion. Biochim. Biophys. Acta, 1376, 147–154. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Heinz, F.X., Mandl, C., Kunz, C. and Harrison, S.C. (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature, 375, 291–298. [DOI] [PubMed] [Google Scholar]
- Rothman J.E. (1994) Mechanisms of intracellular protein transport. Nature, 372, 55–63. [DOI] [PubMed] [Google Scholar]
- Ryan C., Ivanova, L. and Schlesinger, M.J. (1998) Effects of site-directed mutations of transmembrane cysteines in Sindbis virus and E2 glycoproteins on palmitylation and virus replication. Virology, 249, 62–67. [DOI] [PubMed] [Google Scholar]
- Shai Y. (1995) Molecular recognition between membrane-spanning polypeptides. Trends Biochem Sci., 11, 460–464. [DOI] [PubMed] [Google Scholar]
- Shapouri M.R.S., Arella, M. and Silim, A. (1996) Evidence for the multimeric nature and cell binding ability of avian reovirus sigma 3 protein. J. Gen. Virol., 77, 1203–1210. [DOI] [PubMed] [Google Scholar]
- Skehel J.J. and Wiley, D.C. (1998) Coiled coils in both intracellular vesicle and viral membrane fusion. Cell, 95, 871–874. [DOI] [PubMed] [Google Scholar]
- Söllner T., Whiteheart, S.W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P. and Rothman, J.E. (1993) SNAP receptors implicated in vesicle targeting and fusion. Nature, 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Stegmann T., Doms, R.W. and Helenius, A. (1989) Protein-mediated membrane fusion. Annu. Rev. Biophys. Biophys. Chem., 18, 187–211. [DOI] [PubMed] [Google Scholar]
- Strong J.E., Leone, G., Duncan, R., Sharma, R.K. and Lee, P.W.K. (1991) Biochemical and biophysical characterization of the reovirus cell attachment protein σ1: evidence that it is a homotrimer. Virology, 184, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer, D., Jahn, R. and Brunger, A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Theophilos M.B., Huang, J.-A. and Holmes, I.H. (1995) Avian reovirus sigma C protein contains a putative fusion sequence and induces fusion when expressed in mammalian cells. Virology, 208, 678–684. [DOI] [PubMed] [Google Scholar]
- Tiganos E., Friborg, J., Allain, B., Daniel, N.G., Yao, X. and Cohen, É. (1998) Structural and functional analysis of the membrane-spanning domain of the human immunodeficiency virus type 1 Vpu protein. Virology, 251, 96–107. [DOI] [PubMed] [Google Scholar]
- Tollefson A.E., Scaria, A., Hermiston, T.W., Ryerse, J.S., Wold, L.J. and Wold, W.S.M. (1996) The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol., 70, 2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Sato, K. and Wickner, W. (1998) Defining the functions of trans-SNARE pairs. Nature, 396, 543–548. [DOI] [PubMed] [Google Scholar]
- Varela R. and Benavente, J. (1994) Protein coding assignment of avian reovirus strain S1133. J. Virol., 68, 6775–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez M.I., Rivas, G., Cregut, D., Serrano, L. and Esteban, M. (1998) The vaccinia virus 14-kilodalton (A27L) fusion protein forms a triple coiled-coil structure and interacts with the 21-kilodalton (A17L) virus membrane protein through a C-terminal α-helix. J. Virol., 72, 10126–10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M., Reverey, H. and Schmidt, M.F. (1996) Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem. J., 318, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman, B.V., McNew, J.A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T.H. and Rothman, J.E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell, 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Weimbs T., Low, S.H., Chapin, S.J., Mostov, K.E., Bucher, P. and Hoffman, K. (1997) A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl Acad. Sci. USA, 94, 3046–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen, A., Harrison, S.C., Skehel, J.J. and Wiley, D.C. (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature, 387, 426–430. [DOI] [PubMed] [Google Scholar]
- White J.M. (1990) Viral and cellular membrane fusion proteins. Annu. Rev. Physiol., 52, 675–697. [DOI] [PubMed] [Google Scholar]
- Wilkinson B.M., Regnacq, M. and Stirling, C.J. (1997) Protein translocation across the membrane of the endoplasmic reticulum. J. Membr. Biol., 155, 189–197. [DOI] [PubMed] [Google Scholar]
- Yang C., Spies, C.P. and Compans, R.W. (1995) The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl Acad. Sci. USA, 92, 9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. (1992) Bacteriophage lysis: mechanism and regulation. Microbiol. Rev., 56, 430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N. and Gierasch, L.M. (1996) Signal sequences: the same yet different. Cell, 86, 849–852. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J., Vogel, S.S. and Chernomordik, L.V. (1993) Mechanisms of membrane fusion. Annu. Rev. Biophys. Biomol. Struct., 22, 433–466. [DOI] [PubMed] [Google Scholar]