Abstract
Virus-encoded movement protein (MP) mediates cell-to-cell spread of tobacco mosaic virus (TMV) through plant intercellular connections, the plasmodesmata. The molecular pathway by which TMV MP interacts with the host cell is largely unknown. To understand this process better, a cell wall-associated protein that specifically binds the viral MP was purified from tobacco leaf cell walls and identified as pectin methylesterase (PME). In addition to TMV MP, PME is recognized by MPs of turnip vein clearing virus (TVCV) and cauliflower mosaic virus (CaMV). The use of amino acid deletion mutants of TMV MP showed that its domain was necessary and sufficient for association with PME. Deletion of the PME-binding region resulted in inactivation of TMV cell-to-cell movement.
Keywords: movement proteins/pectin methylesterases/plant viruses/plasmodesmata/tobacco
Introduction
Following initial infection, many plant viruses move from cell to cell through plasmodesmata until they reach the vascular system; the viruses are then transported systemically through the vasculature. Presumably, viral spread through the vascular tissue is a passive process, occurring with the flow of photoassimilates (reviewed by Leisner and Howell, 1993); in contrast, cell-to-cell movement is an active function, requiring specific interaction between the invading virus and plasmodesmata. This interaction is mediated by virus-encoded nonstructural movement proteins (MPs) (reviewed by Carrington et al., 1996; Ghoshroy et al., 1997; Lazarowitz and Beachy, 1999).
Tobacco mosaic virus (TMV) MP, one of the best characterized viral MPs, has been shown to localize to plasmodesmata (Tomenius et al., 1987; Atkins et al., 1991; Ding et al., 1992a), increase plasmodesmal permeability (Wolf et al., 1989; Waigmann et al., 1994), cooperatively bind single-strand nucleic acids (Citovsky et al., 1990, 1992a), and interact with cytoskeletal elements (Heinlein et al., 1995; McLean et al., 1995). Based on these observations, MP was proposed to form complexes with the transported genomic TMV RNA, move these complexes throughout the cell using the cytoskeletal network, and target them to and through the enlarged plasmodesmal channels (Citovsky and Zambryski, 1993, 1995; Ghoshroy et al., 1997).
Presently, our knowledge about the cell-to-cell movement of plant viruses derives from studies of its viral components, i.e. MPs. Other than the cytoskeleton (Heinlein et al., 1995; McLean et al., 1995), no cellular components that interact with viral MPs have been identified. Here, we used protein–protein interaction on renatured Western blots to detect a tobacco cell wall protein that binds TMV MP. Following purification on an MP affinity column and microsequencing, this protein was identified as pectin methylesterase (PME). TMV MP–PME binding was confirmed in the yeast two-hybrid system; furthermore, MPs of two other plant viruses, turnip vein clearing virus (TVCV) and cauliflower mosaic virus (CaMV), interacted with PME. Binding experiments using amino acid deletion mutants of TMV MP identified an MP domain required for binding to PME. Deletion of this region blocked the MP ability to mediate the spread of viral infection, suggesting the role of MP–PME binding in the TMV cell-to-cell movement.
Results
Detection of a tobacco cell wall protein that binds MP
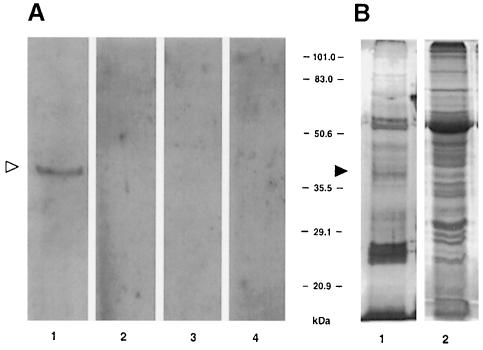
Because TMV MP has been shown to localize to plasmodesmata within plant cell walls (Tomenius et al., 1987; Ding et al., 1992a), it may interact directly with a cell wall-associated receptor. To test this possibility, we developed a renatured blot overlay assay for interaction between TMV MP and cell wall proteins. In this technique, a protein mixture containing a putative receptor is separated by SDS–PAGE (Laemmli, 1970) and electrotransferred onto a PVDF membrane followed by guanidine hydrochloride extraction of SDS from the blotted proteins. The membrane is then renatured, incubated with purified TMV MP, washed and the MP binding is visualized using anti-MP antibodies. Figure 1A, lane 1 shows that MP binding detected a single protein band with a relative electrophoretic mobility of ∼38 kDa. This protein was not detected in the absence of TMV MP (Figure 1A, lane 2) or upon probing an empty PVDF membrane with MP (lane 3). Furthermore, no 38 kDa band or any other TMV MP-interacting proteins were detected in the soluble fractions of tobacco cells (Figure 1A, lane 4).
Fig. 1. Detection of TMV MP-interacting protein in cell wall fraction by immunorecognition of bound MP. (A) Renatured blot overlay assay. Lane 1, cell wall fraction blot incubated with MP; lane 2, cell wall fraction blot incubated with buffer alone; lane 3, empty membrane incubated with TMV MP; lane 4, soluble fraction blot incubated with TMV MP. (B) Protein content of the cell wall and soluble fractions of tobacco leaf tissue. Both lanes represent protein extract derived from 10 mg of fresh tobacco leaf tissue. Lane 1, silver-stained cell wall proteins; lane 2, Coomassie blue-stained soluble proteins. The positions of the 38 kDa TMV MP-interacting protein on the blot (open arrowhead) and on stained SDS–polyacrylamide gels (filled arrowhead) are indicated. The numbers between panels indicate molecular mass standards in kDa.
Overall, the cell wall extract had a significantly lower protein heterogeneity than the soluble cell extract (Figure 1B, lanes 1 and 2, respectively). Among the relatively few cell wall proteins, the 38 kDa protein did not represent a major species (Figure 1B, lane 1, arrowhead) although it was easily visualized with MP as ligand (Figure 1A, lane 1, arrowhead).
Purification and identification of the MP-interacting protein
The tight and selective association between TMV MP and its 38 kDa interactor allowed us to purify this tobacco cell wall protein by affinity chromatography using immobilized MP. Cell wall extracts were loaded on the TMV MP column and, following an extensive wash, the bound protein was eluted at high ionic strength and assayed for its ability to bind MP. Figure 2A shows that the overall protein content of the flow-through fraction (lane 2) was very similar to that of the initial cell wall extract (lane 1). However, while the total cell wall extract contained the TMV MP-binding activity (Figure 2B, lane 1), only traces of MP binding were detected in the flow-through fraction (Figure 2B, lane 2). No MP binding was detected in the initial wash fraction (Figure 2B, lane 3), although its protein content remained high (Figure 2A, lane 3). In contrast, the high-salt eluted fraction contained a single protein band with a relative electrophoretic mobility of 38 kDa as assessed by silver staining (Figure 2A, lane 4). This nearly homogeneous protein preparation exhibited high levels of MP-binding activity (Figure 2B, lane 4). Using this protocol, ∼5 μg of the purified MP-interacting protein was obtained from 500 g of fresh tobacco leaf tissue.
Fig. 2. Purification of TMV MP-interacting protein by affinity chromatography. (A) Protein fractions resolved on a 12.5% SDS–polyacrylamide gel. Protein profile was revealed by silver staining. (B) TMV MP binding on renatured protein blots. Lanes 1, total cell wall extract before loading on the column; lanes 2, flow-through fraction; lanes 3, first 0.5 ml wash fraction; lanes 4, eluted fraction. Arrowhead indicates the position of the 38 kDa TMV MP-interacting protein. The numbers on the left indicate molecular mass standards in kDa.
The purified protein was proteolytically digested with Lys-C, trypsin and/or Glu C and the resulting peptides were separated by high performance liquid chromatography (HPLC) and analyzed by mass spectrometry for peptide purity and mass. The mass spectrometry profiles of the three most abundant peptides contained single peptide peaks (data not shown), indicating the presence of one peptide species in each fraction. These peptides were then subjected to amino acid sequence analysis. The use of single HPLC peaks in these experiments increased the clarity and confidence of obtained peptide sequences, as the observed and predicted masses differed by <0.01% (data not shown). Figure 3 shows the amino acid sequences obtained from the purified peptides; all three sequences matched the conserved C–terminal regions of PME from such diverse plant species as tomato (Gaffe et al., 1997) and citrus (Valencia orange) (Nairn et al., 1998). Probably, because tomato and tobacco are closely related, belonging to the family Solanaceae, the best match of the peptide sequences was observed with the tomato PME (Figure 3). Thus, the tobacco cell wall-associated MP interactor most likely represents a PME.
Fig. 3. Amino acid sequence of three peptides derived from the purified TMV MP-interacting protein and its alignment with full-length PME sequences from tomato (DDBJ/EMBL/GenBank accession No. U49330) and citrus (Valencia orange, DDBJ/EMBL/GenBank accession No. U82977). Alignment was performed by the clustal algorithm. Regions of identity between tomato and citrus PMEs are indicated by boxes, gaps introduced for alignment are indicated by dashes. The arrowhead indicates the N–terminus of the mature form of PME (according to Gaffe et al., 1997; Nairn et al., 1998).
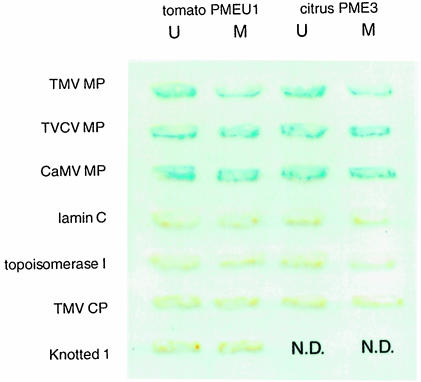
PME proteins are known to undergo post-translational processing (Nairn et al., 1998). For example, size estimates for most plant PMEs are in the range of 32–42 kDa whereas the size of the predicted translation products is >50 kDa (Nairn et al., 1998 and references therein). Thus, PME maturation probably involves a post-translational cleavage that separates the conserved C–terminus of these proteins from their more variable N–terminal region. Indeed, the N-terminal sequence of a mature tomato PME (Markovic and Jornvall, 1986) does not align with the N–terminus of the predicted translation product of the cloned PME gene (Hall et al., 1994); instead it aligns with the conserved C–terminal PME domain (Nairn et al., 1998). The 38 kDa relative electrophoretic mobility of the MP-interacting PME suggests that it represents a post-translationally processed, mature protein. To examine the notion that TMV MP binds to mature forms of PME, we utilized the yeast two-hybrid system (Fields and Song, 1989; Hollenberg et al., 1995) to assay binding between TMV MP and PMEs from tomato (PmeU1; Gaffe et al., 1996, 1997) and Valencia orange (CsPme3; Nairn et al., 1998). Figure 4 shows the positive interaction between TMV MP and proteins expressed from the cDNA clones corresponding to the mature processed tomato and citrus PMEs. Both PMEs did not interact with unrelated proteins, DNA topoisomerase I and lamin C, which are known as non-specific activators in the two-hybrid system (Bartel et al., 1993; Park and Sternglanz, 1998). Furthermore, coat protein (CP) encoded also by TMV but not involved in cell-to-cell movement (Siegal et al., 1962) did not bind PME (Figure 4). Together with the renatured blot overlay results (Figure 1), these observations indicate specific association between TMV MP and mature cellular PMEs.
Fig. 4. Interaction of PME with plant viral MPs in the yeast two-hybrid system. Yeast cells expressing the combination of interacting proteins indicated were grown on tryptophan–leucine double dropout medium and analyzed for β–galactosidase activity. N.D., not done. U, unprocessed form of PME; M, mature form of PME.
Finally, we examined whether or not TMV MP also recognizes a full-length, unprocessed PME that carries an N–terminal signal for translocation into the ER (Gaffe et al., 1997). Figure 4 shows that TMV MP bound unprocessed PME from both tomato and citrus plants. Because mature and unprocessed forms of PME share the C–terminal part of the protein molecule (Gaffe et al., 1997; Nairn et al., 1998), the TMV MP recognition site is likely to reside within this PME region.
PME interacts with other viral MPs
If binding to PME has a role in cell-to-cell movement, MPs of at least some other viruses that traffic between plant cells are expected to recognize this cell wall protein. To test this idea, mature and unprocessed tomato and citrus PMEs were co-expressed in the yeast two-hybrid system with MPs of two unrelated viruses, a crucifer-infecting RNA tobamovirus TVCV (Lartey et al., 1995) and a DNA pararetrovirus CaMV (Hull and Covey, 1985). Figure 4 demonstrates that TVCV and CaMV MPs interacted with both forms of tomato and citrus PMEs. PME binding was confirmed by probing the tobacco cell wall fractions with purified viral MPs in the renatured blot overlay assay (data not shown).
We then examined the potential interaction between PME and a maize homeodomain protein Knotted 1, which is known to move through plasmodesmata in tobacco leaves (Lucas et al., 1995). Interestingly, this protein did not interact with either mature or unprocessed forms of PME in the two-hybrid system (Figure 4). Thus, Knotted 1 may move between cells by a different pathway that does not involve PME. Previously, cell wall extracts were shown to bind gold-conjugated Knotted 1 and MP of cucumber mosaic virus (CMV) (Kragler et al., 1998). Because protein binding was assayed on dot blots, the size or identity of the putative receptor protein(s) was not determined (Kragler et al., 1998); it is possible, however, that the MP-binding protein may also represent PME whereas the Knotted 1-binding activity may derive from a different protein present on the same dot blot.
Immunolocalization of PME
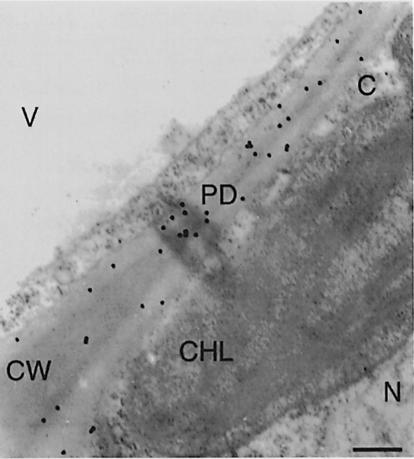
Polyclonal antibodies generated against purified tobacco PME were used to examine the subcellular localization of this protein in tobacco leaves. Figure 5 shows that PME-specific staining was found within the cell wall but not in the cytoplasm, chloroplasts, nucleus, vacuole or mitochondria (not shown) of tobacco leaf mesophyll cells. In addition, PME was also found associated with plasmodesmata embedded in the cell wall. No cell wall-specific staining at all was observed using preimmune antiserum (data not shown). These results support our observation that the MP-interacting protein was found exclusively in the cell wall fraction (Figure 1).
Fig. 5. Immunohistochemical detection of PME in cell walls of tobacco leaf mesophyll cells. CW, cell wall; PD, plasmodesma; N, nucleus, CHL, chloroplast; V, vacuole; C, cytoplasm. Bar, 0.5 μm.
Identification of MP domains involved in binding to PME
Oligonucleotide-directed mutagenesis was used to produce MP derivatives that were then assayed for their ability to interact with PME. Figure 6A shows that the entire open reading frame of TMV MP was saturated with in-frame deletions, resulting in seven mutants (Citovsky et al., 1990, 1992a). These proteins were then used in the renatured blot overlay assay to examine their binding to PME contained within tobacco cell walls. Figure 6B shows that only one deletion mutant, del–4, failed to bind PME, whereas binding of the rest of the mutants appeared unaffected. del–4 eliminates amino acids at positions 130–185, which therefore, may be required for TMV MP–PME interaction. Importantly, del–4 binding to RNA, one of the functional hallmarks of TMV MP (Citovsky et al., 1990), was indistinguishable from that of wild-type MP (data not shown), suggesting that the overall protein structural characteristics were preserved in this MP derivative. Thus, the inability of the del–4 derivative of TMV MP to bind PME may indeed reflect the loss of its PME-binding region. It is possible, however, that this major binding domain is augmented by the context of the native protein.
Fig. 6. Analysis of MP deletion mutants for their ability to bind tobacco PME on renatured blot overlays. (A) Summary of the deletion mutants tested. Ovals indicate retained amino acids; lines indicate deleted amino acids. The numbers in the scale refer to amino acids of the wild-type (wt) TMV MP (Goelet et al., 1982). (B) Renatured blot overlay binding assay using tobacco cell wall fractions. Each lane is designated with the TMV MP derivative whose binding was tested.
Involvement of the TMV MP region between amino acid residues 130–185 in binding to PME was confirmed using an independent assay for protein–protein interaction, i.e. the yeast two-hybrid system. Tomato cDNA clones corresponding to mature and unprocessed PME (Gaffe et al., 1997) were chosen for these experiments. Figure 7 shows that all deletion mutants of TMV MP except for del–4 interacted with both forms of tomato PME expressed in yeast cells.
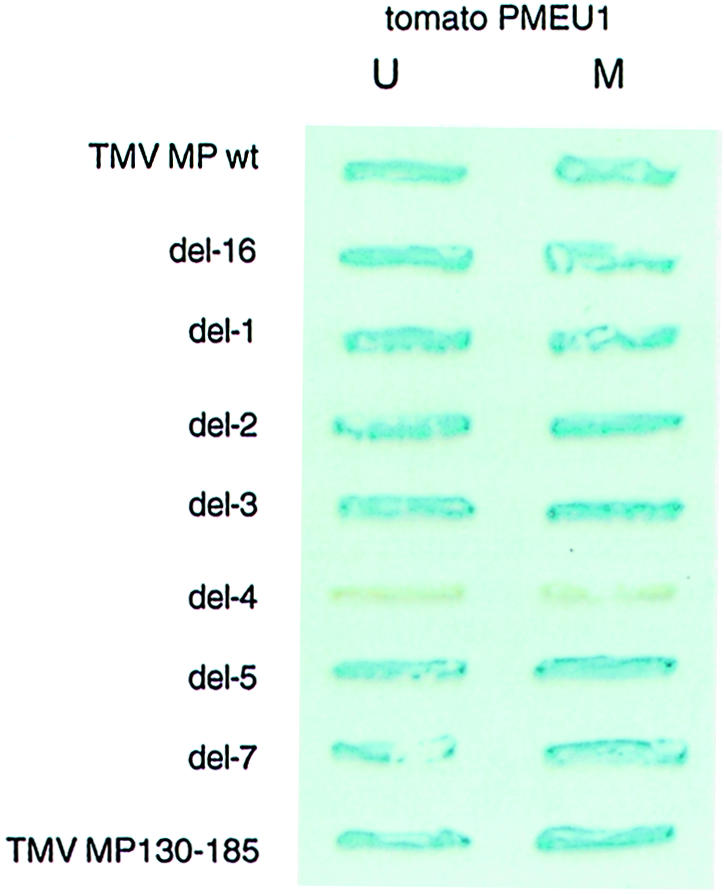
Fig. 7. Analysis of MP deletion mutants for their ability to bind tomato PME in a yeast two-hybrid system. For description of TMV MP deletion mutants see Figure 6. TMV MP wt, full-length MP; TMV MP130–185, MP fragment composed of amino acid residues 130–185. U, unprocessed form of PME; M, mature form of PME.
Next, the 55 amino acid-long TMV MP region deleted in the del–4 mutant was examined for its ability to bind PME in the absence of the rest of the MP sequence. Because we were unable to overexpress this relatively small protein fragment to use in the renatured blot overlay assay, its binding to PME was tested in the yeast two-hybrid system. Figure 7 demonstrates that the TMV MP fragment composed of amino acid residues 130–185 (MP130–185) interacted with mature and unprocessed forms of tomato PME, inducing the β–galactosidase reporter gene expression. The fact that removal of the TMV MP amino acid residues between positions 130 and 185 abolished PME binding while the same region alone was able to interact with PME suggests that this MP domain is necessary and sufficient for recognition and binding of PME. Interestingly, deletion of amino acid residues 130–185 has been shown also to block the ability of TMV MP to dilate plasmodesmata (Waigmann et al., 1994). Involvement of this MP domain in PME binding may explain this observation, implicating MP–PME interaction in the TMV MP function.
Deletion of the PME-binding region of TMV MP blocks viral cell-to-cell movement
To examine further the role of PME binding in TMV movement in vivo, the del–4 derivative was introduced into the infectious cDNA clone of TMV in place of the wild-type MP. The resulting construct was transcribed in vitro and the viral RNA was inoculated onto the local lesion host Nicotiana tabacum cv. Xanthi NN plants. TMV cell-to-cell movement and infectivity was quantified by the number and size of necrotic lesions induced on the inoculated leaves (Gafny et al., 1992). Table I shows that local lesions were observed after inoculation with TMV RNA transcripts carrying the wild-type MP gene. In contrast, no necrotic lesions and, by implication, viral cell-to-cell movement occurred when plants were inoculated with TMV RNA containing the del–4 mutant of MP (Table I). The lack of del–4 movement was confirmed using a CP assay (Ghoshroy et al., 1998), in which TMV presence is detected by appearance of the viral CP within the inoculated tissues of the systemic N.tabacum cv. Turk host (data not shown). However, del–4 TMV RNA became infectious on transgenic N.tabacum cv. Xanthi NN plants expressing the wild-type MP (Table I), indicating that its failure to infect the wild-type local lesion host plants is the result of non-functional MP. These results suggest that disrupting TMV MP–PME interaction blocks viral cell-to-cell spread in wild-type plants but does not interfere with replication of viral genomes or their ability to move when the functional MP is provided in trans in MP-transgenic plants.
Table I. Effect of del-4 on TMV infectiona.
| Lesions |
|||
|---|---|---|---|
| Number/leaf ± SE | Diameter ± SE (mm) | % of wild-type control (number/diameter) | |
| Wild-type plants inoculated with: | |||
| wild-type TMV | 85.0 ± 10.2 | 3.7 ± 1.3 | 100/100 |
| TMV del-4 | 0 | 0 | 0 |
| MP-transgenic plants inoculated with: | |||
| wild-type TMV | 80.2 ± 14.3 | 3.4 ± 1.1 | 94/92 |
| TMV del-4 | 84.1 ± 15.2 | 3.6 ± 1.2 | 99/97 |
aThe number and size of local lesions are the average of four infected wild-type or TMV MP-transgenic N.tabacum cv. Xanthi NN plants in which lesions on four inoculated leaves per plant were counted and measured 5 days after inoculation. SE, standard error. Wild-type TMV, TMV RNA carrying an intact TMV MP; TMV del-4, TMV RNA carrying the del-4 derivative of TMV MP.
Discussion
Cell-to-cell spread of TMV, as well as many other plant viruses, is mediated by a specialized viral MP, which presumably associates with the transported viral genomic nucleic acid molecule, shapes it into a transferable form, and targets it to and through plasmodesmata (reviewed by Carrington et al., 1996; Ghoshroy et al., 1997; Lazarowitz and Beachy, 1999). The molecular pathway by which TMV MP promotes cell-to-cell transport of viral genomes is unknown. To understand this process better, it would be useful to isolate plant proteins that interact directly with the viral MP in the host cell. Here, we have purified a 38 kDa tobacco cell wall protein, identified as a mature form of PME, which specifically binds TMV MP. TMV MP–PME interaction was confirmed and further studied in the yeast two-hybrid system. Using this approach, we identified an MP domain responsible for binding to PME. TMV genomic RNA carrying MP derivative lacking this protein region was unable to spread within tobacco tissue in vivo. The same viral RNA remained infectious in transgenic tobacco hosts that express intact TMV MP, indicating that blocking PME binding did not interfere with replication and assembly activities of the viral genomes. Thus, association with the cellular PME may contribute to the ability of TMV MP to transport the viral genomic RNA between the host plant cells. The fact that removal of the PME-binding domain blocks the ability of TMV MP to increase plasmodesmal permeability (Waigmann et al., 1994) further supports this hypothesis.
The members of the PME multigene family are involved in cell wall turnover and appear to have a role in plant growth and development. PME activity is thought to modulate pH and ion balance and affect cell wall porosity (Pressey, 1984; Nairn et al., 1998 and references therein). In addition, PME has been implicated in more specialized cellular processes such as plant response to pathogen attack (Markovic and Jornvall, 1986). Here we present evidence that PME may also function during cell-to-cell spread of TMV and, potentially, several other plant viruses. TMV MP binding to PME may facilitate viral movement by several mechanisms. First, MP may bind unprocessed PME, which carries the ER translocation signal and is destined to be transported to the cell wall. In cell fractionation studies, TMV MP behaved as a highly hydrophobic protein and copurified with enzyme markers common to the ER (Moore et al., 1992), suggesting that it utilizes the ER for transport from the site of viral synthesis to plasmodesmata (Heinlein et al., 1998), which are also known to contain the ER membranes (Ding et al., 1992b). However, the fact that TMV MP lacks an apparent ER signal sequence (Atkins et al., 1991; Deom et al., 1991) and does not associate with membranes when expressed in vitro [unpublished results described by Heinlein et al. (1998)] contradicts this hypothesis. TMV MP association with unprocessed PME may solve this inconsistency; potentially, binding to PME may provide the ER signal in trans, resulting in a ‘piggyback’ transport of TMV MP through the ER secretory pathway. In this model, the transported PME molecule spans the ER membrane so that its C–terminal part can interact with TMV MP and attach it to the cytoplasmic face of the ER. Following transport to the cell wall, PME may become secreted while TMV MP is retained at the cell wall. The ability of TMV MP to recognize and bind the unprocessed form of PME in the two-hybrid system supports this hypothesis. On the other hand, the fact that the renatured blot overlay experiments did not detect TMV MP binding to unprocessed PME may be due to the relatively low abundance of this protein form and/or its proteolytic cleavage during the cell fractionation procedure.
Secondly, PME may simply function as a cell wall receptor for TMV MP. Our immunoelectron microscopy studies suggest that PME is localized throughout the cell wall, including plasmodesmata. Potentially, binding to PME may initially target and/or anchor TMV MP to the host cell wall. In this scenario, TMV MP association with PME in the vicinity of plasmodesmata will commence the cell-to-cell transport process. In contrast, binding to PME in the cell wall areas that do not contain plasmodesmata will result in abortive movement, with TMV MP either being degraded or redirected back into the cell cytoplasm. This model assumes that TMV MP targeting to the cell periphery may occur irrespective of the presence of plasmodesmata. Indeed, recent data suggest that TMV MP expressed in tobacco protoplasts that do not possess plasmodesmata forms protrusions on the cell surface (Heinlein et al., 1998); in these cells, TMV MP may recognize the cell surface via binding to PME likely to be present within the residual cell wall of the protoplasts.
Finally, a more active role for PME in viral movement cannot be excluded. For example, TMV MP binding may interfere with the PME activity, altering the cell wall ion balance and, consequently, inducing changes in plasmodesmal permeability. Regardless of the exact molecular mechanism by which PME acts, it is probably not limited to the cell-to-cell movement of TMV. Indeed, MPs of TVCV and CaMV also bound PME.
Materials and methods
Plant material
Nicotiana tabacum cvs. Turk and Xanthi NN were used. Transgenic N.tabacum cv. Xanthi NN plants expressing TMV MP were generated as described (Citovsky et al., 1992b).
Mutagenesis of MP
Deletion mutants of TMV MP lacking amino acid residues from 1 to 63 (del-16), 65 to 86 (del-1), 88 to 113 (del-2), 111 to 125 (del-3), 130 to 185 (del-4), 185 to 225 (del-5) and 225 to 268 (del-7) were generated as described previously (Citovsky et al., 1990, 1992a). TMV MP and its deletion derivatives were produced in Escherichia coli, purified to near homogeneity, and verified by Western blot analysis as described (Citovsky et al., 1990, 1992a, 1993).
TMV cDNA clone carrying MP del–4 was produced directly from a full-length infectious cDNA of TMV under the control of SP6 promoter (pTMV304, kindly provided by Dr W.Dawson, University of Florida, Lake Alfred) using the TransformerTM Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Clontech Laboratories, Inc.), followed by dideoxynucleotide sequencing (Kraft et al., 1988).
Plant cell wall fractions
Wild-type tobacco plants (N.tabacum cv. Turk) were used as the source of plant tissue. Cell wall fractions were prepared as described (Citovsky et al., 1993). Briefly, fresh plant tissue was ground to a fine powder and homogenized at 4°C in 1 vol of buffer H [0.1M N–(2–hydroxyethyl)piperazine-N′–(2–ethanesulfonic acid) (HEPES) pH 7.4, 5 mM dithiothreitol (DTT), 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM leupeptin, 1 mM pepstatin]. After centrifugation (1000 g for 5 min at 4°C), the cell walls were further homogenized at 4°C in 10 vol of buffer H with 2% Triton X-100 and centrifuged again. This procedure was repeated twice, followed by six washes (1000 g for 5 min at 4°C) in buffer H with 2% Triton X-100 and two washes in buffer H without the detergent. The resulting white insoluble material was resuspended in 0.5vol of buffer H, quick-frozen in liquid nitrogen, slowly thawed on ice, and centrifuged (20 000 g for 20 min at 4°C). This freeze–thaw cycle released ∼50% of the TMV MP-binding activity. The supernatant containing the solubilized TMV MP-interacting protein was immediately processed for further purification and characterization.
Renatured blot overlay assay
Cell wall extracts (10 μg protein) were resolved on a 12.5% SDS–polyacrylamide gel (Laemmli, 1970) and electroblotted (135 mA for 1 h at room temperature) onto a 7.5 × 15 cm PVDF Immobilon P membrane in transfer buffer (50 mM Tris–base, 192 mM glycine, 20% methanol, 0.01% SDS). The membrane was then washed for 15 min with gentle shaking in buffer B (30 mM Tris–HCl pH 7.4, 0.05% Tween-20). The washed membrane was denatured for 2h at room temperature in 50 ml of denaturation buffer [7 M guanidine hydrochloride (ICN Pharmaceuticals, Inc.), 2mM EDTA, 50mM DTT, 50mM Tris–HCl pH 8.3]. After a 5 min wash in TBS (140 mM NaCl, 30 mM Tris–HCl pH 7.4), the blotted proteins were renatured by an overnight incubation at 4°C in 250 ml of renaturation buffer [140 mM NaCl, 10 mM Tris–HCl pH 7.4, 2 mM EDTA, 1% bovine serum albumin (BSA), 0.1% Tween-20, 2 mM DTT] with gentle shaking (10 r.p.m.). Renaturation in the presence of 1% BSA also blocked potential non-specific protein adsorption sites on the membrane. For TMV MP binding, the renatured blot was incubated for 90 min at room temperature with 10 ml of 10 μg/ml TMV MP in the renaturation buffer. Following incubation, the membrane was washed four times (5 min at room temperature) with TBST (TBS plus 0.1% Tween-20) and incubated for 1 h at room temperature with rabbit anti-TMV MP antibody (1:2000 dilution in TBST with 1% reconstituted milk as blocking agent) for 1 h. After four washes with TBST, TMV MP binding was detected with the ECL Western blotting kit (Amersham).
Purification of PME
TMV MP (100 μg) was immobilized on Affi-Gel 10 (Bio-Rad, CA) according to the manufacturer's instructions and packed into a 1 ml column equilibrated in binding buffer (140 mM NaCl, 10 mM Tris–HCl pH 7.4, 2 mM EDTA, 0.1% Tween-20, 2 mM DTT). Cell wall extract derived from 500 g of tobacco leaves was concentrated to 5 ml by ultrafiltration, mixed with one vol. of double-strength denaturation buffer, and incubated for 2 h at 4°C. After incubation, the protein extract was renatured by overnight dialysis against 2 l of the binding buffer. The resulting sample was loaded on the TMV MP column at a flow rate of 0.5 ml/min. The column was then washed with 50 ml of binding buffer and eluted with 1 ml of 2 M NaCl in the binding buffer. The eluted fraction was dialyzed against buffer H and concentrated to 0.25 ml by ultrafiltration.
Peptide sequence analysis
Purified 38 kDa MP-interacting protein was electrophoresed on a 12.5% SDS–polyacrylamide gel (Laemmli, 1970), the band was cut out and subjected to in-gel proteolysis by Lys-C, trypsin and/or Glu-C as described (Hwang et al., 1996). The digested peptides were fractionated by HPLC (Applied Biosystems, CA) and subjected to MALDI-TOF mass analysis (Hewlett-Packard, CA). Following mass spectrometry, selected peptide fractions were analyzed by amino acid sequencing using the Edman degradation method (Applied Biosystems, CA). All these procedures were performed at the Protein and Nucleic Acid Facility (Beckman Center, Stanford University).
Two-hybrid protein–protein interaction assay
TMV MP, TVCV MP and CaMV MP derived from pETP30 (Citovsky et al., 1990), pTVCV50 (this plasmid contains the full-length TVCV cDNA described in Lartey et al., 1995) and pCaMV10 (Gardner et al., 1981), respectively, were amplified by PCR as BamHI–PstI fragments and cloned into BamHI–PstI sites of pBTM116 (TRP1+; Hollenberg et al., 1995), producing MP fusions with LexA. High fidelity pfu DNA polymerase (Stratagene) was used in all PCRs. Because Knotted 1 itself activated the lacZ reporter gene of the two-hybrid system when fused to LexA (data not shown), it was cloned as a PCR-amplified EcoRI–SalI fragment of pZY226–12 (kindly provided by Dr D.Jackson, Cold Spring Harbor Laboratory, NY) into EcoRI–SalI sites of pGAD424 (LEU3+; Clontech); the resulting translational fusion between GAL4 activation domain and Knotted 1 did not activate the reporter gene expression (data not shown).
Tomato PME cDNA, PmeU1 (Gaffe et al., 1997), was subcloned as a PCR-amplified EcoRI–BamHI fragment into EcoRI–BamHI sites of pGAD424 and Valencia orange PME cDNA, CsPme3 (kindly provided by Dr D.Lewandowski, University of Florida, Lake Alfred, FL) (Nairn et al., 1998), was subcloned as a PCR-amplified BamHI–BglII fragment into the BamHI site of pGAD424, producing PME fusions with the GAL4 activation domain. For subcloning of the unprocessed form of PME, the entire cDNA sequence was used, whereas for subcloning of the mature forms, the PCR primers were designed to amplify only the cDNA sequences corresponding to the processed protein, starting from amino acid residues at position 270 for PmeU1 (Gaffe et al., 1997) and 200 for CsPme3 (Nairn et al., 1998). In addition, for experiments with Knotted 1, unprocessed and mature forms of tomato PME were cloned as PCR-amplified EcoRI–BamHI fragments into EcoRI–BamHI sites of pBTM116 to produce LexA–PME fusions. All DNA constructs were verified by dideoxynucleotide sequencing (Kraft et al., 1988).
For the two-hybrid assay, the potential interactors, e.g. viral MPs, and various forms of PMEs, were introduced into yeast strain L40 (Hollenberg et al., 1995) and grown for 2 days at 30°C on a leucine- and tryptophan-deficient medium. The resulting colonies were analyzed by the β–galactosidase assay as described (Hollenberg et al., 1995). For negative controls, we utilized several unrelated proteins such as a fragment of the human lamin C gene (Bartel et al., 1993) or eukaryotic DNA topoisomerase I (Park and Sternglanz, 1998) in pBTM116 and TMV CP cloned as a PCR-amplified EcoRI–BamHI fragment of pTMV304 cloned into EcoRI–BamHI sites of pBTM116.
Electron microscopy
Leaf samples (0.5–1.0 cm in diameter) from N.tabacum cv. Turk plants were processed for electron microscopy using the Durcupan ACM embedding protocol (Ghoshroy and Citovsky, 1998). Ultra-thin sections (70–80 nm) were reacted with polyclonal antibody raised against the purified tobacco cell wall PME, followed by anti-rabbit IgG conjugated to 15 nm colloidal gold as described (Ghoshroy and Citovsky, 1998), and examined under a JEOL 100C transmission electron microscope.
Assay of TMV infectivity by inoculation on a local lesion host
TMV cDNA clones carrying wild-type MP or del-4 in the pTMV304 construct were linearized with KpnI and their infection transcripts were produced from the SP6 promoter and capped with 7m-diguanosine triphosphate using the RiboMax large scale RNA Production System kit according to the manufacturer's directions (Promega). Nicotiana tabacum cv. Xanthi NN plants were mechanically inoculated with 10 μl of the resulting 100 μg/ml RNA solution supplemented with 3 mg/ml celite. Four individual plants were inoculated with each viral transcript. Five days after inoculation, necrotic lesions on each inoculated leaf were counted and their individual sizes measured.
Acknowledgments
Acknowledgements
We thank Yoon Rhee for stimulating discussions and Dick Winant (Stanford PAN facility) for his help with protein sequencing. This work was supported by grants from National Institutes of Health (Grant No. GM50224), U.S. Department of Agriculture (Grant No. 94–02564), National Science Foundation (Grant No. DBI 9975717), and U.S.–Israel Binational Research and Development Fund (BARD) (Grant No. US-2247–93) to V.C. G.H. was supported by funding from the Division of Energy Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy.
Note added in proof
After submission of this manuscript and in support of our findings, the TMV MP interaction with PME was also described by Dorokhov et al. (FEBS Lett., 1999, 461, 223–228).
References
- Atkins D., Hull, R., Wells, B., Roberts, K., Moore, P. and Beachy, R.N. (1991) The tobacco mosaic virus 30 K movement protein in transgenic tobacco plants is localized to plasmodesmata. J. Gen. Virol., 72, 209–211. [DOI] [PubMed] [Google Scholar]
- Bartel P., Chien, C., Sternglanz, R. and Fields, S. (1993) Elimination of false positives that arise in using the two-hybrid system. BioTechniques, 14, 920–924. [PubMed] [Google Scholar]
- Carrington J.C., Kassachau, K.D., Mahajan, S.K. and Schaad, M.C. (1996) Cell-to-cell and long-distance transport of viruses in plants. Plant Cell, 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V. and Zambryski, P. (1993) Transport of nucleic acids through membrane channels: Snaking through small holes. Annu. Rev. Microbiol., 47, 167–197. [DOI] [PubMed] [Google Scholar]
- Citovsky V. and Zambryski, P. (1995) Transport of protein–nucleic acid complexes within and between plant cells. Membr. Protein Transp., 1, 39–57. [Google Scholar]
- Citovsky V., Knorr, D., Schuster, G. and Zambryski, P. (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell, 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Wong, M.L., Shaw, A., Prasad, B.V.V. and Zambryski, P. (1992a) Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell, 4, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan, J., Warnick, D. and Zambryski, P. (1992b) Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science, 256, 1803–1805. [DOI] [PubMed] [Google Scholar]
- Citovsky V., McLean, B.G., Zupan, J. and Zambryski, P. (1993) Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally-regulated plant cell wall-associated protein kinase. Genes Dev., 7, 904–910. [DOI] [PubMed] [Google Scholar]
- Deom C.M., Wolf, S., Holt, C.A., Lucas, W.J. and Beachy, R.N. (1991) Altered function of the tobacco mosaic virus movement protein in a hypersensitive host. Virology, 180, 251–256. [DOI] [PubMed] [Google Scholar]
- Ding B., Haudenshield, J.S., Hull, R.J., Wolf, S., Beachy, R.N. and Lucas, W.J. (1992a) Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell, 4, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Turgeon, R. and Parthasarathy, M.V. (1992b) Substructure of freeze-substituted plasmodesmata. Protoplasma, 169, 28–41. [Google Scholar]
- Fields S. and Song, O.-K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Gaffe J., Tizando, M.E. and Handa, A.K. (1996) Cloning and nucleotide sequence of a pectin methylesterase cDNA homologue (Accession No. U49330) from tomato leaves (PGR 96-017). Plant Physiol., 110, 1436. [Google Scholar]
- Gaffe J., Tiznado, M.E. and Handa, A.K. (1997) Characterization and functional expression of a ubiquitously expressed tomato pectin methylesterase. Plant Physiol., 114, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafny R., Lapidot, M., Berna, A., Holt, C.A., Deom, C.M. and Beachy, R.N. (1992) Effects of terminal deletion mutations on function of the movement protein of tobacco mosaic virus. Virology, 187, 499–507. [DOI] [PubMed] [Google Scholar]
- Gardner R.C., Howarth, A.J., Hahn, P., Brown-Luedi, M., Shepherd, R.J. and Messing, J. (1981) The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res., 9, 2871–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshroy S. and Citovsky, V. (1998) Preservation of plant cell ultrastructure during immunolocalization of virus particles. J. Virol. Methods, 74, 223–229. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S., Lartey, R., Sheng, J. and Citovsky, V. (1997) Transport of proteins and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol., 48, 27–49. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S., Freedman, K., Lartey, R. and Citovsky, V. (1998) Inhibition of plant viral systemic infection by non-toxic concentrations of cadmium. Plant J., 13, 591–602. [DOI] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff, G.P., Butler, P.J.G., Akam, M.E., Gait, M.J. and Karn, J. (1982) Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl Acad. Sci. USA, 79, 5818–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.N., Bird, C.R., Picton, S., Tucker, G.A., Seymour, G.B. and Grierson, D. (1994) Molecular characterisation of cDNA clones representing pectin esterase isozymes from tomato. Plant Mol. Biol., 25, 313–318. [DOI] [PubMed] [Google Scholar]
- Heinlein M., Epel, B.L., Padgett, H.S. and Beachy, R.N. (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science, 270, 1983–1985. [DOI] [PubMed] [Google Scholar]
- Heinlein M., Padgett, H.S., Gens, J.S., Pickard, B.G., Casper, S.J., Epel, B.L. and Beachy, R.N. (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell, 10, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg S.M., Sternglanz, R., Cheng, P.F. and Weintraub, H. (1995) Identification of a new family of tissue-specific basic helix–loop–helix proteins with a two-hybrid system. Mol. Cell. Biol., 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. and Covey, S.N. (1985) Cauliflower mosaic virus: pathways of infection. BioEssays, 3, 160–163. [Google Scholar]
- Hwang B.J., Smith, A.J. and Chu, G. (1996) Internal sequence analysis of proteins eluted from polyacrylamide gels. J. Chromatogr. B Biomed. Appl., 686, 165–175. [DOI] [PubMed] [Google Scholar]
- Kraft R., Tardiff, J., Kranter, K.S. and Leinwand, L.A. (1988) Using miniprep plasmid DNA for sequencing double stranded templates with Sequenase. BioTechniques, 6, 544–547. [PubMed] [Google Scholar]
- Kragler F., Monzer, J., Shash, K., Xoconostle-Cazares, B. and Lucas, W.J. (1998) Cell-to-cell transport of proteins: requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J., 15, 367–381. [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 277, 680–685. [DOI] [PubMed] [Google Scholar]
- Lartey R.T., Voss, T.C. and Melcher, U. (1995) Completion of a cDNA sequence from a tobamovirus pathogenic to crucifers. Gene, 166, 331–332. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S.G. and Beachy, R.N. (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell, 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner S.M. and Howell, S.H. (1993) Long-distance movement of viruses in plants. Trends Microbiol., 1, 314–317. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B. and Hake, S. (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science, 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Markovic O. and Jornvall, H. (1986) Pectinesterase. The primary structure of the tomato enzyme. Eur. J. Biochem., 158, 455–462. [DOI] [PubMed] [Google Scholar]
- McLean B.G., Zupan, J. and Zambryski, P. (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell, 7, 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.J., Fenczik, C.A. and Beachy, R.N. (1992) Developmental changes in plasmodesmata in transgenic plants expressing the movement protein of tobacco mosaic virus. Protoplasma, 170, 115–127. [Google Scholar]
- Nairn C.J., Lewandowski, D.J. and Burns, J.K. (1998) Genetics and expression of two pectinesterase genes in Valencia orange. Physiol. Plant., 102, 226–235. [Google Scholar]
- Park H. and Sternglanz, R. (1998) Two separate conserved domains of eukaryotic DNA topoisomerase I bind to each other and reconstitute enzymatic activity. Chromosoma, 107, 211–215. [DOI] [PubMed] [Google Scholar]
- Pressey R. (1984) Role of pectinesterase in pH-dependent interactions between pea cell wall polymers. Plant Physiol., 76, 547–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal A., Zaitlin, M. and Sehgal, O.P. (1962) The isolation of defective tobacco mosaic virus strains. Proc. Natl Acad. Sci. USA, 48, 1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenius K., Clapham, D. and Meshi, T. (1987) Localization by immunogold cytochemistry of the virus coded 30 K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology, 160, 363–371. [DOI] [PubMed] [Google Scholar]
- Waigmann E., Lucas, W., Citovsky, V. and Zambryski, P. (1994) Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc. Natl Acad. Sci. USA, 91, 1433–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom, C.M., Beachy, R.N. and Lucas, W.J. (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science, 246, 377–379. [DOI] [PubMed] [Google Scholar]