SUMMARY
mTor kinase is involved in cell growth, proliferation, and differentiation. The roles of mTor activators, Rheb1 and Rheb2, have not been established in vivo. Here, we report that Rheb1, but not Rheb2, is critical for embryonic survival and mTORC1 signaling. Embryonic deletion of Rheb1 in neural progenitor cells abolishes mTORC1 signaling in developing brain and increases mTORC2 signaling. Remarkably, embryonic and early postnatal brain development appears grossly normal in these Rheb1f/f, Nes-cre mice with the notable exception of deficits of myelination. Conditional expression of Rheb1 transgene in neural progenitors increases mTORC1 activity and promotes myelination in the brain. In addition, the Rheb1 transgene rescues mTORC1 signaling and hypomyelination in the Rheb1f/f, Nes-cre mice. Our study demonstrates that Rheb1 is essential for mTORC1 signaling and myelination in the brain, and suggests that mTORC1 signaling plays a role in selective cellular adaptations, rather than general cellular viability.
INTRODUCTION
mTor kinase regulates multiple intracellular processes in response to extracellular signals and nutrient availability, and is an important therapeutic target in neurological disorders, brain tumors, and immunology and diabetes research. mTor functions as part of two distinct signaling complexes—mTORC1 and 2, with mTORC1 consisting of mTor, mLST8, and Raptor, and mTORC2 consisting of mTor, mLST8, Sin1, and Rictor (Jacinto et al., 2006; Sarbassov et al., 2005a; Sarbassov et al., 2005b). mTORC1 phosphorylates proteins involved in protein synthesis and cell growth, including 4E-BP1 and p70S6 kinase 1 (S6K1), which phosphorylates the ribosomal protein S6. mTORC2 phosphorylates AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase subfamilies such as the AKT/PKBs and all conventional and atypical PKCs (Ikenoue et al., 2008), and is involved in the regulation of actin cytoskeleton (Jacinto et al., 2004).
mTORC1 and 2 are activated by multiple growth factors including insulin and insulin-like growth factor1 (IGF1). For example, insulin binding to the insulin growth factor receptor initiates a signaling cascade that sequentially includes insulin receptor substrate 1 and 2 (IRS1/2), phosphoinositide 3-OH kinase (PI3K), PI3K-regulated serine/threonine kinase PKB (also known as AKT), tuberous sclerosis complex 1/2 (TSC1/2), Rheb (Ras homolog enriched in brain) (Avruch et al., 2006; Garami et al., 2003), while PKB/AKT phosphorylates PRAS40—a Raptor binding protein and releases its inhibitory effect on mTORC1 (Sancak et al., 2007; Vander Haar et al., 2007) . mTORC1 can also be activated by nutrients, e.g. amino acids (Hara et al., 1998; Nobukuni et al., 2005) via Rag GTPase (Sancak et al., 2010; Sancak et al., 2008).
Rheb was identified based on its regulation as a cellular immediate early gene in the brain (Yamagata et al., 1994). Based on sequence homology, another Rheb-related family member Rheb2 was identified (Tabancay et al., 2003); therefore, the original Rheb is now known as Rheb1. Rheb1 mRNA is expressed in all tissues, whereas Rheb2 is detectable in several peripheral tissues but primarily expressed in brain, particularly in the regions involved in higher cognitive function (Saito et al., 2005). A role of Rheb in Tor signaling was revealed in functional screen for genes that regulate the size of cells in Drosophila, and was placed upstream of Tor and downstream of TSC1/2 (Kwon et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Zhang et al., 2003). TSC1/2 exhibits GTPase activating protein activity towards Rheb, thus inhibiting Rheb activity (Inoki et al., 2003; Zhang et al., 2003). For example, TSC1 and TSC2 inhibit Rheb1 and mTORC1 signaling in the insulin pathway (Garami et al., 2003). Rheb1 appears to activate mTORC1 via direct binding to mTor (Long et al., 2005a) but the molecular basis remains controversial. Both Rheb1 and Rheb2 transgenes can activate mTor in heterologous cells.
Rheb has been linked to mTORC2 activity. Increasing Rheb1 activity by either overexpresson of Rheb1 transgene or by loss of function mutation of TSC1 or 2 increases mTORC1 activity, but decreases mTORC2 activity (Garami et al., 2003; Yang et al., 2006). This functional link between mTORC1 and mTORC2 is mediated by a negative feedback mechanism involving mTORC1/S6K1 activity inhibiting the transcription of IRS1/2 and promoting degradation of IRS1/2 (Harrington et al., 2004; Shah et al., 2004). Recently, S6K1 has been shown to directly phosphoryalte Rictor at T1135 and inhibits mTORC2 activity (Dibble et al., 2009; Gao et al.; Julien et al., 2010; Treins et al.). Studies of Rictor-and mLST8-difficient mice, however, indicate that mTORC2 activity is not required for mTORC1 activation by serum/growth factor stimulation (Guertin et al., 2006).
Despite progress in the understanding of mTor signaling, there are no in vivo studies that indicate whether Rheb1 or Rheb2 is necessary to activate mTor signaling. The importance of mTor signaling in diverse aspects of cellular function, including protein translation, suggests that it may be essential for organ development and organismal survival. Indeed, mutations of genes that are part of mTORC1 signaling complex, including mTor and Raptor, have been reported to cause embryonic lethality (Gangloff et al., 2004; Guertin et al., 2006; Murakami et al., 2004). To examine the contribution of Rheb in mTor signaling in vivo, and to assess its role in tissue development, we established mouse models that afford conditional genetic deletion of Rheb1 or Rheb2, or conditional expression of Rheb1 transgene, and examined brain development. Our studies demonstrate an essential role of Rheb1, but not Rheb2, in mTORC1 signaling in vivo, and confirm crosstalk between mTORC1 and mTORC2. Additionally, we establish myelination as an important target of Rheb1/mTORC1 signaling in brain.
RESULTS
Requirement of Rheb1, but not Rheb2, for Organism Viability
We generated mice with conditional genetic deletions of Rheb1 (Figures S1A to S1C) and Rheb2 (Figures S1D and S1E). Germline deletion of Rheb1 (Actin-cre) results in embryonic death between E10.5 and E11.5. No viable homozygous Rheb1 mutant embryos (Rheb1−/−) were identified after E11.5. Heterozygous Rheb1 (Rheb1+/−) embryos develop to term, mature and grow normally, and are fertile. Germline deletion of Rheb2 does not affect embryonic viability, and mutant mice mature to adulthood without obvious physical deficits. Deletion of Rheb2 does not alter the survival of Rheb1−/− or Rheb1+/− mice, as no offspring with the genotype of Rheb1−/−Rheb2−/− were identified from inter-crossing of Rheb1 and Rheb2 double heterozygous mice, while offspring with the genotype of Rheb1+/−Rheb2−/− were obtained at the expected Mendelian ratio. Thus, Rheb1, but not Rheb2, is essential for organism viability.
Rheb1 is Essential for Growth Factor and Amino Acid Stimulated mTORC1 Signaling
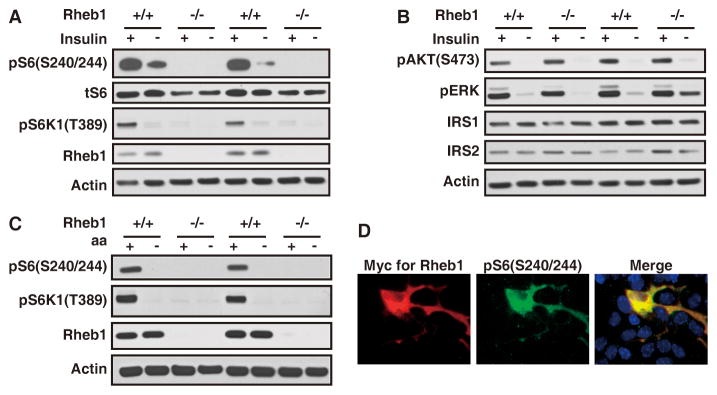
To examine Rheb1-dependent signaling, we cultured mouse embryonic fibroblasts (MEFs) from individual E10.5 embryos obtained by inter-crossing of Rheb1+/− mice. Cell lysates were assayed for Rheb1 protein and phospho-S6 (pS6); an indicator of mTORC1 activation to phosphorylate S6 kinase (p70S6K1). Rheb1 protein migrates at 17kDa, as confirmed by its absence in Rheb1−/− MEFs. While pS6 (S240/244) antibody reliably detected phosphorylation in WT MEFs, it did not detect phosphorylation of S6 in primary Rheb1−/− MEFs cultured with 0.5% serum (Figure 1A). Insulin induced a robust increase of phosphorylation of p70S6 kinase and S6 in WT MEFs, but not in Rheb1−/− MEFs (Figure 1A). This was not due to reduced expression of IRS1 or IRS2 (Figure 1B). While the mTORC1 pathway was affected in the Rheb1−/− MEFs, other insulin-dependent signaling was preserved. Thus, insulin strongly increased pERK (Figure 1B) in Rheb1−/− MEFs. Moreover, insulin increased pAKT(S473) (Figure 1B), which is selectively phosphorylated by mTORC2 (Sarbassov et al., 2005b). Rheb2 mRNA was detected in the MEFs by RT-PCR (data not shown). The absence of mTORC1 activity in Rheb1−/− MEFs suggests that Rheb2 is not able to compensate for the loss of Rheb1 in the activation of mTORC1.
Figure 1.
Rheb1 is Essential for Growth Factor Stimulated mTORC1 Signaling.
(A) Western blots show the failure of serum-starved primary Rheb1−/− MEFs to activate mTORC1 as indicated by pS6K and pS6 in response to insulin stimulation (200nM, 15min).
(B) Western blots reveal normal response of serum-starved Rheb1−/− MEFs to insulin stimulation (200nM, 15min) to activate mTORC2 (pAKT S473) and ERK kinase. IRS1 and 2 are not upregulated in the Rheb1−/− MEFs.
(C) Western blots show the failure of Rheb1−/− primary MEFs to activate mTORC1 activity with AA stimulation (see Supplemental Information).
(D) Immunostaining shows the restoration of the mTORC1 activity in the Rheb1−/− MEFs by myc-Rheb1. DAPI staining (blue) reveals Rheb1−/− MEFs not transfected with Rheb1 transgene.
Amino acids potently activate mTor signaling (Hara et al., 1998; Nobukuni et al., 2005), and the mTor pathway is considered critical for integration of growth factor signaling and nutrient availability. Studies of signaling pathway(s) that mediate amino acid-dependent activation of mTor are conflicting (Long et al., 2005b; Smith et al., 2005). To examine the role of Rheb, MEFs were grown in 0.5% FBS overnight and then subjected to complete nutrient starvation (dPBS) for 2 hours, and subsequently supplemented with amino acids for 1 hour. Amino acids stimulated robust activation of mTORC1 in WT MEFs as indicated by phosphorylation of S6 and S6K1 (Figure 1C). In contrast to WT MEFS, Rheb1−/− MEFs failed to show any induction of pS6 or pS6K1 under the same condition (Figure 1C). Serum-stimulated S6 phosphorylation was rescued in Rheb−/− MEFs by expression of myc-tagged Rheb1 (Figure 1D). These findings indicate that Rheb1 is essential for growth factor and amino acid stimulated mTORC1 signaling in MEFs, but is not required for mTORC2.
mTORC1 Activity is Selectively Impaired by Genetic Deletion of Rheb1
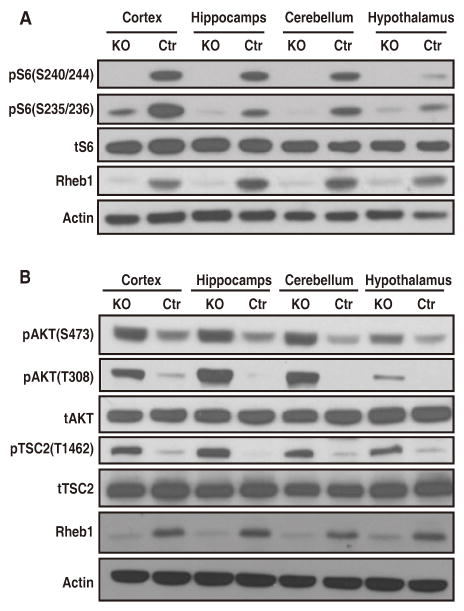
To examine the roles of Rheb1 in mTor signaling in vivo, we generated a mouse model in which Rheb1 was conditionally excised in neural progenitor cells using Nestin-cre driver line (Rheb1f/f, Nestin-cre, referred to as Rheb1 KO in figure legends). Nestin-cre (Nes-cre) transgenic mice (Tronche et al., 1999) express Cre activity as early as E10.5 that is typically restricted to neural stem/progenitor cells that give rise to both neurons and glial cells in the CNS. Nes-cre has been reported to excise floxed targets in peripheral tissues including excisions in germ cells.We confirmed deletion of Rheb1 by E15.5 (Figure S2A). Rheb1f/f, Nes-cre mice were born at expected Mendelian ratios, and appeared normal at birth but showed postnatal growth retardation (Figure S2B). Rheb1f/f, Nes-cre mice survived up to 6 weeks after birth. Consistent with data from MEFs, we found that in the 3~4-wk mice, phosphorylation of S6 was robust in the WT brains, but was nearly absent in Rheb1f/f, Nes-cre brains, despite normal levels of total S6 and S6K (Figure 2A). Nestin-cre provided highly efficient excision of Rheb1, however a trace amount of Rheb1 immunoreactivity remained in brain samples and may underlie low residual pS6 at S235/236 in some brain regions. Alternatively, residual activity may be consistent with the report that S6 kinase accounts for about 90% of phosphorylation of S6 in S6K1 and 2 double KO mice (Pende et al., 2004).
Figure 2.
mTORC1 is Selectively Impaired in the Rheb1f/f, Nes-cre KO Brain.
(A) Western blots show a drastic reduction in mTORC1 activity in the 4-wk old Rheb1f/f, Nes-cre (hereafter, KO) brain while the total level of S6 is not altered.
(B) Western blots show significant upregulation of mTORC2 activity in the 4-wk old Rheb1 KO brain and concurrent upregulation of the activity of signaling components upstream of Rheb1 (pAKT308 and pTSC2).
While mTORC1 activity was reduced in the Rheb1f/f, Nes-cre brain, mTORC2 activity was upregulated. mTORC2 activity was assayed by phosphorylation of AKT at S473 and PKCα at S657. Both phosphorylation indices were robust without changes in the total levels of AKT and PKCα in Rheb1f/f, Nes-cre brain (Figure 2B; Figure S2C), indicating preservation of mTORC2 signaling. Indeed, comparisons with WT indicated that mTORC2 signaling was substantially increased, consistent with reported crosstalk between mTORC1 and mTORC2 in heterologous cells (Julien et al., 2010; Rosner et al., 2009). An examination of other signaling molecules that function in the mTORC1 pathway revealed phosphorylation of AKT (T308) and TSC2 (T1462) were increased, while the levels of AKT and TSC2 expression were not altered (Figure 2B). These data are consistent with homestatic adaptations of AKT and TSC signaling, as they normally function to increase Rheb activity (Harrington et al., 2004; Shah et al., 2004).
While Rheb1 is essential for mTORC1 signaling, we found that Rheb2 is not essential for mTor signaling in vivo. Although Rheb2 is expressed in multiple brain regions implicated in cognitive function, we found that deletion of Rheb2 did not alter the level of pS6, pAKT S473 or any other components in the mTor pathway (Figures S2E to S2H). Deletion of Rheb2 in Rheb1+/− mice (Rheb2−/−Rheb1+/−, 4wk-old) appeared to trend toward a modest reduction in pS6 and pAKT in the cortex and hippocampus compared to Rheb2−/−, Rheb1+/−, or WT mice, but did not reach statistical significance (Figures S2F and S2H). Accordingly, this analysis failed to detect a role for Rheb2 in mTORC1 signaling in vivo.
Rheb1 is not Essential for Embryonic Brain Development
The deletion of Rheb1 in neural progenitor cells by Nestin-cre afforded an opportunity to examine the role of Rheb1/mTORC1 in brain development. We confirmed the embryonic excision of Rheb1 in the Rheb1f/f, Nes-cre mice by Western blots showing that Rheb1 protein level was reduced by >90% in the forebrain of E15.5, E17.5, and P1 and P5 postnatal brains (Figure 3A; Figure S2A). Consistent with the Rheb1 protein reduction, the level of pS6 (S235/236, and S240/244) in the forebrains was markedly reduced at P1 and P5, compared to age-matched controls (Figure 3A). Nissl staining revealed that at P1 and P5, the gross morphology of the brain of the Rheb1f/f, Nes-cre mice was preserved. The brain weight at P1 was identical between WT and Rheb1f/f, Nes-cre mice (Figure S3A). However, by P5 to P9, the weight of Rheb1f/f, Nes-cre brains was reduced, and this difference increased through subsequent postnatal development (Figure 3B; Figure S3A). By 2~3 wks, the weight of the Rheb1f/f, Nes-cre brain was ~50% of that of WT controls (Figure S3A). The cortical thickness of the Rheb1f/f, Nes-cre brain was ~65% of WT controls at 4-wk old (Figure S3B). There is no loss of neurons in the Rheb1f/f, Nes-cre brain, as Nissl and NeuN staining of P1, P5 and P28 did not reveal a reduction in neuronal counts (Figure 3B; Figures S3C and S3D). Biochemical analysis revealed normal expression of neuronal markers NeuN, Tau1, Tuj1 and MAP2 (Figure 3A), and the astrocyte marker GFAP in the early postnatal Rheb1f/f, Nes-cre brains (Figure 3A). GFAP staining of early postnatal (P0 and P2) and young adult (3~4-wk old) Rheb1f/f, Nes-cre brains showed normal appearance of astrocytes (Figure S3E). The number of oligodendrocyte precursor cells indicated by NG2 and PDGFRα in the P2 brain was comparable to that in the control (Figure 3C), suggesting the commitment of neural stem cells into oligodendrocyte lineage cells was not impaired.
Figure 3.
Rheb1 is not Essential for the Formation of the Brain During Embryonic Development.
(A) Western blots show no changes in neuronal markers (NeuN, Tuj1, Tau1, and MAP2), and astrocyte marker (GFAP) in E17.5 to P5 Rheb1 KO brains. A representative blot from three independent experiments is shown.
(B) Nissl staining reveals the preservation of the gross morphology of the brain structure in the P1 and P5 Rheb1 KO mice, albeit at a reduced size. Bar, 500 μm. Note the reduction in the thickness of the cortex. Bar, 100 μm.
(C) Immunostaining shows normal genesis of oligodendrocyte precursor cells in the corpus callosum of P2 KO brain. Insets show higher magnification of the cells indicated by arrows. Bar, 20 μm. Data represent mean ± SEM. Results are averages of two independent experiments.
Loss of Rheb1 Affects Postnatal Myelination in the Brain
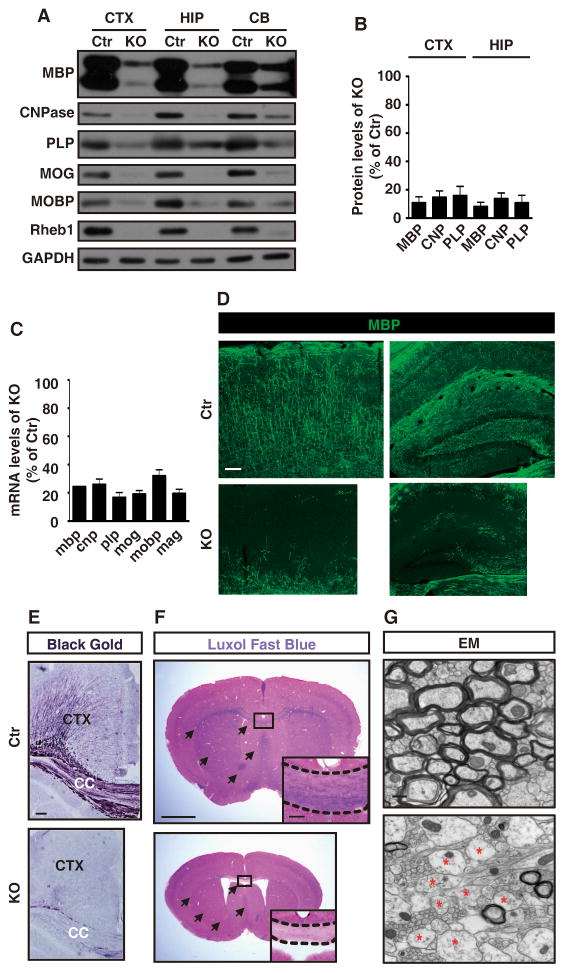
TSC patients and mouse models show white matter/myelin abnormalities (Meikle et al., 2007; Ridler et al., 2001); therefore, we examined the myelination in the Rheb1f/f, Nes-cre mice. Rheb1f/f, Nes-cre mice exhibited prominent hypomyelination. Western blots of 4-wk old brain revealed reductions in myelin proteins including MBP (myelin basic protein), CNP (2', 3'-cyclic nucleotide 3'-phosphodiesterase), PLP (phospholipoprotein), MAG (myelin-associated glycoprotein), MOG (myelin oligodendrocyte glycoprotein), and MOBP (myelin-associated oligodendrocyte basic protein) in Rheb1f/f, Nes-cre mice compared with controls (WT, Rheb1f/f or Rheb1+/+, Nes-cre) (Figures 4A and 4B). Similar reductions were evident in cortex, hippocampus and cerebellum. mRNA levels for these myelin proteins were also reduced in the Rheb1f/f, Nes-cre brain (Figure 4C). A reduction in the myelin proteins in the Rheb1f/f, Nes-cre was also noted in 2-wk old brain (Figure S4A), an early stage of myelination in the brain. Most Rheb1f/f, Nes-cre mice die when they are 5~ 6-wk old, making it impossible to examine myelination at later stages. Immunostaining of brain from 3-wk old Rheb1f/f, Nes-cre mice with anti-MBP antibody revealed wide spread loss of fine MBP reactive fibers in all cortical layers and in neuropil regions of hippocampus (Figure 4D). Black Gold (Savaskan et al., 2009) and Luxol Fast Blue (LFB) staining confirmed a reduction in myelination in regions with dense white matter tracts as well, e.g. the cingulum bundles and corpus callosum, striatum, and cerebellum (Figures 4E and 4F, ; Figures S4D and S4E). Electron microscopy analysis of the corpus callosum revealed a marked reduction in the number of myelinated axons (Figure 4G). However, the few remaining myelinated axons in the Rheb1f/f, Nes-cre brain exhibited comparable thickness of myelin sheath, as measured by g-ratio (data not shown). Myelination in the optic nerves of the Rheb1f/f, Nes-cre mice was similarly reduced as shown by Western blots and electron microscopy (Figures S4B and S4F).
Figure 4.
Rheb1 Regulates Postnatal Myelinaton in the Brain.
(A) Western blots show the reduction in myelin proteins in the 4-wk old Rheb1 KO brain. CTX: cortex; HIP: hippocampus; CB: cerebellum.
(B) Histograms show quantification of the reduction in myelin proteins in the cortex and hippocampus of 4-wk old Rheb1 KO brain. Data represent mean ± SEM. Results are averages of three independent experiments.
(C) Quantitative RT-PCR shows the reduction in mRNA of myelin genes in the cortex of the 3-wk old Rheb1 KO brain. Data represent mean ± SEM. Results are averages of three independent experiments.
(D) Immunostaining with MBP antibody shows wide-spread hypomyelination in the cortex and hippocampus of the 3-wk old Rheb1 KO brain. Bar, 100 μm.
(E) Black Gold staining shows hypomyelination in the cortex and corpus callosum of 3-wk old KO brain. Bar, 100 μm.
(F) LFB staining shows dramatic hypomyelination in the corpus callosum (CC) and striatum of 3-wk old Rheb1 KO brain. Arrows indicate the LFB positive areas in control and comparable areas in Rheb1 KO brain. Bar, 1mm. Insets show higher magnification of the boxed regions. Bar, 200 μm.
(G) Electron microscopy shows the reduction of myelinated axons of the corpus callosum in the 3-wk old Rheb1 KO. Unmyelinated axons are indicated by red asterisk. Bar, 500 nm.
Myelin is generated by mature oligodendrocytes that express APC (Adenomatous Polyposis Coli) or known as CC1 marker. The number of CC1+Olig2+ cells was reduced by 60 to 70% in multiple regions, including corpus callosum, cortex and hippocampus of Rheb1f/f, Nes-cre mice (Figures 5A and 5B; Figures S5A and S5B), suggesting that lack of mature oligodendrocytes could account for the hypomyelination in the Rheb1f/f, Nes-cre brain. The number of oligodendrocyte precursor cells (OPCs), which are identified as NG2+/PDGFRα+, increased in the cortex and hippocampus (Figures 5C and 5D and data not shown), but not in the corpus callosum (Figures 5C and 5D). NG2 protein expression increased in cortex, hippocampus and cerebellum (Figure 5E). The number of Olig2 cells (representing all oligodendrocyte lineage cells) is reduced in the Rheb1f/f, Nes-cre brain, particularly in the corpus callosum (50% reduction), less so in the cortex and hippocampus (Figures 5A and 5B; Figures S5A and S5B). Consistent with the reduction in the number of Olig2+ cells, we found that Olig2 protein level is reduced by 50% in the cortex and hippocampus (data not shown). While oligodendrocyte progenitor cells express Olig2, their increase in number appears not sufficient to make up for the loss of postmitotic oligodendrocytes that are Olig2- and CC1-double positive. These data indicate that hypomyelination in the Rheb1f/f, Nes-cre brain is linked to reduced numbers of mature oligodendrocytes that produce myelin proteins, but not due to a reduction of OPCs. It is possible that that the loss of Rheb1 affects the differentiation of OPCs into postmitotic, myelinating oligodendrocytes.
Figure 5.
Differentiation of OPCs into Postmitotic Oligodendrocytes is Impaired in the Rheb1 KO.
(A) and (B) Immunostaining shows the reduction in the number of postmitotic oligodendrocytes (CC1+Olig2+) in the corpus callosum of 2~3-wk old Rheb1 KO brain (n=3). Bar, 20 μm. Data represent mean ± SEM. **p<0.01.
(C) and (D) Immunostaining shows increased number of OPCs in the cortex, but normal number of OPCs in the corpus callosum of 3-wk old Rheb1 KO brain (n=3). Arrows indicate the NG2/PDGFRα double positive cells. Bar, 20 μm. Data represent mean ± SEM. **p<0.01.
(E) Western blots show increasing amount of NG2 proteins in the cortex, hippocampus, and cerebellum of 3~4-wk old Rheb1 KO mice.
Rheb1 Transgene Promotes Myelination
The observation of hypomyelination in the Rheb1f/f, Nes-cre brain raises an intriguing question whether increasing Rheb1/mTORC1 activity promotes myelination, or causes hypomyelination, as predicted by the hypomyelination noted in TSC1 or TSC2 KO mouse (Meikle et al., 2007; Way et al., 2009) and attributed to increased mTORC1 activity (Meikle et al., 2007). To answer this question, we generated a Cre recombinase-dependent conditional Rheb1 transgenic mouse, in which a myc-tagged mutant form of Rheb1 (S16H) was knocked into Rosa26 locus (Figures S6A and S6B). Rheb1 S16H is relatively resistant to TSC GTPase activating activity (Yan et al., 2006). In this Rosa26-Rheb1 S16H knockin mouse (Rheb1k/+, Nes-cre), the myc-tagged Rheb1 cDNA is preceded by a floxed transcriptional —stop signal and transcription of Rheb1 is dependent on removal of the —stop by cre- recombinase. The expression of Rheb1 S16H transgene was validated by anti-myc and anti-Rheb1 antibody (Figure S6C). The expression of Rheb1 S16H transgene increased mTORC1 activity and reduced mTORC2 activity (Figure S6D). Mice heterozygous or homozygous for Rheb1 S16H transgene (with the genotype of either k/+, Nes-cre or k/k, Nes-cre, respectively) are born with predicted Mendelian frequency and were viable through adulthood (>2 months).
The Rosa26-Rheb1k/k, Nes-cre mice exhibited enlarged brain size with increased brain weight, increased thickness of cortex and an increase in size of the soma of cortical neurons (layers 1 to 6) (Figure S6E to S6H). Expression of myelin proteins was prominently increased during early postnatal stage (P8 to P14) in cortex and hippocampus (Figures 6A, 6B and 6C; Figure S6I)., but changes in myelin protein expression were not observed in the young (4- to 6- wk old) and old (3-month old) adult mice (Figures 6A, 6B, 6C, and 6D; Figure S6J and S6K). This enhanced myelination is associated with an increase in the number of Olig2+/CC1+ mature oligodendrocytes at P7 (Figures 6E and 6F). By P14 there was no difference between Rheb1k/k, Nes-cre versus control (Rheb1k/k) (Figure 6G) suggesting enhanced mTOR1 signaling results in precocious myelination. Importantly, increasing Rheb1/mTORC1 did not cause hypomyelination in the brain. This point is further supported by electron microscopy revealing comparable myelination in the corpus callosum of 2-month old mice (Figure S6L).
Figure 6.
Rheb1 Transgene Promotes Myelination.
(A) Western blots show increasing amount of myelin proteins in the brain of 2-wk old Rheb1 transgenic (Tg, k/k, Nes-cre) mice relative to controls (k/k or k/+) and comparable amount of myelin proteins in the 3-month old Rheb1 Tg mice.
(B) and (C) Histograms show quantifications of myelin protein and mTORC1 activity in the cortex of Rheb1 Tg mice. Data represent mean ± SEM. Results are averages of three independent experiments.
(D) Immunostaining with MBP antibody shows enhanced myelination in the corpus callosum (CC), cingulum bundles (Cing) and cortex (CTX) at P7 and P14 and normal myelination at P28. Arrows indicate the MBP positive labeling. Bar, 20 μm.
(E) and (F) Immunostaining with CC1 and Olig2 antibody shows increased postmitotic oligodendrocytes in the 1-wk old Rheb1 Tg mice compared with controls (k/k; k/+). Arrows indicate the NG2/PDGFRα double positive cells. Bar, 50 μm. Data represent mean ± SEM. Results are averages of three independent experiments.
(G) Histogram shows comparable number of postmitotic oligodendrocytes in the 2-wk old Rheb1 Tg mice revealed by immunostaing. Data represent mean ± SEM. Results are averages of three independent experiments.
Rheb1 S16H Transgene Rescues mTORC1 Signaling and Hypomyelination in Rheb1f/f, Nes-cre Brain
We crossed Rheb1 S16H transgene onto Rheb1f/f, Nes-cre background and obtained mice with the genotype of Rheb1f/f, k/+, Nes-cre (—rescued Rheb1f/f, Nes-cre) mice. In these mice, Rheb1 transgene expression was detected, but not native Rheb1 protein using Rheb1 specific antibody (Figure 7A). The expression of a single copy of Rheb1 S16H transgene more than compensated the loss of native Rheb1 as indicated by higher pS6 and lower pAKT level than in the control (Rheb1f/f) (Figure 7A). Hypomyelination in the Rheb1f/f, Nes-cre mice was rescued as indicated by normal levels of myelin proteins and mRNA in the brain (Figures 7B and 7C), and MBP and Black Gold staining (Figures 7D and 7E). The number of CC1+ cells in the corpus callosum was also restored (Figure 7G). Consistent with the restoration of mTORC1 function in the Rheb1f/f, Nes-cre brain, the brain size and weight of the —rescued Rheb1f/f, Nes-cre mice was comparable to that in the control (f/+ or f/f, k/+) (Figures 7E and 7F). These findings verify that Rheb1 is the causal agent in regulating mTORC1 activity and myelin formation in brain.
Figure 7.
Conditional Expression of Rheb1 Transgene Restores mTORC1 activity and Myelin Formation in the Rheb1 KO Brain.
(A) Western blots show the expression of one copy of Rheb1 S16H transgene (indicated by an asterisk) is more than sufficient to restore pS6 level in the cortex of 4-wk old Rheb1 KO brain. The pAKT level in the rescued Rheb1 KO (Rheb1f/f, k/+, Nes-cre) brain is lower than that in the control.
(B) Western blots show restoration of myelin proteins in the cortex of 4-wk old rescued Rheb1 KO (f/f, k/+, Nes-cre) brain.
(C) Quantitative RT-PCR shows normal levels of mRNA of myelin genes in the cortex of 3~4-wk old, rescued Rheb1 KO (f/f, k/+, Nes-cre) mouse. Data represent mean ± SEM. Results are averages of four independent experiments.
(D) and (E) MBP and Black Gold staining show restored myelination in the hippocampus (D), cortex, corpus callosum and striatum (E) of the 6~10-wk old, rescued Rheb1 KO (f/f, k/+, Nes-cre). Bars, 100 μm in (D) and 1mm in (E).
(F) Histogram shows the weight of brain and body in the 3 to5 month old rescued Rheb1 KO (f/f, k/+, Nes-cre) is comparable to that of the controls. Data represent mean ± SEM.
(G) Immunostaining shows normal number of postmitotic oligodendrocytes. (CC1+Olig2+) in the corpus callosum of 6~10-wk old rescued Rheb1 KO (f/f, k/+, Nes-cre) brain. Bar, 20 μm. Data represent mean ± SEM. Results are averages of two independent experiments.
DISCUSSION
Rheb and mTor signaling
The present study uses genetic approaches to demonstrate that Rheb1 is necessary and sufficient for mTORC1 activation in vivo. Rheb1−/− MEFs do not respond to growth factor or amino acid stimulation to activate mTORC1, and mTORC1 activity was nearly absent in the brain of Rheb1f/f, Nes-cre mice. mTORC1 activity was restored to the normal level by the expression of a single copy of Rheb1(S16H) transgene (Figure 7). Demonstration of the role of Rheb1 in the activation of mTORC1 in vivo verifies earlier studies using alternative approaches in heterologous cells (Garami et al., 2003; Yang et al., 2006); for example, the knockdown of Rheb1 by siRNA reduces insulin stimulated activation of mTor/S6K/4E-BP1 signaling (Garami et al., 2003). The observation that Rheb1 is essential for amino acid dependent activation of mTORC1 is consistent with a recent report that amino acids promote the localization of mTor to a cellular compartment that contains Rheb1 (Sancak et al., 2010; Sancak et al., 2008). Together with the observation that amino acid stimulation increases the amount of GTP-bound Rheb in TSC2 mutant cells (Gulati and Thomas, 2007; Roccio et al., 2006), our data support the notion that amino acid sensing for mTORC1 occurs at a level upstream of TSC2. However, mTORC1 is also reported to be activated by amino acids in cells deficient for TSC1–TSC2 (Smith et al., 2005), suggesting that the nutrient input to mTORC1 may also occur independent of TSC1–TSC2. Future studies will need to resolve how these mechanisms are dependent on Rheb1.
Rheb1 is not required to activate mTORC2, consistent with the notion that mTORC1 and 2 are regulated by distinct mechanisms, as indicated by studies of Rictor or mLST8 mutant MEFs. In these MEFs, mTORC2 activity is apparently absent, however, mTORC1 activity can still be evoked by serum or insulin stimulation (Guertin et al., 2006). We note that Rheb1f/f, Nes-cre KO reduces the output of mTORC1 but increases mTORC2 (pAKT S473) and IRS1 or 2 (Figure 2B; Figure S2D). The upregulation of IRS1/2 may not fully account for the increased mTORC2 in the Rheb1f/f, Nes-cre brain, since S6K1 has been shown to phosphorylate Rictor at T1135 and results in a downregulation of mTORC2 activity (Dibble et al., 2009; Gao et al.; Julien et al., 2010; Treins et al.). Therefore, we envision that loss of S6K1 activity in Rheb1f/f, Nes-cre KO would release the negative effect of phosphorylation of phosphor-Rictor (T1135) on mTORC2 activity. Reciprocal adaptations occur in TSC1/2 KO (Harrington et al., 2004; Shah et al., 2004). Together, these observations suggest that the mTor signaling network is subject to homeostatic adaptations that may be important in understanding the pathogenesis of diseases that impact mTor signaling in vivo.
The basis of postnatal growth defect of Rheb1f/f, Nes-cre mice is likely due to mTORC1 signaling in neural tissues, since we found that mTORC1 signaling in hypothalamus was reduced (figure 2A), which may subsequently cause a defect in the growth hormone production/secretion from anterior pituitary. It is also possible that cre-activity is expressed in the pituitary stem cells (Gleiberman et al., 2008) and deletion of Rheb1 in these stem cells could affect the production/secretion of growth hormones. Nestin-cre has been reported to result in occasional excision of floxed genes in peripheral tissues (Betz et al., 1996) . But in view of the essential role for Rheb1 in embryonic development this is unlikely to underlie the highly penetrate growth defect in Rheb1f/f, Nes-cre mice. Additional studies that conditionally delete Rheb1 in selective populations of neurons will be informative
Our studies do not reveal the function of Rheb2. Although Rheb2 can activate mTor when over expressed in vitro (Tabancay et al., 2003), Rheb2 is not required for organismal viability, and is not essential for mTORC1 or mTORC2 signaling, or myelination (Figure S2E to S2G and S4C) in brain. Accordingly, alternative functions for Rheb2 should be considered.
Rheb1 and Embryonic Brain Development
Rheb1 is required for the mid-stage embryonic viability. This is consistent with the essential role of mTORC1 components mTor and Raptor in the early embryonic development (Gangloff et al., 2004; Guertin et al., 2006; Murakami et al., 2004) (Gangloff et al., 2004; Guertin et al., 2006; Murakami et al., 2004), and is not surprising in view of the many critical cellular functions such as cell growth, proliferation and differentiation ascribed to mTORC1 signaling. What is notable is that a complex organ such as brain can develop in the near absence of Rheb1/mTORC1 signaling. Nestin-cre activity is expressed as early as E10.5 and results in effective excision in all types of neural cells that populate the brain (Tronche et al., 1999). We have confirmed that Rheb1 is reduced by >90% by Western blotting with parallel reduction of mTORC1 signaling by E17.5 (Figure 3). At this developmental time, neural cells (neurons and glia) populating the brain are still being generated. This suggests that Rheb1/mTORC1 may not be essential for neural stem cells to differentiate into neurons, or for neurons to migrate and populate brain structures during embryonic development and to function sufficiently to support feeding and locomotion and survival of 5~6 weeks after birth. Since a reduction in mTor activity has been implicated in forebrain development in studies of the —flat-top mouse mutant created by ethyl-nitroso-urea (ENU) mutagenesis showing severe forebrain defect and lack of telecephalon (Hentges et al., 1999; Hentges et al., 2001). The contrast with the present observations may be related to the conditional deletion of Rheb or a role for mTORC2 in forebrain development.
Rheb1 and Postnatal Myelination in the Brain
Rheb1f/f, Nes-cre mice display a prominent defect in myelination during development. Previous studies have implicated mTor signaling in the regulation of oligodendrocyte differentiation and myelination. For example, inhibition of mTor signaling by rapamycin or siRNA for Raptor (mTORC1) or Rictor (mTORC2) inhibits the differentiation of cultured OPCs into postmitotic oligodendrocytes expressing myelin genes such as MBP and PLP (Tyler et al., 2009). Additionally, the differentiation of OPCs is arrested at the late progenitor stage (O4+) by rapamycin treatment and the general cellular viability is not affected during the course of rapamycin treatment, and chronic administration of rapamycin in vivo counteracts the effect of transgenic expression of active AKT on promoting myelination in adult mice (Narayanan et al., 2009). In the present study, effect of Rheb1 deletion on myelination is confirmed to be due to loss of Rheb1 protein by genetic rescue, and is likely dependent on mTORC1 since this is the only validated function of Rheb1. The reduction in the number of mature oligodendrocytes together with the increase in the number of OPCs suggests that Rheb1 is important in OPC differentiation. The Rosa-Rheb1 S16H transgenic mouse further supports this notion, since at P7, the total number of oligodendrocyte lineage cells (Olig2+) is not altered but the number of postmitotic oligodendrocytes (CC1+) is significantly increased (Figures 6E and 6F). The reduction in the number of CC1+ cells does not seem to be due to increased cell death; we did not detect any increase in cell death in 2-wk and 4-wk old Rheb1f/f, Nes-cre brain by TUNEL staining (data not shown). It is also possible that in the Rheb1f/f, Nes-cre brain, OPCs can still differentiate into postmitotic, non-myelinating oligodendrocytes that express APC and Olig2 at a reduced level and fail to mature into oligodendrocytes that are able to myelinate. While it is highly probable that Rheb1 loss in oligodendrocyte lineage cells is crucial for the impaired oligodendrocyte differentiation and myelin formation in the Rheb1 f/f, Nes-cre mice, it is likely that Rheb1 loss in neurons also contributes to some extent. This notion is supported by the recent report using neuronal SRF (serum response factor) knockout to show paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression (Stritt et al., 2009). The present genetic models will afford lineage and kinetic studies required to firmly establish how Rheb1 affects myelination in vivo.
Rosa26-Rheb1 S16H transgenic mice exhibit ~6 fold increase of mTORC1, which appears comparable to that reported in TSC KO mice (Way et al., 2009; Zeng et al., 2008), and share certain phenotypes related to increased imTORC1 activity similar to Pten knockout. All show increased brain size and enlargement of neuronal soma (Kwon et al., 2003; Tavazoie et al., 2005; Zeng et al., 2008), which is likely mediated by increased S6K1 (Pende et al., 2004). However, Rosa 26-Rheb1 S16H mice do not show hypomyelination in either adolescent or adult animals (Figure 6A to 6D; Figure S6I to S6L). This is surprising since TSC1 or 2 KO mice show hypomyelination that is thought to be due to increased mTOR since it is partially rescued by rapamycin (Meikle et al., 2008) In this study, rapamycin was administered for >5 wks, starting at early postnatal stage (P5 to P7) and did not inhibit myelination in control mice (Meikle et al., 2008). However, another study reported that rapamycin treatment of wild type P21 mice for 3 weeks caused hypomyelination (Narayanan et al., 2009). Our finding that Rosa-Rheb1 S16H transgenic mice do not exhibit hypomyelination suggests that mechanisms independent of Rheb1/mTORC1 may contribute to the myelin deficit caused by TSC1 or 2 mutation.
Concluding Remarks
We report the generation and characterization of Rheb1 and 2 knockout, and Rheb1 transgenic mice. We show that Rheb1, but not Rheb2, is required for mTORC1 activation in vivo, and neither Rheb1 nor 2 is essential for mTORC2 activation. While Rheb1-mediated mTORC1 activity is critical for the embryonic survival, it is not critical for the embryonic brain formation, since neurons, astrocytes and OPCs are formed in the absence of Rheb1. Instead, Rheb1 plays a critical role in the regulation of their subsequent cell-type specific adaptations, e.g. myelination during postnatal brain development.
EXPERIMENTAL PROCEDURES
Generation of Rheb1 and Rheb2 Knockout Mice
The exon 3 of Rheb1 is —floxed and the first two coding exons of Rheb2 are —floxed in the corresponding targeting vector. In both cases, the mutant protein lacks the N-terminal region that contains GTP binding domain that is critical for the normal functioning of Rheb. The Rheb1 and 2 gene targeting vectors illustrated in Figures S1A and S1D were constructed using standard molecular biology. The targeting vector was electroporated into embryonic stem (ES) cells and the transfected ES clones were selected for G418 resistance according to standard protocols. The G418-resistant ES clones with targeted homologous recombination were screened by PCR and further confirmed by Southern blotting (Figure S1B). Confirmed ES clones were injected into blastocyst mouse embryos to generate chimeric mice. The chimeric mice were crossed with C57BL/6 mice to validate germline transmission. The offspring with —floxed Rheb1 or 2 were crossed with Actin-cre or Nestin-Cre transgenic mice to generate germline or CNS-specific deletion of Rheb1 or 2. The genotype of the —floxed mice was determined by PCR on tail genomic DNA using DNA primers as follows: Rheb1 5’-GCC CAG AAC ATC TGT TCC AT-3’ and 5’-GGT ACC CAC AAC CTG ACA CC-3’ to amplify wt and 5’ floxed allele, wt: 650bp, floxed allele: 850bp; Rheb2 5’-TTT TGG TTT TCC TTG GC T TG-3’and 5’-GTG ATT CGA TGC ATT CAG TGA-3’, wt:570bp, floxed: 861bp). The Cre-mediated excision in Rheb1 and 2 knockout mice were confirmed by PCR amplification of genomic DNA using primers (5’-ATA GCT GGA GCC ACC AAC AC-3’, and 5’-GCC TCA GCT TCT CAA GCA AC -3’for Rheb1 to amplify a 750bp product; and 5’-TTT GGT TTT CCT TGG CTT GC GGG-3’ and 5’-TGC ACA CTG CTA ACA GCT CAA TGC-3’ for Rheb2 to amplify a 540bp product) and subsequently by amplification and sequencing of mutant Rheb1 and 2 transcripts. The primers for the amplication of Cre are 5’-TGC CAC GAC CAA GTG ACA GCA ATG -3’, and 5’-ACC AGA GAC GGA AAT CCA TGG CTC -3’ with the amplicon of 400bp.All mouse work was done in accordance with the Animal Care and Use Committee guidelines of Johns Hopkins University School of Medicine and Sichuan University West-China Hospital.
Generation of Conditional Rosa26-mycRheb1S16H Transgenic Mice
The targeting vector to knock a cassette that permits conditional expression of mycRheb1 S16H cDNA into Rosa26 locus was illustrated by the diagram (Figure S7H). It was made using components of the Rosa26 targeting system provided by Philippe Soriano (Soriano, 1999). The original PGK-neo sequence was replaced by PGK-EM7-neo expression cassette in plasmid PL452 (Liu et al., 2003) to allow kanamycin and neomycin selection in E.coli and ES cells, respectively. A CMV β-actin enhancer-promoter (Okada et al., 1999) was placed upstream of the —floxed PGK-EM7-neo-tPA sequence to drive Rheb1 cDNA transcription once the floxed tPA (transcriptional stop) is removed by Cre recombinase. This CMV- β-actin enhancer-promoter has been shown to be efficient in driving transgene expression when knocked into Rosa26 locus (Zong et al., 2005). The targeting vector was electroporated into ES cells and ES clones with targeted homologous recombination was identified by two sets of PCR primers coupled with sequencing of PCR products. Confirmed ES clones were injected into blastocyst mouse embryos to generate chimeric mice. The chimeric mice were crossed with C57BL/6 mice to validate germline transmission. The genotype of the transgenic mice was determined by PCR with the following primers to distinguish wt or knockin allele and Cre-mediated excision of the —stop signal: WTF1(forward): 5- GCA CTT GCT CTC CCA AAG TC-3’;WTR1(reverse): 5’-GCG GGA GAA ATG GAT ATG AA-3’) to amplify wt allele (596bp); FloxF (forward): 5’-GCA ACG TGC TGG TTA TTG TG-3’; FloxR(reverse):5’-GGG GAA CTT CCT GAC TAG GG-3’ to amplify the knockin allele (395bp); and ExF(Forward): 5’-CAG CCA TTG CCT TTT ATG GT-3’; ExR(reverse): 5’-ACC ACC ACC ACC ATT GAG AT-3’ to amplify an excision band of 609 bp, confirming the Cre-mediated excision of the —floxed PGK-neo and —stop signal.
Insulin and Amino Acid Stimulation
Primary MEFs routinely cultured with DMEM culture medium with 10% FBS were subjected to serum starvation (0.5% FBS) for 16 hours before insulin stimulation (200nM) for 15 minutes. Before amino acid stimulation, the serum-starved MEFs were washed once with dPBS (Dulbecco’s phosphate-buffered saline, Gibco) and then incubated with dPBS for 2 hours before adding a mixture of amino acids prepared with dPBS. The amino acid stimulation lasted for 1 hour before cells were harvested for Western blotting. The components of the amino acid mixture are as follows (mg/liter): L-Arg, 84; L-Cys, 48; L- Glu, 584; L-His, 42; L-Ile, 105; L-Leu, 105; L-Lys, 146; L-Met, 30; L-Phe, 66; L-Thr, 95; L-Trp, 16; L-Tyr, 72; L-Val, 94.
Western Blotting on Brain Lysates (Provided in SUPPLEMENTAL INFORMATION)
Transfection and Immunostaining
Primary MEFs were grown on glass coverslips with DMEM containing 10% FBS and were transfected with myc-tagged Rheb1 expression plasmid using calcium-phosphat-DNA co-precipitation method. Immunostaining was performed by standard procedure, as described in the SUPPLEMENTAL INFORMATION.
Tissue Processing, Immunohistochemistry and Electron Microscopy
Standard immuo-histochemical procedures were performed as detailed in the SUPPLEMENTAL INFORMATION. For quantification of immunopositive cells on tissue sections, three to six sections from each 3 mouse per genotype were quantified using Image-pro Plus software. The statistic analysis of the data--paired t test was performed using Graph-Pad Prism software. For electron microscopy, mice were perfused with 2% glutaraldehyde/2% paraformaldehyde in 0.1 M cacodylate buffer. Corpus callosum and optic nerves were dissected and postfixed overnight in the same fixative buffer. Tissues were then processed as standard procedure before photographing.
RNA Extraction and RT-PCR (Provided in SUPPLEMENTAL INFORMATION)
Supplementary Material
Acknowledgments
We thank Chip Hawkins, Holly Wellington, Johnisha Witherspoon, and Ann Lawler at the Transgenic Facility of the Johns Hopkins University School of Medicine for help with the generation of Rheb1 and 2 knockout and Rosa26-Rheb1 S16H transgenic mice and with the preparation of mouse embryonic fibroblasts. We also thank Liheng Guo of Sichuan University, Chengdu, for artistic work. This work was supported in part by grants from National Institutes of Health (MH068830-05 to P.F.W. and DA00266-36 to P.F.W.); the National Natural Science Foundation of China (Grant #30800388 to X.X. Du); and the National Basic Research Program of China (2004CB518800 to X. Z.); DOD TSC New Concept Award (W81XWH0510139, W81XWH0510106, and W81XWH0510200 to B.X.); and National 973 Basic Research Program of China (20009CB941400 to B.X.). We declare that we have no conflict of interest.
Footnotes
Supplemental Information includes seven figures and Supplemental Procedures and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERECES
- Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Betz UA, Vosshenrich CA, Rajewsky K, Muller W. Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr Biol. 1996;6:1307–1316. doi: 10.1016/s0960-9822(02)70717-3. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wan L, Inuzuka H, Berg AH, Tseng A, Zhai B, Shaik S, Bennett E, Tron AE, Gasser JA, et al. Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction. Mol Cell. 39:797–808. doi: 10.1016/j.molcel.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Gleiberman AS, Michurina T, Encinas JM, Roig JL, Krasnov P, Balordi F, Fishell G, Rosenfeld MG, Enikolopov G. Genetic approaches identify adult pituitary stem cells. Proc Natl Acad Sci U S A. 2008;105:6332–6337. doi: 10.1073/pnas.0801644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gulati P, Thomas G. Nutrient sensing in the mTOR/S6K1 signalling pathway. Biochem Soc Trans. 2007;35:236–238. doi: 10.1042/BST0350236. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges K, Thompson K, Peterson A. The flat-top gene is required for the expansion and regionalization of the telencephalic primordium. Development. 1999;126:1601–1609. doi: 10.1242/dev.126.8.1601. [DOI] [PubMed] [Google Scholar]
- Hentges KE, Sirry B, Gingeras AC, Sarbassov D, Sonenberg N, Sabatini D, Peterson AS. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc Natl Acad Sci U S A. 2001;98:13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. Embo J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Julien LA, Carriere A, Moreau J, Roux PP. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol. 2010;30:908–921. doi: 10.1128/MCB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Baker SJ. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci U S A. 2003;100:12923–12928. doi: 10.1073/pnas.2132711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005a;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005b;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- Meikle L, Pollizzi K, Egnor A, Kramvis I, Lane H, Sahin M, Kwiatkowski DJ. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci. 2008;28:5422–5432. doi: 10.1523/JNEUROSCI.0955-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan SP, Flores AI, Wang F, Macklin WB. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J Neurosci. 2009;29:6860–6870. doi: 10.1523/JNEUROSCI.0232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Lansford R, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridler K, Bullmore ET, De Vries PJ, Suckling J, Barker GJ, Meara SJ, Williams SC, Bolton PF. Widespread anatomical abnormalities of grey and white matter structure in tuberous sclerosis. Psychol Med. 2001;31:1437–1446. doi: 10.1017/s0033291701004561. [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–664. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- Rosner M, Fuchs C, Siegel N, Valli A, Hengstschlager M. Functional interaction of mammalian target of rapamycin complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet. 2009;18:3298–3310. doi: 10.1093/hmg/ddp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005a;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005b;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Savaskan NE, Weinmann O, Heimrich B, Eyupoglu IY. High resolution neurochemical gold staining method for myelin in peripheral and central nervous system at the light- and electron-microscopic level. Cell Tissue Res. 2009;337:213–221. doi: 10.1007/s00441-009-0815-9. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Stritt C, Stern S, Harting K, Manke T, Sinske D, Schwarz H, Vingron M, Nordheim A, Knoll B. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat Neurosci. 2009;12:418–427. doi: 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- Tabancay AP, Jr, Gau CL, Machado IM, Uhlmann EJ, Gutmann DH, Guo L, Tamanoi F. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J Biol Chem. 2003;278:39921–39930. doi: 10.1074/jbc.M306553200. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- Treins C, Warne PH, Magnuson MA, Pende M, Downward J. Rictor is a novel target of p70 S6 kinase-1. Oncogene. 29:1003–1016. doi: 10.1038/onc.2009.401. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tyler WA, Gangoli N, Gokina P, Kim HA, Covey M, Levison SW, Wood TL. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci. 2009;29:6367–6378. doi: 10.1523/JNEUROSCI.0234-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Way SW, McKenna J, 3rd, Mietzsch U, Reith RM, Wu HC, Gambello MJ. Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum Mol Genet. 2009;18:1252–1265. doi: 10.1093/hmg/ddp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- Yan L, Findlay GM, Jones R, Procter J, Cao Y, Lamb RF. Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J Biol Chem. 2006;281:19793–19797. doi: 10.1074/jbc.C600028200. [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Kim E, Guan KL. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci U S A. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LH, Xu L, Gutmann DH, Wong M. Rapamycin prevents epilepsy in a mouse model of tuberous sclerosis complex. Ann Neurol. 2008;63:444–453. doi: 10.1002/ana.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.