Abstract
The antigen-binding site of the camel heavy-chain antibodies devoid of light chain consists of a single variable domain (VHH) that obviously lacks the VH–VL combinatorial diversity. To evaluate the extent of the VHH antigen-binding repertoire, a germline database was constructed from PCR-amplified VHH/VH segments of a single specimen of Camelus dromedarius. A total of 33 VHH and 39 VH unique sequences were identified, encoded by 42 and 50 different genes, respectively. Sequence comparison indicates that the VHHs evolved within the VH subgroup III. Nevertheless, the VHH germline segments are highly diverse, leading to a broad structural repertoire of the antigen-binding loops. Seven VHH subfamilies were recognized, of which five were confirmed to be expressed in vivo. Comparison of germline and cDNA sequences demonstrates that the rearranged VHHs are extensively diversified by somatic mutation processes, leading to an additional hypervariable region and a high incidence of nucleotide insertions or deletions. These diversification processes are driven by hypermutation and recombination hotspots embedded in the VHH germline genes at the regions affecting the structure of the antigen-binding loops.
Keywords: antigen-binding repertoire/camel heavy-chain antibody/gene replacement/germline VH/hypermutation hotspots
Introduction
The emergence of the Camelidae (camels and llamas) within the Artiodactyls is accompanied by a unique event in immunoglobulin (Ig) evolution, namely the appearance of additional classes of functional antibodies (Abs) composed solely of heavy chains (Hamers-Casterman et al., 1993). These heavy-chain antibodies (HCAbs) lack the first domain of the constant region (CH1), which is present in the genome but is spliced out during mRNA processing (Nguyen et al., 1999; Woolven et al., 1999). The antigen (Ag)-binding site of these HCAbs is composed of a single variable domain (referred to as VHH). The VHH structure resembles that of the heavy chain variable domain (VH) of the conventional Abs. However, there are remarkable sequence differences at the second framework (FR2) and the third complementarity-determining region (CDR3) (Muyldermans et al., 1994; Vu et al., 1997). Most striking are the amino acid substitutions V37F (Val at position 37 in the VH to Phe in the VHH), or V37Y, G44E, L45R or L45C, and W47 most often to G [numbers refer to the amino acid positions numbered according to Kabat et al. (1991)]. In the conventional VHs, these FR2 amino acids interact with the variable domain of the light chain (VL), and are conserved during evolution (Kabat et al., 1991). The CDR3 of the VHH is longer on average than that of a VH domain (Vu et al., 1997), and is often constrained by an interloop disulfide bond (Davies and Riechmann, 1996; Desmyter et al., 1996).
A high titre and a complex repertoire of HCAbs can be obtained from immunized or infected dromedaries or llamas (Hamers-Casterman et al., 1993; Ghahroudi et al., 1997). Many HCAbs raised against enzymes are competitive inhibitors (Lauwereys et al., 1998). This is surprising, since the active site of enzymes has a low antigenicity for conventional Abs (Novotny, 1991). Thus, the HCAbs recognize a broad range of epitopes, some of which differ from those for conventional Abs.
Previously, we identified germline VH and VHH segments indicating that the variable domain of the HCAbs is encoded by a distinct set of V genes (Nguyen et al., 1998). In this study, we investigate the potential VHH germline repertoire to gain insight into the ways by which the dromedary HCAbs acquire a complex repertoire of Ag-binding sites. In conventional Abs, the diversity of the Ag-binding site is generated at multiple levels. The VH is generated by assembling variable (V), diversity (D) and joining (J) elements (Tonegawa, 1983), in which the V-gene segment encodes the CDR1 and CDR2; the CDR3 is generated by the V–D–J joining. In this joining process, great sequence variation is introduced by non-template addition of nucleotides at the V–D and D–J junctions (junctional diversity). Random association of a VH and a VL (combinatorial diversity) generates an immensely diverse Ag-binding repertoire. Additional diversification of the Ag-binding repertoire could be achieved by somatic hypermutation (Berek et al., 1991) and gene conversion (Reynaud et al., 1987; Becker and Knight, 1990). Thus, the primary Ag-binding repertoire of the HCAbs lacking the VH–VL combinatorial diversity relies on the innate number and sequence diversity of the VHH germline segments and the junctional diversity.
The identification of the germline VHH genes is not only of fundamental interest but also has a potential biotechnological benefit. At the moment, HCAbs with enzyme inhibiting activity can only be obtained after immunizing camels or llamas. Techniques have been developed to retrieve various binders from synthetic libraries of Ab fragments (Hoogenboom and Winter, 1992; Winter et al., 1994). Single-domain Ab libraries have been constructed by adding a synthetic CDR3 region to the known human VH elements (Davies and Riechmann, 1995; Reiter et al., 1999). It would be an asset if similar libraries of VHHs were available to retrieve binders or inhibitors of interest. To be as successful as the dromedary in making good Ag binders we should start from a library that covers all potential VHH segments. In addition, analysis of the amino acids that are mutated during the in vivo affinity maturation would provide a rational strategy for increasing the repertoire of the VHH library or to improve the affinity of binders.
We cloned from a single dromedary the germline VHH gene segments to analyse their complexity. The comparisons of the germline and cDNA VH/VHH sequences reveal the somatic diversification mechanisms used by the camelids to enlarge the primary Ag-binding repertoire of the HCAbs. The involvement of DNA signal sequences in these diversification processes is discussed.
Results
Southern blot analysis of the genomic DNA
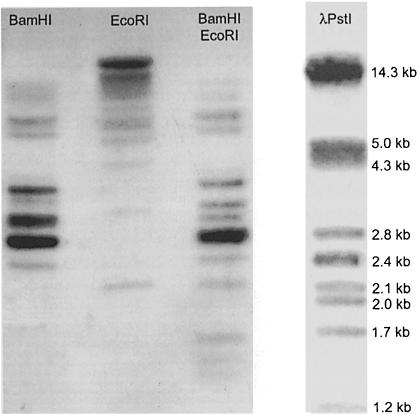
A rough estimate of the VH/VHH germline repertoire was first obtained by Southern blot analysis of dromedary liver DNA, probed by the PCR fragments from the upstream conserved octamer sequence to the FR3 of camel germline VH or VHH clones (Nguyen et al., 1998). The hybridization patterns shown by these two different probes were identical. In single digestions with BamHI or EcoRI, 11 bands of different intensity and ranging from 1.7 to >14 kb could be visualized (Figure 1). The double digest revealed at least 15 bands. Since we never observed an EcoRI or BamHI restriction site between the octamer and FR3 sequences of VH/VHH (this work), we conclude that the dromedary VH/VHH germline gene segments are spread over a minimum of 15 different EcoRI–BamHI size families.
Fig. 1. Southern blot analysis of dromedary liver genomic DNA hybridized with a camel VHH probe. Dromedary DNA was digested with BamHI, EcoRI and BamHI–EcoRI, as indicated on top of the lanes. Phage λ PstI restriction fragments are used as size marker.
Determination of dromedary germline VH/VHH sequences
A germline V-gene database was obtained by cloning and sequencing PCR fragments of VH/VHH elements obtained from liver DNA of a single dromedary. Out of 255 sequences we found 145 different sequences, which formed an initial germline VH/VHH database. Assuming a PCR error rate of 2×10–4 errors/base (Cha and Thilly, 1993), we estimated that up to four base errors might have accumulated within each 600 bp sequence. Hence, we only considered clones differing by at least 5 nucleotides (nt), which reduced the VH/VHH germline database from 145 to 94 sequences. The nucleotide sequences of these 94 clones have been submitted to the DDBJ/EMBL/GenBank databank (accession Nos AJ245107–AJ245200).
In an alternative approach, we screened the dromedary genomic library cloned in phage λ (Nguyen et al., 1998). A total of 55 different phage clones were isolated and their VH or VHH regions were subcloned and sequenced. However, no new VH/VHH sequences were discovered, supporting that our database of 94 VH/VHH sequences obtained by PCR is representative to evaluate the potential VHH repertoire.
Characterization of VH/VHH sequences and VHH subfamilies
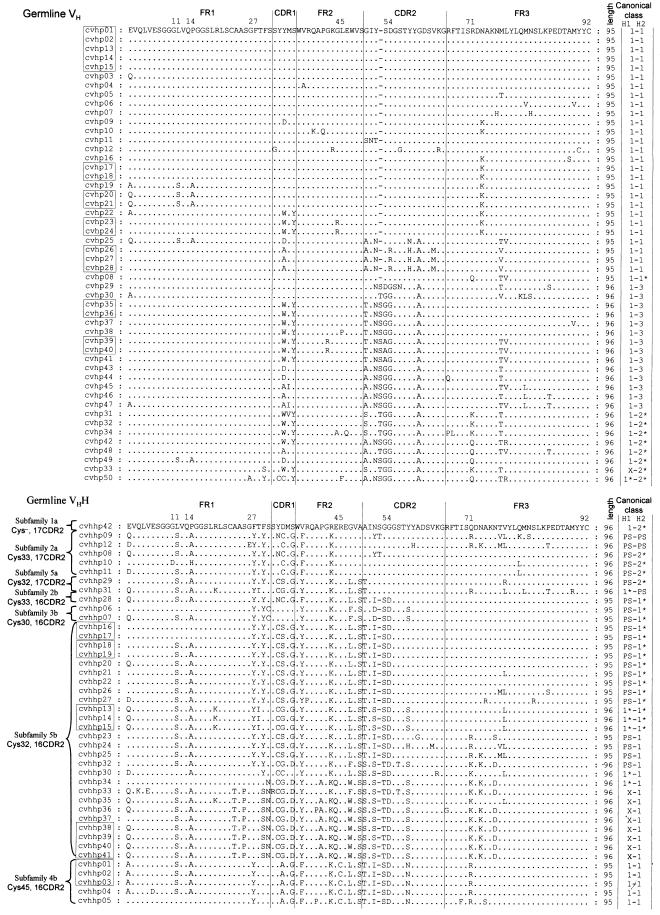
All 94 genomic VH/VHH sequences span from the conserved octamer to the Cys92 of FR3. They contain a TATA box 105 nt upstream of the ATG initiation codon. This ATG starts an open reading frame (ORF) of conserved sequences encoding a leader signal peptide, which is interrupted by an intron of 104 bp. The remaining sequence encodes the VH or VHH segment. Two sequences (cvhp51 and cvhp52) are interrupted by stop codons. These clones could be pseudo-genes and were excluded from further analyses. The remaining 92 clones probably encode functional genes as no aberrant amino acid occurs in the ORF at the critical positions for the Ig fold (Chothia et al., 1988).
Identification of VH/VHH sequences. The nucleotide identity between any pair of the 92 VH/VHH encoding sequences is >80%. The amino acid sequences of FR1 and the beginning of FR3 are homologous to each other (Figure 2) and to the human or mouse VH of family 3. Therefore, according to the family definitions (Brodeur and Riblet, 1984; Schroeder et al., 1990), all these camel sequences could be assigned to the VH family 3. Based on crucial amino acid differences in the FR2 region, we have previously distinguished the camel VH3 family (used in the conventional Abs) and the VH3H family (for the HCAbs) (Nguyen et al., 1998). The current database can also be divided into these two families according to the presence of V37, G44, L45 and W47 (VH3), and F/Y37, Q/E44 and R/C45 (VH3H) (Figure 2). Out of 92 VHs/VHHs, 50 sequences were clearly assigned to VH3 (cvhp01–cvhp50) and 42 to the VH3H family (cvhhp01–cvhhp42). This VH/VHH ratio (1.2/1) was also found among sequences obtained from the phage λ-isolated clones. We therefore conclude that a similar number of VH3 and VH3H genes reside in the dromedary genome.
Fig. 2. The deduced amino acid sequences of the dromedary germline VH (upper) and VHH segments (lower). Braces group the VHH sequences in subfamilies as defined by their CDR2 length and the position of the additional Cys at indicated positions. Boxes indicate V genes that differ by at least 5 nt, but encode an identical amino acid sequence. The amino acid length is given in the column following the primary structure. The types of the predicted H1 and H2 canonical structures are in the last columns. Asterisks denote where the predicted loop may deviate from loop type defined by Chothia et al. (1992) due to novel residue at the key-site. X, unpredictable; PS, potentiality to switch (see text).
Comparison of amino acid sequences revealed that a number of clones have an identical coding capacity (boxed in Figure 2) although their nucleotide sequence differed by >4 nt. As a result, the 50 VH and 42 VHH gene segments code respectively for 39 VH and 33 VHH unique amino acid sequences.
VHH subfamilies. Multiple alignment of the VHH amino acid sequences revealed the clustering of sequences with an additional cysteine at position 30, 32, 33 or 45, and with a CDR2 length of 16 or 17 amino acids (Figure 2). Based on these hallmarks, we further classified the 42 clones of the VHH into seven subfamilies named 1a, 2a, 2b, 3b, 4b, 5a and 5b (Table I; Figure 2).
Table I. VHH germline sub-families and their cDNA counterparts.
| Subfamily | Cys/CDR2a | Members | Total cDNA found | Binder found | Agsb |
|---|---|---|---|---|---|
| VH3H–1a | Cys–/17 | 1 | 0 | 0 | |
| VH3H–2a | Cys33/17 | 5 | 54 | 18 | lysozyme, carbonic and anhydrase, cutinase, tetanus toxoid, RNase phenyl oxazolone |
| VH3H–2b | Cys33/16 | 1 | 2 | 2 | lysozyme, carbonic anhydrase |
| VH3H–3b | Cys30/16 | 2 | 5 | 2 | amylase |
| VH3H–4b | Cys45/16 | 5 | 8 | 3 | amylase, tetanus toxoid |
| VH3H–5a | Cys32/17 | 2 | 0 | 0 | |
| VH3H–5b | Cys32/16 | 26 | 3 | 1 | amylase |
| Total | 42 | 72 | 26 |
aThe presence of an additional Cys at indicated position/the length of CDR2 (in amino acids).
bReference from Desmyter et al. (1996), Ghahroudi et al. (1997), Lauwereys et al. (1998), Decanniere et al. (1999) and other unpublished
Ag-binders provided by M.Lauwereys, K.Silence, T.Laeremans and M.T.M.C.Serrao.
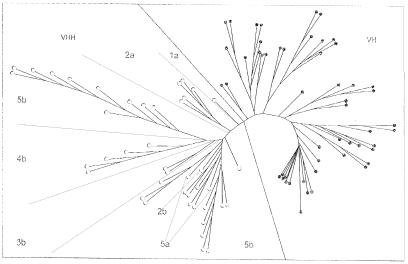
A neighbour-joining tree was constructed using the VH/VHH coding sequences to analyse the VH and VHH relationship. The dendrogram (Figure 3) shows clearly two clusters, one cluster contains only the VH3 and the other the VH3H genes, despite the fact that the pairwise sequence identity among VHs or among VHHs may be lower than that of a VH–VHH pair. Moreover, the branches appear to correspond to the VHH subfamilies defined above.
Fig. 3. Neighbour-joining phylogenetic tree of the dromedary germline VH and VHH segments. The tree was constructed by using clustalW, phylip packages with 1000 replicates for Bootstrap of nucleotide sequences encoding the VH/VHH portions. Filled and open circles denote a VH and VHH member, respectively. The two filled rectangles indicate pseudogenes. VHH subfamilies are indicated.
Determination of expressed VHH gene segments
We assembled a VHH cDNA database containing 103 sequences, 32 of which have a known Ag specificity. Thirty-one of the 103 cDNAs have a length deviating from any of the germline VHH segments. While these clones could originate from a germline VHH that has not yet been discovered, they could also originate from unequal DNA recombination or gene conversion (Reynaud et al., 1987). The remaining 72 VHH cDNA sequences (26 with known Ag specificity) were compared with all 42 germline VHH sequences to reveal the closest sequence identity. The combination of the sequence identity score and the two VHH subfamily hallmarks defined earlier leaves no doubt as to the identification of the germline subfamily. Among the 72 VHH cDNA clones, there is no indication for the in vivo expression of members of subfamilies 1a and 5a (Table I). The result shows that at least five out of the seven VHH subfamilies are used to generate productive VHH domains found in HCAbs.
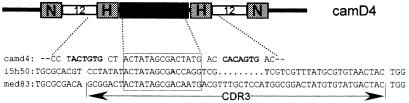
Identical D and JH segments in VH and VHH cDNAs. A germline D mini-gene (similar to human D4) was identified with an RSS at both ends. We noted that some VH and VHH cDNAs employ this D element (Figure 4). In addition, our cDNA database also contains VHs and VHHs that make use of the same JH element. This would indicate that VH and VHH genes are within the same functional V region of the dromedary genome.
Fig. 4. The common usage of the D element in VH and VHH: a genomic D element (camD4) is flanked at both sides with the RSS containing a nonamer (N) separated from a heptamer (H) by a 12-nt spacer. The sequences derived from the germline camD4 (upper line), VH-cDNA clone i5h50 (middle line) and VHH-cDNA clone med83 (lower line) are boxed. The CDR3 region between the FR3 and FR4 is indicated.
Structural repertoire of Ag-binding loops of camel germline VH/VHH
The structure of the Ag-binding loops of the camel germline VHs and VHHs was predicted by algorithms of Chothia and Martin (http://www.biochem.ucl.ac.uk/~martin/abs/chothia.html) (Chothia et al., 1989, 1992; Martin and Thornton, 1996), and by taking into account the crystallographic structures of dromedary and llama VHHs (Desmyter et al., 1996; Spinelli et al., 1996; Decanniere et al., 1999) (Table II; Figure 2).
Table II. Structural repertoire of the camel germline VH and VHH segments.
| VH | VHH | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | VHCSC members | H1 | H2 | VHCSC members | ||||||||
| 24 | 26 | 34 | 52 | 71 | 24 | 26 | 34 | 52 | 71 | ||||
| A | GFTFSGY | M | Y-SDGG | R | 1–1 | 27 | A | GYTFSSY | M | I-SDGS | R | 1–1 | 5 |
| A | GFTFSSY | M | Y-SDGS | Q | 1–1* | 1 | A | GFT@@CS | M | S-TDGS | K | 1*–1 | 2 |
| A | GFTFSSY | @ | NSDGSN | R | 1–3 | 14 | A | GYIFSCS | M | S-SDGS | Q | 1*–1* | 3 |
| A | GFTFSSY | @ | @TGGGS | K/Q | 1–2* | 6 | A | GYTYSSC | M | @-@DG@ | K | PS-1 | 4 |
| A | GFTSSSY | M | YTGGGS | K | X-2* | 1 | A | GYTY@S@ | M | D-SDGS | Q | PS-1* | 12 |
| A | AFTYSSC | M | NSGGGS | Q | 1*–2* | 1 | A | GFTSN@C | M | S-TDG@ | K | X-1 | 8 |
| A | GFTFSSY | M | NSGGGS | Q | 1–2* | 1 | |||||||
| A | GYIFSCS | M | NSGGGS | R | 1*-PS | 1 | |||||||
| A | GYTYSS@ | M | @@GGGS | Q | PS-2* | 4 | |||||||
| A | @YTYSS@ | M | @@GGGS | R | PS-PS | 2 | |||||||
VHCSC or VHHCSC: canonical structure class of VH or VHH, respectively. Asterisks denote that the predicted loop may deviate from the loop-type as defined by Chothia et al. (1992) due to a novel residue (underlined) at the key site. The @ denotes more than one residue occurring at that site. X, unpredictable; PS, potentiality to switch (see text).
The Ag-binding loops of the dromedary VHs are predicted to conform to the known canonical structures characterized by well-defined key amino acids. In contrast, the Ag-binding loops of the VHHs are expected to deviate frequently from the known canonical structures, mainly due to the substitution of key amino acids. Notable substitutions are G26E and F29S in the H1 loop (the solvent-exposed part of the loop overlapping with the CDR1) and R71Q dictating the conformation of the H2 loop (the exposed part of the CDR2).
From crystallographic data on the VHH structures, we infer that the combination Y27–Y29 (found in 22 germline VHHs) can adopt either a type-1 or type-4 canonical structure, depending on the nature of the residue 31 (Decanniere et al., 1999). Similarly, the six-residue H2 loops associated with R71 reveal either a H2 type-2 (Spinelli et al., 1996) or a novel H2 structure (Decanniere et al., 1999). These loops are therefore denoted as ‘PS’ in Table II for ‘potentiality to switch’.
Thus, the dromedary VH germline sequences have a structural repertoire of six different H1–H2 combinations (Table II), whereas the structural repertoire of the germline VHHs is far more diverse, up to 10 different combinations of H1 and H2 loop conformations are observed. Moreover, it seems that both the H1 and H2 loop conformations of the VHHs are apt to reshape due to minor modifications, thereby further enlarging the VHH structural repertoire.
Variability of camel VH/VHH sequences
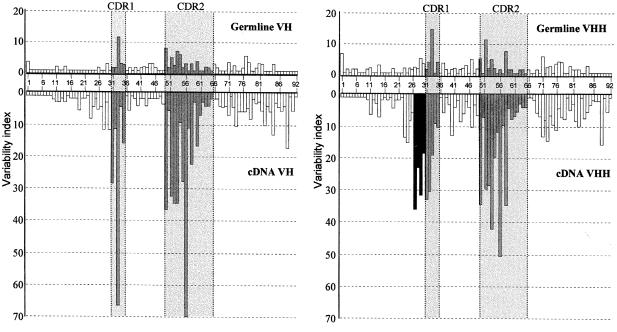
The variability plots (Kabat et al., 1991) were constructed for the germline VHs and VHHs (Figure 5, upper panels). Both histograms delineate the conventional CDR1 and CDR2 regions as the sites of greater variability.
Fig. 5. Amino acid sequence variability plots for the VH and VHH germline (upper panels) and cDNA sequences (lower panels). The variability index (bars) was calculated from FR1 to FR3, as described in the results. The grey and open bars are for amino acid positions of the CDR and FR regions, respectively, and the black bars are for the extra hypervariable region in the VHH. The CDR1 and CDR2, as defined by Kabat et al. (1991), are shaded and placed between dotted lines.
Hypervariability in the H1 region of VHHs cDNA. The variability of the VH and VHH cDNAs was plotted using sets of 50 VH and 42 VHH cDNAs, and compared with that of the germline sequences (Figure 5). The overall variability is much higher in cDNAs than in the germline, indicating that somatic mutation plays an important role in the generation of the camel VH/VHH repertoire. The most striking observation is the presence, exclusively in the VHH cDNAs, of an additional hypervariable region (residues 27–30) located upstream of the conventional CDR1 region.
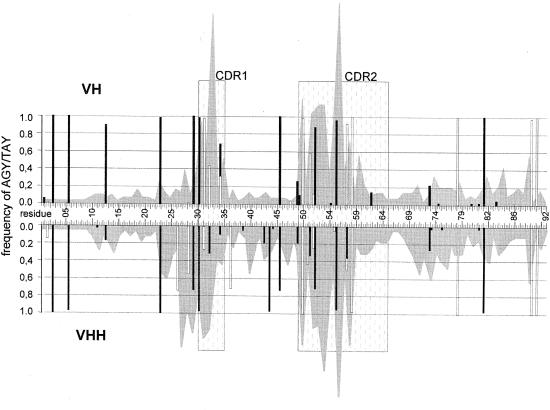
Hypermutational hotspots imprinted in the germline. Somatic diversification results from different mechanisms including gene conversion, and somatic hypermutation (Wagner and Neuberger, 1996; Neuberger et al., 1998). The latter was proposed to be driven by hypermutational hotspots such as the AGY and TAY (Y = C or T) sequences (Yelamos et al., 1995; Milstein et al., 1998). The occurrence of these hotspots in the germline VHs and VHHs was superimposed on the cDNA variability plots (Figure 6). Clearly, the occurrence of hotspots corresponds to the highest variability at the CDR1 and CDR2 of cDNA sequences. The hypervariable region found exclusively in the VHH cDNA (residues 27–30) could result from the TAY hotspots that are present in the germline VHHs, but absent in the germline VHs.
Fig. 6. Distribution and frequency of AGY (filled bars) and TAY (open bars) triplets in the germline VH (upper) and VHH (lower) segments. The occurrence of AGY and TAY triplets were scored in all reading frames. The numbers in between refer to the position of VH codons (Kabat numbering). The shaded background originated from the cDNA variability plots shown in Figure 5. The dotted regions denote the CDR1 and CDR2, as defined by Kabat et al. (1991).
Putative recombination
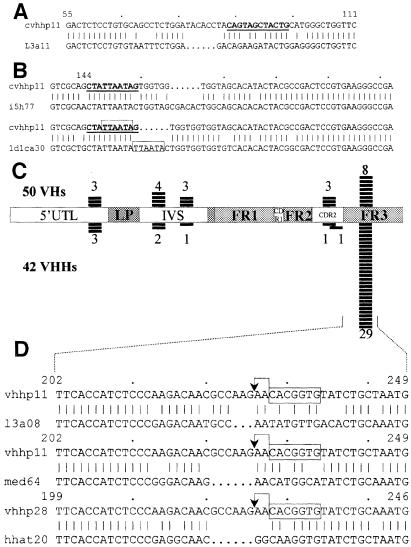
As mentioned previously, the 31 VHH cDNAs having lengths deviating from that of the germline could originate from unequal DNA recombination, gene conversion or gene replacement. Comparison of the nucleotide sequences of these off-size cDNA with germline VHHs revealed four distinct regions in which nucleotides were inserted or deleted (Ins/Del). The four regions surround the residues 24 ± 1 (1 Del and 5 Ins), 30 ± 3 (9 Dels and 6 Ins), 54 ± 3 (2 Dels and 8 Ins) and 74 ± 1 (3 Dels). We noted that most of these Ins/Del regions are within or at the border of peculiar DNA sequences in the germline VHHs, suggesting an active involvement of these sequences in the Ins/Del events. For example, a palindromic sequence ‘CAGTAGCTACTG’ (corresponding to residues 30–33) borders the Del/Ins region 30 ± 3 (example in Figure 7A). Similarly, the Ins/Dels at region 54 ± 3 appear to coincide with the presence of another palindrome ‘CTATTAATAG’ (codons 51–52a) (Figure 7B). Furthermore, two different types of nucleotide insertions can be discerned. Among 17 insertion events, eight are clearly duplications of the bordering sequences, while nine are non-templated nucleotide insertions. An example of each case is shown in Figure 7B.
Fig. 7. (A) Nucleotide alignment of cDNA clone l3a11 (lower line) and its putative germline gene (cvhhp11, upper line), revealing a deletion of 6 nt (dotted). The palindromic sequence is in bold and underlined. Numbers indicate nucleotide positions of the germline VHH element. (B) Alignment of the cDNA (lower lines) and the closest germline sequences (upper lines) indicates insertions, which are non-templated (pair 1) or are duplicated (boxed in pair 2). (C) Frequency and distribution of the RSS-like elements in the camel germline VHs and VHHs. Numbers indicate the incidence of RSS found in 50 VHs (upper) and 42 VHHs (lower). (D) Improper joining adjacent to the RSS signal resulting in clones with a deletion: the heptameric sequences are boxed and the expected cleavage site is indicated by an arrow. The three camel VHH cDNA sequences (l3a08, med64 and hhat20) show a codon deletion when aligned with their putative corresponding genes (cvhhp28 and cvhhp11).
The high incidence of the heptamer-like sequence, a component of the Ig recombination signal (RSS) is another peculiar feature of the VHH sequence (Nguyen et al., 1998). The RSS occurrence is almost twice as high in the germline VHHs (37/42) as it is in the VHs (21/50) (Figure 7C). The majority of these RSS in the VHH are concentrated at the FR3 site corresponding to residues 76–78. Interestingly, this RSS location is downstream of a region with a modest increased variability (Figure 5), and it also abuts a deletion at position 74–75 found in three individual VHH cDNA clones, indicating a possible causal relationship with the heptamer-like sequence in these deletion events (Figure 7D).
Discussion
Dromedary VH/VHH gene segments
To evaluate the potential Ag-binding repertoire of the dromedary HCAbs, we established representative databases of the germline and cDNA VH/VHH sequences. The germline database contains 94 V-gene segments, of which the majority were found in two independent PCR experiments and confirmed by sequences derived from a genomic library. Statistical analysis taking into account the total number of sequenced clones and the number of doubles, triples, etc. also indicated that the dromedary may contain a total of 110 ± 20 VH and VHH genes. This estimation is compatible with the Southern blot analysis (Figure 1), in which the intensively labelled bands may correspond to VH/VHH genes residing in multiple fragments of similar size that overlap in our blot, and the longer fragments may contain more than one V element. The V-gene segments in our database have a high degree of sequence homology, several encoding an identical amino acid sequence. Some of these could be the result of a recent gene duplication, or alternatively, they might be allelic variants as the sequences were derived from a diploid genome. However, the allelic location of the genes has little relevance to our goal to evaluate the potential VHH repertoire.
Clearly, the dromedary contains two distinct sets of V genes (∼40 VHHs and ∼50 VHs), encoding the V domain of the conventional Abs and HCAbs, respectively. The VH and VHH genes appear to be within the same functional V region of the dromedary genome. The identification of an identical D mini-gene in a VH cDNA and a VHH cDNA (Figure 4) suggests the common use of the D segments for VH and VHH. Moreover, preliminary evidence shows that rearranged VH and VHH genes bear identical 3′ sequences downstream of the DJH segments (unpublished results). However, within this functional V region it is still unclear whether the germline VHs and VHHs are interspersed or clustered.
Diverse germline VHHs
All germline VHHs reported here belong to a single family. However, the intrinsic structural repertoire of their Ag-binding loops is quite diverse. The prediction of the canonical structure shows that 10 combinations of the H1–H2 loops are possible in the VHHs, which is higher than what is observed in the VH of family 3, e.g. six in the dromedary, five in human and three in mouse (Tomlinson et al., 1992; Almagro et al., 1997). A total of 10 combinations of the H1–H2 loop structures were found in human and mouse germline VHs of all families (Almagro et al., 1997). The higher complexity of the germline VHHs within one family is due to the presence of novel residues at key sites for the loop conformation, the variable length of the CDR2 and the presence of an additional Cys located at position 30, 32, 33 or 45.
VHH usage in the dromedary
A total of 42 VHH gene segments were identified, which encode 33 unique sequences. In contrast to rabbits where one VH germline is predominantly rearranged (Knight, 1992), members of at least five out of seven VHH subfamilies are used to generate HCAbs in the dromedary (Table I). It also appears that the larger subfamilies were not necessarily the most frequently encountered in the cDNA clones. On the contrary, five members of subfamily 2a accounted for 75% of the inspected cDNAs, while 26 sequences of subfamily 5b matched only three cDNA clones (Table I). This preferential usage of particular VHH segments could be due to their proximal location to the D-gene cluster, as observed in other mammals e.g. the human VH3–23 (Stewart et al., 1992) and mouse VH10 (Schiff et al., 1988).
We note that the Ag-binding loop structures of the VHH subfamily 2a are predicted to switch readily. This subfamily appears to be the most prevalent to generate Ag binders, indicating that the paratope repertoire derived from these VHH members could cope with various Ags. Therefore, the selection of these germline VHHs as a scaffold to construct a synthetic single domain library would generate a sufficiently diverse repertoire to retrieve good Ag binders.
Somatic diversification of VHHs
From a limited number of germline V genes, many species can generate a large Ag-binding repertoire through a somatic diversification process (Reynaud et al., 1989; Knight, 1992; Sun et al., 1994; Dufour et al., 1996; Sinclair et al., 1997). We note that the dromedary VHH structural repertoire is readily diversified by the introduction of an additional disulfide bridge, the high incidence of nucleotide Ins/Dels, gene replacement and the extensive somatic hyperpoint mutations.
Extra disulfide bridge. The germline VHH possesses a Cys in addition to the conserved Cys22 and Cys92 that form a disulfide bond in all Ig domains. This additional Cys is maintained in the vast majority of the VHH cDNAs, which also acquired an additional Cys within the CDR3 (Muyldermans et al., 1994; Vu et al., 1997). The disulfide bonds between these additional cysteines cross-links the Ag-binding loops (Davies and Riechmann, 1996; Desmyter et al., 1996). This has two consequences: it stabilizes the domain, and it induces constraints in the CDR1 or CDR3, which could lead to novel loop conformations and thereby increasing the paratope repertoire.
High incidence of insertions and deletions. The Ins/Dels of nucleotides found in 31 VHH cDNAs cluster in four regions. The high incidence (30%) and the clustering feature of Ins/Dels suggest that they (at least in part) could be relics of a gene conversion-like event as described for chicken, rabbit and cattle (Reynaud et al., 1987; Becker and Knight, 1990; Parng et al., 1996).
The equal presence of two different types of nucleotide insertions indicates that there are at least two different modes of action leading to these sequence length variations. Half of the insertions are nucleotide duplications, indicating that these Ins/Dels could be by-products of somatic hypermutation (Wilson et al., 1998). In the other half of the events, the inserted nucleotide sequences are different from their flanking sequences. Therefore, this later type could be referred to as non-templated nucleotide insertions as often found at the VD or DJ joints. Though the actual mechanism is still unknown, this observation opens the possibility that the nucleotide addition process could occur during the rejoining of double strand DNA breaks of the VHHs.
Under any hypothesis, it is important to note that the Ins/Dels are not randomly distributed, but often occur near or within the paratope. Therefore, they substantially reshape the VHH loop structure. This is true for both the conventional VH and the VHH. However, in dromedary the incidence of off-size clones is much higher in the VHH cDNA than in the conventional VH cDNA (30 versus 1.5%), indicating that the VHH is more prone to these changes. Moreover, whereas in human most Ins/Dels were found in non-functional Ig genes (Klein et al., 1998), in the dromedary, six out of 31 off-size cDNAs have a known Ag specificity.
Gene replacement. While the exact Ins/Del mechanism involving palindromes remains unknown, the high incidence of the embedded RSS at VHH-FR3 abutting the Del/Ins region 74 ± 1 [Figure 7D and Ins at position 75A of two llama VHHs (Vu et al., 1997)] supports the gene replacement mechanism (Reth et al., 1986) involving recombination-activating gene (RAG) proteins as observed in human and mouse (Kleinfield et al., 1986; Komori et al., 1993). The involvement of a RAG-like activity to initiate the gene replacement predicts a DNA cleavage two bases upstream of the heptamer sequence (Ramsden et al., 1996). This is exactly the position where the deletions are observed in these individual clones (Figure 7D). It is possible that improper joining (Melek et al., 1998) causes these Ins/Dels, and proper joining retains the sequence length but induces nucleotide replacements (Weinstein et al., 1994). The latter possibly leads to the increased variability observed in this region of the VHH (Figure 5). The region around position 75 lies in a solvent exposed loop immediately adjacent to the H2 loop in the folded Ig domain. In monomeric T-cell receptors, the corresponding region is also hypervariable and reported to interact with ligand (Howell et al., 1991). By analogy, we suppose that this VHH region can be involved in the Ag–Ab interaction by either directly contacting the Ag or inducing structural changes in the Ag-binding site. Taken together, it is plausible that gene replacement at this region, in which a new VHH rearranges to the pre-existing VHH–D–JH, results in a novel Ag-binding site composed of the incoming VHH and the existing CDR3.
Extended hypervariable CDR1 region. The patterns of the cDNA variability plots (Figure 5) are basically consistent with the classification of FR- and CDR-regions as defined by Kabat et al. (1991). However, the extra hypervariable region (residues 27–30) present exclusively in the VHH is most remarkable. Crystallographic studies of VHH–antigen complexes demonstrated that amino acids located in this area interact with Ag (Desmyter et al., 1996; Decanniere et al., 1999). Obviously, the VHH uses this region together with a long CDR3 to increase the surface area interacting with Ags. Surprisingly, to attain new amino acids at these positions the germline VHHs accumulated two new hypermutation hotspots (Figure 6) by two single point mutations. The non-hotspot triplet TTY in the VH is substituted by TAY in the VHH, a well-known hotspot for hypermutation (Milstein et al., 1998). These mutations in the germline lead to the amino acid substitutions F27Y and F29Y, two key-elements for the H1 loop conformation (Chothia et al., 1992). Further mutations of Y27 or Y29 in the VHHs has two pronounced effects. Some mutations will lead to complete new loop conformations and thereby will expand the structural repertoire of the VHHs. Other mutations might lead to amino acid substitutions provoking subtle surface modifications that might improve the VHH–antigen fit. For these reasons, we propose to vary these amino acids in libraries of synthetic single domain VHHs or human VHs (Davies and Riechmann, 1996; Reiter et al., 1999) to increase the potential repertoire and to search for more potent Ag binders.
In conclusion, the data presented here demonstrate that the Ag-binding repertoire of HCAb, derived from ∼40 germline VHHs, is largely diversified by specific mechanisms. These include the introduction of a variety of interloop disulfide bridges, the increased surface area of hypervariable regions and a high rate of paratope reshaping. The use of these specific mechanisms also results in novel paratopes different from those of the conventional Abs. This could explain the large proportion of HCAbs acting as competitive enzyme inhibitors. It is clear that particular DNA signals with a high incidence in the VHHs trigger these somatic diversification processes. It is possible that the absence of the light chain provides the freedom to make these changes possible and to allow the VHH to explore new structures.
Materials and methods
Southern blots
High MW genomic DNA was prepared from liver tissue of a single camel (Camelus dromedarius) from Morocco. Dromedary genomic DNA (10 μg) was digested with BamHI, EcoRI or BamHI–EcoRI and electrophoresed in a 0.8% agarose gel. The DNA was transferred to Hybond-N (Amersham) membranes. Two homologous VH probes of 0.6 kb corresponding to the dromedary germlines VH and VHH (Nguyen et al., 1998) were obtained by PCR and labelled with 32P (Radprime, Gibco-BRL). After hybridization, the membranes were washed at 65°C for 15 min/wash (twice with 2× SSC, 0.1% SDS, once with 2× SSC, 1% SDS, and finally with 0.1× SSC), and autoradiographed.
Genomic DNA amplification, cloning and DNA sequencing
The primers used to amplify VH/VHH genes were derived from the VH and VHH genomic sequences (Nguyen et al., 1998). One specific primer of 26 nt corresponds to the upstream conserved Ig octamer sequence (VHOCTB: 5′-TCTATATATCTAGATGACATGCAAAT-3′). The other primer anneals at the FR3 sequence and starts from the Cys92 codon (VHFR3F: 5′-ACAGTAATACATGGCCGTGTCCTC-3′). PCRs (50 μl) were performed on 0.5 μg liver genomic DNA with 2.5 U of Taq DNA polymerase (ROCHE) (30 cycles of 30 s at 94°C, 30 s at 55°C and 60 s at 72°C with a final incubation of 10 min at 72°C). PCR products were gel-purified, ligated into TA-PCRII cloning vector (Invitrogen), and subsequently transformed into DH5α cells. Randomly chosen clones with inserts of the expected size were sequenced from both ends. Two sets of PCR and cloning were carried out independently to exclude possible PCR errors.
Screening of a genomic library
A camel liver genomic library (Nguyen et al., 1998) was screened by plaque hybridization with the same probes used in Southern blot experiment. The VH/VHH element from the putative positive phage clone was amplified using the VHFR3F primer and a primer that corresponds to the leader-peptide sequence: VHLB 5′-GGCTGAGCTCGGTGGTCCTGGCT-3′. The phage-derived PCR fragments were cloned, and two clones were sequenced for each experiment.
Camel V databases
Germline VH DNA sequences were aligned and grouped according to sequence identity. DNA sequences (600 bp) differing by no more than four bases were considered identical as these differences might be due to PCR or sequencing errors. The camel germline VH/VHH database contains the remaining unique sequences. The camel cDNA data were collected from the VH and VHH cDNA sequences available within our group.
Sequence analysis
All DNA and protein sequence analyses were performed with version 10 of the University of Wisconsin Genetics Computer Group package (Devereux et al., 1984).
Acknowledgments
Acknowledgements
We thank C.Bouton, M.De Kerpel, Y.Hou and L.M.Tam for technical assistance, Dr K.Decanniere and D.Maes for discussions, Dr W.van der Loo for help with the evolutionary analysis, and all colleagues for their binders' sequences. N.V.K. was supported by a VLIR-ABOS training grant. This work was granted by VLIR, VIB and FWO.
References
- Almagro J.C., Hernandez, I., del Carmen, R. and Vargas-Madrazo, E. (1997) The differences between the structural repertoires of VH germ-line gene segments of mice and humans: implication for the molecular mechanism of the immune response. Mol. Immunol., 34, 1199–1214. [DOI] [PubMed] [Google Scholar]
- Becker R.S. and Knight, K.L. (1990) Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell, 63, 987–997. [DOI] [PubMed] [Google Scholar]
- Berek C., Berger, A. and Apel, M. (1991) Maturation of the immune response in germinal centers. Cell, 67, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Brodeur P.H. and Riblet, R. (1984) The immunoglobulin heavy chain variable region (Igh-V) locus in the mouse. I. One hundred Igh-V genes comprise seven families of homologous genes. Eur. J. Immunol., 14, 922–930. [DOI] [PubMed] [Google Scholar]
- Cha R.S. and Thilly, W.G. (1993) Specificity, efficiency and fidelity of PCR. PCR Methods Appl., 3, S18–S29. [DOI] [PubMed] [Google Scholar]
- Chothia C., Boswell, D.R. and Lesk, A.M. (1988) The outline structure of the T-cell αβ receptor. EMBO J., 7, 3745–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., et al. (1989) Conformations of immunoglobulin hypervariable regions. Nature, 342, 877–883. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk, A.M., Gherardi, E., Tomlinson, I.M., Walter, G., Marks, J.D., Llewelyn, M.B. and Winter, G. (1992) Structural repertoire of the human VH segments. J. Mol. Biol., 227, 799–817. [DOI] [PubMed] [Google Scholar]
- Davies J. and Riechmann, L. (1995) Antibody VH domains as small recognition units. Biotechnology, 13, 475–479. [DOI] [PubMed] [Google Scholar]
- Davies J. and Riechmann, L. (1996) Single antibody domains as small recognition units: design and in vitro antigen selection of camelized, human VH domains with improved protein stability. Protein Eng., 9, 531–537. [DOI] [PubMed] [Google Scholar]
- Decanniere K., Desmyter, A., Lauwereys, M., Ghahroudi, M.A., Muyldermans, S. and Wyns, L. (1999) A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure, 7, 361–370. [DOI] [PubMed] [Google Scholar]
- Desmyter A., Transue, T.R., Ghahroudi, M.A., Thi, M.H., Poortmans, F., Hamers, R., Muyldermans, S. and Wyns, L. (1996) Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nature Struct. Biol., 3, 803–811. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli, P. and Smithies, O. (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res., 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour V., Malinge, S. and Nau, F. (1996) The sheep Ig variable region repertoire consists of a single VH family. J. Immunol., 156, 2163–2170. [PubMed] [Google Scholar]
- Ghahroudi M.A., Desmyter, A., Wyns, L., Hamers, R. and Muyldermans, S. (1997) Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett., 414, 521–526. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E.B., Bendahman, N. and Hamers, R. (1993) Naturally occurring antibodies devoid of light chains. Nature, 363, 446–448. [DOI] [PubMed] [Google Scholar]
- Hoogenboom H.R. and Winter, G. (1992) By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J. Mol. Biol., 227, 381–388. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Diveley, J.P., Lundeen, K.A., Esty, A., Winters, S.T., Carlo, D.J. and Brostoff, S.W. (1991) Limited T-cell receptor β-chain heterogeneity among interleukin 2 receptor-positive synovial T cells suggests a role for superantigen in rheumatoid arthritis. Proc. Natl Acad. Sci. USA, 88, 10921–10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat E.A., Wu,T.T., Perry,H.M., Gottesman,K.S. and Foeller,C. (1991) Sequences of proteins of immunological interest. US Public Health Services, NIH Bethesda, MD, Publication No. 91-3242.
- Klein U., Goossens, T., Fischer, M., Kanzler, H., Braeuninger, A., Rajewsky, K. and Kuppers, R. (1998) Somatic hypermutation in normal and transformed human B cells. Immunol. Rev., 162, 261–280. [DOI] [PubMed] [Google Scholar]
- Kleinfield R., Hardy, R.R., Tarlinton, D., Dangl, J., Herzenberg, L.A. and Weigert, M. (1986) Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature, 322, 843–846. [DOI] [PubMed] [Google Scholar]
- Knight K.L. (1992) Restricted VH gene usage and generation of antibody diversity in rabbit. Ann. Rev. Immunol., 10, 593–616. [DOI] [PubMed] [Google Scholar]
- Komori T., Minami, Y., Sakato, N. and Sugiyama, H. (1993) Biased usage of two restricted VH gene segments in VH replacement. Eur. J. Immunol., 23, 517–522. [DOI] [PubMed] [Google Scholar]
- Lauwereys M., Ghahroudi, M.A., Desmyter, A., Kinne, J., Holzer, W., De Genst, E., Wyns, L. and Muyldermans, S. (1998) Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J., 17, 3512–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C. and Thornton, J.M. (1996) Structural families in loops of homologous proteins: automatic classification, modelling and application to antibodies. J. Mol. Biol., 263, 800–815. [DOI] [PubMed] [Google Scholar]
- Melek M., Gellert, M. and van Gent, D.C. (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science, 280, 301–303. [DOI] [PubMed] [Google Scholar]
- Milstein C., Neuberger, M.S. and Staden, R. (1998) Both DNA strands of antibody genes are hypermutation targets. Proc. Natl Acad. Sci. USA, 95, 8791–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S., Atarhouch, T., Saldanha, J., Barbosa, J.A. and Hamers, R. (1994) Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng., 7, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Neuberger M.S., Ehrenstein, M.R., Klix, N., Jolly, C.J., Yelamos, J., Rada, C. and Milstein, C. (1998) Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol. Rev., 162, 107–116. [DOI] [PubMed] [Google Scholar]
- Nguyen V.K., Muyldermans, S. and Hamers, R. (1998) The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J. Mol. Biol., 275, 413–418. [DOI] [PubMed] [Google Scholar]
- Nguyen V.K., Hamers, R., Wyns, L. and Muyldermans, S. (1999) Loss of splice consensus signal is responsible for the removal of the entire CH1 domain of the functional camel IgG2a heavy-chain antibodies. Mol. Immunol., 36, 515–524. [DOI] [PubMed] [Google Scholar]
- Novotny J. (1991) Protein antigenicity: a thermodynamic approach. Mol. Immunol., 28, 201–207. [DOI] [PubMed] [Google Scholar]
- Parng C.L., Hansal, S., Goldsby, R.A. and Osborne, B.A. (1996) Gene conversion contributes to Ig light chain diversity in cattle. J. Immunol., 157, 5478–5486. [PubMed] [Google Scholar]
- Ramsden D.A., McBlane, J.F., van Gent, D.C. and Gellert, M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- Reiter Y., Schuck, P., Boyd, L.F. and Plaksin, D. (1999) An antibody single-domain phage display library of a native heavy chain variable region: isolation of functional single-domain VH molecules with a unique interface. J. Mol. Biol., 290, 685–698. [DOI] [PubMed] [Google Scholar]
- Reth M., Gehrmann, P., Petrac, E. and Wiese, P. (1986) A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature, 322, 840–842. [DOI] [PubMed] [Google Scholar]
- Reynaud C.A., Anquez, V., Grimal, H. and Weill, J.C. (1987) A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell, 48, 379–388. [DOI] [PubMed] [Google Scholar]
- Reynaud C.A., Dahan, A., Anquez, V. and Weill, J.C. (1989) Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell, 59, 171–183. [DOI] [PubMed] [Google Scholar]
- Schiff C., Corbet, S. and Fougereau, M. (1988) The Ig germline gene repertoire: economy or wastage?Immunol. Today, 9, 10–14. [DOI] [PubMed] [Google Scholar]
- Schroeder H.W.J., Hillson, J.L. and Perlmutter, R.M. (1990) Structure and evolution of mammalian VH families. Int. Immunol., 2, 41–50. [DOI] [PubMed] [Google Scholar]
- Sinclair M.C., Gilchrist, J. and Aitken, R. (1997) Bovine IgG repertoire is dominated by a single diversified VH gene family. J. Immunol., 159, 3883–3889. [PubMed] [Google Scholar]
- Spinelli S., Frenken, L., Bourgeois, D., de Ron, L., Bos, W., Verrips, T., Anguille, C., Cambillau, C. and Tegoni, M. (1996) The crystal structure of a llama heavy chain variable domain. Nature Struct. Biol., 3, 752–757. [DOI] [PubMed] [Google Scholar]
- Stewart A.K., Huang, C., Long, A.A., Stollar, B.D. and Schwartz, R.S. (1992) VH-gene representation in autoantibodies reflects the normal human B-cell repertoire. Immunol. Rev., 128, 101–122. [DOI] [PubMed] [Google Scholar]
- Sun J., Kacskovics, I., Brown, W.R. and Butler, J.E. (1994) Expressed swine VH genes belong to a small VH gene family homologous to human VHIII. J. Immunol., 153, 5618–5627. [PubMed] [Google Scholar]
- Tomlinson I.M., Walter, G., Marks, J.D., Llewelyn, M.B. and Winter, G. (1992) The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol., 227, 776–798. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. (1983) Somatic generation of antibody diversity. Nature, 302, 575–581. [DOI] [PubMed] [Google Scholar]
- Vu K.B., Ghahroudi, M.A., Wyns, L. and Muyldermans, S. (1997) Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol., 34, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Wagner S.D. and Neuberger, M.S. (1996) Somatic hypermutation of immunoglobulin genes. Ann. Rev. Immunol., 14, 441–457. [DOI] [PubMed] [Google Scholar]
- Weinstein P.D., Anderson, A.O. and Mage, R.G. (1994) Rabbit IgH sequences in appendix germinal centers: VH diversification by gene conversion-like and hypermutation mechanisms. Immunity, 1, 647–659. [DOI] [PubMed] [Google Scholar]
- Wilson P.C., de Bouteiller, O., Liu, Y.J., Potter, K., Banchereau, J., Capra, J.D. and Pascual, V. (1998) Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J. Exp. Med., 187, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G., Griffiths, A.D., Hawkins, R.E. and Hoogenboom, H.R. (1994) Making antibodies by phage display technology. Ann. Rev. Immunol., 12, 433–455. [DOI] [PubMed] [Google Scholar]
- Woolven B.P., Frenken, L.G., van der Logt, P. and Nicholls, P.J. (1999) The structure of the llama heavy chain constant genes reveals a mechanism for heavy-chain antibody formation. Immunogenetics, 50, 98–101. [DOI] [PubMed] [Google Scholar]
- Yelamos J., Klix, N., Goyenechea, B., Lozano, F., Chui, Y.L., Gonzalez, F.A., Pannell, R., Neuberger, M.S. and Milstein, C. (1995) Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature, 376, 225–229. [DOI] [PubMed] [Google Scholar]