Abstract
Each viral particle of HIV-1, the infectious agent of AIDS, contains two copies of the full-length viral genomic RNA. Encapsidating two copies of genomic RNA is one of the characteristics of the retrovirus family. The two RNA molecules are both positive-sense and often identical; furthermore, each RNA encodes the full complement of genetic information required for viral replication. The two strands of RNA are intricately entwined within the core of the mature infectious virus as a ribonuclear complex with the viral proteins, including nucleocapsid. Multiple steps in the biogenesis of the genomic full-length RNA are involved in achieving this location and dimeric state. The viral sequences and proteins involved in the process of RNA dimerization, both for the initial interstrand contact and subsequent steps that result in the condensed, stable conformation of the genomic RNA, are outlined in this review. In addition, the impact of the dimeric state of HIV-1 viral RNA is discussed with respect to its importance in efficient viral replication and, consequently, the potential development of antiviral strategies designed to disrupt the formation of dimeric RNA.
Keywords: HIV-1, Genome, RNA, Dimerization, Recombination, Maturation
Introduction
During the replication cycle of HIV-1, its genetic information progresses through two types of nucleic acid: DNA and RNA. The extracellular virion contains two copies of the HIV-1 genome in RNA form, which, upon entering a new cell, act as the template for the production of a DNA copy through the process of reverse transcription. The nascent DNA is integrated into the host cell genome to form the provirus, which serves, in turn, as a template for the production of multiple copies of the RNA genome. Two of the full-length, 9-kb RNA genomes are subsequently packaged into each budding virus particle to complete the cycle of replication. Thus, HIV-1 exists both as a stable DNA element within the infected cell’s genome, to be replicated along with the cell using host machinery, and a mobile RNA element capable of transmission to uninfected cells. The fate and conformation of the HIV-1 virion RNA genome are the focus of this review.
Early during HIV-1 infection, most of the nascent RNA is multiply spliced within the nucleus to yield a number of different species. These sub-genomic RNA are constitutively exported from the nucleus via the NXF1/NXT-mediated transport pathway to serve as the templates for the translation of the viral accessory proteins Tat, Rev, and Nef. Both the full-length and singly spliced products (used for expression of Vif, Vpr, Vpu, and Env) are incapable of exiting the nucleus using this pathway, due, in part, to the presence of unspliced intronic sequences and through the presence of HIV-1-specific inhibitory sequences within these RNA molecules16,94. To overcome these blocks to nuclear export, the viral protein Rev specifically binds a complex motif only present in the full-length and singly spliced RNA, called the Rev response element16,84. Rev bridges the Rev response element to the chromosome region maintenance 1 nuclear export factor, which directs these RNA out of the nucleus71. Within the cytoplasm, the full-length genomic RNA has the potential to act as the mRNA for translation of the viral structural proteins Gag and Gag-Pol, for which the RNA must traffic to the correct site for the translated products to function properly105,106. Alternatively, the full-length RNA may be packaged into a budding virion to act as the genomic template for the next round of infection. In contrast to murine leukemia virus, which has two separate pools of RNA, one for translation and the other for RNA packaging55,57, HIV-1 genomic RNA has the potential to perform both functions9,20. The cellular and viral factors involved in the regulation of translation and packaging of full-length genomic RNA are relatively poorly understood, though their correct coordination is vital to the successful replication of HIV-1 and are potentially regulated by RNA dimerization.
The two RNA present within each HIV-1 virion are positive-sense, single-stranded copies of the provirus from the R region of the 5′ long terminal repeat to the R region of the 3′ long terminal repeat. As such, both RNA contain sufficient genetic information to initiate nascent infections alone. However, there appears to be a requirement to package two copies of RNA for the successful completion of the retroviral replication cycle. The two strands of genomic RNA are not independent within the virion, but instead are found tightly associated as a compact dimer. The sequence requirements, firstly for the initial contacts, subsequently for those that stabilize the interaction, will be discussed, as will the potential involvement of viral proteins in the dimerization process.
HIV-1 virions contain dimeric RNA
The dimeric nature of retroviruses was suggested early on by results showing that retroviral particles contain RNA of twice the expected buoyant density compared with a single copy of the full-length viral genome86. Furthermore, the RNA could easily be resolved to the correct monomeric buoyant density upon heat treatment21. Similarly, when analyzed by non-denaturing electrophoresis, the viral RNA runs as a single band (proposed dimer), which upon heat denaturation alters its migration pattern to a faster-moving species (proposed monomer)48. Electron microscopy (EM) has also been used to determine the conformation of the genomic RNA. Early EM work performed on RNA isolated from an endogenous feline retrovirus identified dimers held together at one end; this conformation was also termed rabbit ears or Y-shaped50. Further EM studies, using SV40 DNA circles linked to poly(A) probes, demonstrated that the dimeric RNA was oriented with two free 3′ ends6. Therefore, the interface between the two molecules was located within the two 5′ ends, a region termed the dimer linkage structure (DLS). Much of this early work used gamma retroviruses and was performed prior to the identification of HIV-1 as the causative agent of AIDS. Subsequently, studies using HIV-1 RNA have demonstrated similar features13. However, studies from two different groups have noted that the RNA isolated from HIV-1 virions does not form a Y-shape; instead, a small loop region at the 5′ end was detected, suggesting two major interactions within this region4,38. It is worth noting that the morphology of viral RNA evaluated by EM was highly dependent on the stringency of the RNA treatment conditions prior to the analysis. A reduction in the stringency (from 50% formamide, 2.5 M urea to 40% formamide, 2 M urea) lead to tight coils of RNA that were too condensed for structural evaluation, and a slight increase (up to 60% formamide, 3 M urea) resulted in the majority of RNA being visualized as monomers38. Therefore, the morphology observed by EM reveals important RNA contact points, probably the most stable ones. However, it is likely that other interactions exist that are less stable and, therefore, not observed by the EM analyses35,76. The same caveat probably also applies to most biochemical RNA analyses, such as native gel electrophoresis, during which weak interactions of potential biological relevance may not survive.
Points of contact: RNA element(s) involved in dimerization
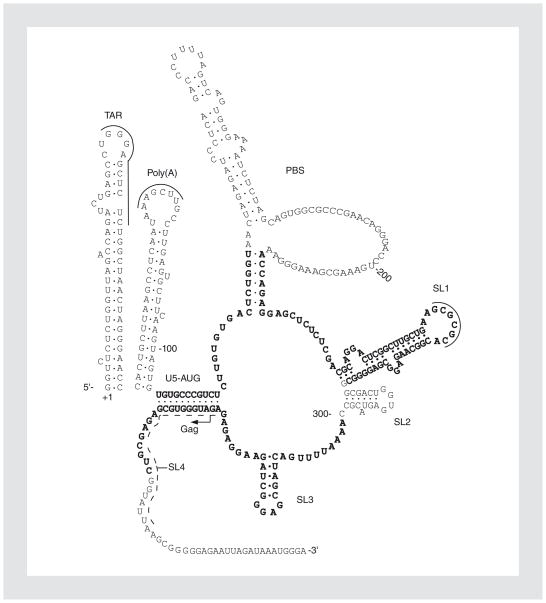
Many studies have attempted to identify the regions of the RNA molecule that are responsible for the observed RNA-RNA interaction(s). The EM structural data pointed to the main sequences involved being within the 5′ end of the genome, namely the DLS. Using in vitro-transcribed short RNA corresponding to various lengths of HIV-1 genomic RNA, it was demonstrated that the DLS included the first 311 nucleotides61, although downstream bases (311–415) had previously been thought sufficient19. The 5′ untranslated region of the viral genome, which includes the DLS, is predicted to fold into a highly ordered secondary structure that has been experimentally modeled by computational, phylogenetic, biochemical, and mutational probing studies18,45,78. Although minor variations exist, the consensus structure involves six hairpin stem loops (SL), each with their own functional or alphanumerical designation (Fig. 1). A seventh hairpin, containing the AUG start codon of Gag, was originally proposed as SL4 (Fig. 1, sequences intermittently underlined), but the weight of data suggests that this region is linear and involved in interactions with the junction between the poly(A) and primer binding site hairpins1,18. All six proposed hairpins play vital roles during HIV-1 replication, including, but not limited to, transcriptional regulation (transactivation response element or TAR), reverse transcription (primer binding site), dimerization (SL1), splicing of the full-length RNA (SL2), and genomic packaging (SL3).
Figure 1.
RNA sequences and conformation at the 5′ end of the HIV-1 genome. Nucleotide sequences are based on the HIV-1HXB2 RNA transcript and the first base is marked +1; nt 100, 200, and 300 are marked. The palindromic sequences proposed to pair with the counterpart palindrome of the RNA partner are marked by solid lines, whereas sequences in the originally proposed SL4 are marked by dashes. The refined dimer linkage structure sequence by Suguraki, et al.92 is shown in boldface. TAR: transactivation response element; PBS: primer binding site; SL: stem loop.
After the initial contact, the two RNA molecules go through a number of conformational alterations that enhance the stability of the dimer and condense it into that which is extracted from isolated infectious particles. In this review, we have divided the two processes into initiation and maturation; one section details the requirements for the initial RNA-RNA contact and the second section describes the sequential stabilization of the preformed dimer into its final conformation.
Initiation of RNA dimerization
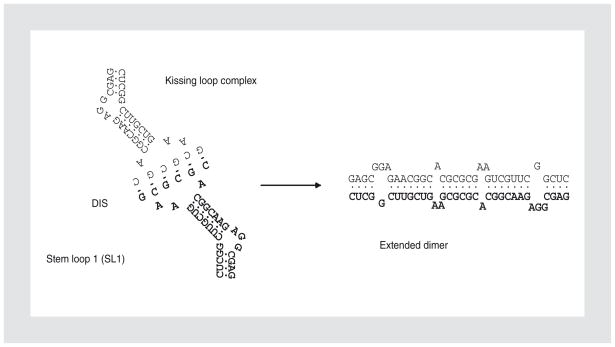
Within the identified DLS, a single stem-loop structure (SL1) appeared to be responsible for the majority of the dimerization potential of the full-length HIV genomic RNA81. The SL1 is a highly evolutionarily conserved hairpin from nt 236 to nt 282 (based on numbering with respect to the cap site of HIV-1HXB2 genomic transcripts), containing two internal bulges and an apical loop consisting of nine bases, of which six form a palindrome. Deletion of this motif prevents in vitro dimerization of the nt 1–707 fragment of HIV-1 genomic RNA102, and antisense oligonucleotides targeting the palindrome also prevent dimerization of sequences in this region70. Thus, this motif is widely accepted as the dimerization initiation site (DIS). The palindromic nature of the DIS allows it to form classic Watson-Crick base pairs with another RNA containing the same DIS sequence in an antiparallel manner (Fig. 2)13,69. This interaction, the result of which is termed the “kissing loop” dimer, has been extensively studied in vitro and its structure has been solved using nuclear magnetic resonance and X-ray crystallographic techniques25,49,65. The purine residues of the loop, surrounding the palindromic sequence, are also essential to the dimerization process82. The molecular structure predicted by nuclear magnetic resonance suggests that these bases form noncanonical interactions82. In support of this suggestion, the identities of these bases have profound effects on the stability of the kissing loop complex60. Also, the bulge halfway up SL1 is metastable, a property that is important for the capacity of SL1 to form dimers and their resulting stability33,99.
Figure 2.
Proposed interactions between stem loop 1 (SL1) of two RNA molecules for dimerization. The palindromic sequences in two dimerization initiation site (DIS) regions form base-pairing to generate a kissing loop complex. Base-pairing could proceed down the SL1 stem to form the extended dimer.
Genetic evidence indicating that genomic RNA from HIV-1 strains with the same DIS are co-packaged more efficiently than when their DIS sequences are incompatible, further supports the role of the DIS in the initiation of RNA dimerization11,64. Interestingly, it would appear that not all palindromes are equally capable of performing this function; most of the circulating strains of HIV-1 contain one of two DIS sequences: GCGCGC (subtypes B and D) or GUGCAC (all other subtypes, group N, and group O)104. Cell-based experiments indicated that only a few other palindromes can successfully substitute the two canonical sequences53. Moreover, the size of the palindrome is constrained to 6 nt, with numerical alterations quickly reverting to wild-type7. The highly restricted nature of the DIS, when considering there are a possible 64 palindromes of six nucleotides, indicates either that the dynamics of the kissing loop dimerization process are very specific or that other properties of the DIS, outside of its role in dimerization, are sequence specific77. Alternatively, the DIS may simply be evolutionarily constrained due to the nature of its role in virus replication; a single change within the DIS would destroy its palindromic nature and decrease dimerization efficiency. If, by chance, multiple mutations did occur to generate a different palindrome at the DIS, the mutated virus would dimerize less efficiently with the parent virus11. This finding suggests an interesting hypothesis that the DIS is not only the dimerization initiation site, but also could have been pivotal in initiating divergence of some of the subtypes.
Although the DIS is clearly required for efficient dimerization, it is not sufficient in vivo62. The originally identified DLS encompassing bases 1–311, and including the DIS, has since been refined. By inserting a second ectopic DLS downstream of the original sequence, Sakaguri, et al. generated a virus with two DLS regions that produced a subset of virions with apparent monomeric genomic RNA, presumably due to the substitution of intermolecular DLS interaction(s) with an intramolecular one(s)91. By mutating the ectopic DLS region and monitoring the generation of monomeric RNA in the released virions, they further characterized the sequence requirements of the DLS to a noncontiguous 144 nt region, consisting of sequences from the junction between the R/U5 and U5/L stem loops to the end of the putative SL4 – more specifically, sequences 105–131, 217–281, and 301–352 (Fig. 1, sequences in bold). In agreement with this study, mutations within these sequences had previously been shown to be deleterious to the dimerization of HIV-1 genomic RNA87,88. Currently, the roles of these sequences in the RNA interactions are not well defined. It is possible that they are not directly involved in the interstrand contacts, but are required to ensure correct orientation and stability of SL1 to allow the DIS to interact with its counterpart in another RNA62.
Although the important role of the DIS as the central element for initiation of RNA-RNA contact has been well demonstrated, one major caveat is that HIV-1 can replicate without the DIS, or even without the entire SL1, albeit at reduced levels in most cell types36,104. Furthermore, the viral RNA extracted from such DIS mutants is dimeric, even though the dimers display heterogeneous migration by native gel electrophoresis7,64,103. Interestingly, when DIS mutant viruses were placed in long-term culture, the compensatory mutations that arose were within Gag and resulted in an improved ability of Gag to distinguish full-length viral RNA from spliced RNA58,59,90. This observation suggests that the DIS is not required for replication per se. Rather, it enhances dimerization and thereby potentially allows for better recognition of the genomic RNA species by Gag, whereas, in the absence of the DIS, dimerization can still occur, but with reduced kinetics and presumably through currently unidentified redundant mechanisms64,103.
Other RNA elements that may contribute to the stabilization of the RNA dimer
Using short RNA containing the SL1 motif, in vitro experiments demonstrated that after the initial contact at the DIS to generate the kissing loop form, under certain conditions base-pairing could progress down the stems of the SL1 hairpin and convert intramolecular bonds into intermolecular ones, thereby forming an extended dimer with greater thermodynamic stability than the kissing loop form (Fig. 2)67,68. Although it is an attractive model, and both kissing loop and extended dimer structures have been solved using nuclear magnetic resonance and X-ray crystallography25,32,66, to date there is no direct evidence that supports the proposed extension from the kissing loop to the extended dimer in vivo109. Biochemical probing of the area is incapable of distinguishing between inter- and intramolecular interactions, as they are both hypothetically in the same local environment. Additionally, viral mutations designed to prevent extension of the kissing loop dimer to the extended dimer by swapping or inverting the stem of SL1 have no observable impact on replication or dimerization12,64. Moreover, there is conflicting in vitro data as to whether longer HIV-1 RNA can carry out this conformational switch80.
RNA elements outside the SL1 have also been proposed to be involved in the stabilization of the RNA dimer93. For example, a ~ 100 nt region downstream of the splice donor site in SL2 has been shown to enhance the stability of dimeric RNA, albeit in a DIS sequence-dependent manner51,82. In addition, the aforementioned EM studies of the HIV-1 genomic RNA dimer indicated that, under the isolation conditions used, at least two major points of contact existed between the two strands4,38. Although both groups point to the DIS as the 3′ component of these interactions, they differ in their interpretation of the 5′ interaction. Hoglund, et al. proposed that the 5′ interaction involves a palindrome at the apical loop of the R-U5 stem loop, whereas Andersen, et al. hypothesized that it involves the two TAR elements, which also contain a palindromic sequence (Fig. 1, palindromic sequences are underlined). The R-U5 region has since been shown not to affect dimerization, but instead to be involved in a long-distance interaction with sequences encoding the matrix domain52,79. Also, the palindrome of the TAR element is partially occluded, reducing its potential to interact in the proposed manner. However, it has been demonstrated that this palindrome can become available for a kissing loop-type interaction after incubation of the RNA with nucleocapsid4. Although it is more than likely that other intermolecular contact points exist, not all of their identities, molecular interactions, and contributions to the formation and stability of the mature dimer are known at this time.
Protein elements involved in RNA dimerization
Viral proteins play important roles in the stabilization (maturation) of the viral RNA dimer. During or soon after virus assembly, the viral-encoded protease cleaves the Gag and Gag-Pol polyproteins to generate the mature products. These proteolytic events are also linked with an alteration in viral morphology, including the appearance of a cone-shaped core in a process referred to as virus maturation3,5,44,83,108. The dimeric RNA also undergoes changes during this process. The RNA isolated from immature viral particles, either recently released (rapid-harvest) virions or protease defective (PR−) mutant viruses, are different from RNA from infectious mature virions30,31. Mature viral RNA are found mostly as dimers, which are fairly thermostable. In contrast, a majority of the RNA isolated from immature viruses migrate as monomers; the remaining dimers migrate more slowly and dissociate at much lower temperatures compared with RNA extracted from mature virions30. This observation led to the proposed model that immature virions contain loose dimers that are induced to mature through the action of nucleocapsid, released during the virion maturation process. By integrating the in vitro RNA data into this model, it was proposed that the loose dimer corresponds to the interaction at the DIS (kissing loop dimer) and the mature dimer consists of nucleocapsid-mediated extension of the base pairing down the stem of SL1 (extended dimer).
However, recent studies of rapid-harvest viruses indicated that the maturation of RNA dimers is more complicated than previously envisioned103. It was shown that although RNA samples isolated from PR− viruses or rapid-harvest wild-type viruses both contain slow-moving weak dimers and monomers, dimeric RNA was not observed in RNA isolated from rapid-harvested PR− viruses103. Instead, the characteristic immature dimers of PR− virus were only detected after incubation of the rapid-harvested PR− virions. Additionally, after incubation of the rapid-harvest wild-type viral RNA, an intermediate dimer conformation was identified, which migrated between that of the loose immature dimer and the stable, condensed, mature RNA. Based on these and other observations, the original model was modified and it was postulated that RNA dimerization undergoes three distinct steps: first, the dimer is initiated through the DIS interaction (kissing loop form); proteolytic cleavage of the viral proteins then induces the kissing loop form to be extended into the extended dimer form; and finally, other RNA contact points are established to achieve a fully matured dimer. Furthermore, in the absence of viral maturation, the immature dimers that eventually form within PR− viruses do so in a DIS and proteolytic cleavage independent manner.
The mature products of proteolytic cleavage of the Gag and Gag-Pol polyproteins are likely to be important in the stabilization of viral RNA dimers. Nucleocapsid has both RNA binding and chaperone activities56,85. Many studies have demonstrated the effects of nucleocapsid on the conformation of RNA in vitro27,47,67,68; for example, it was shown that nucleocapsid can induce the conformational switch from the kissing loop dimer to the extended dimer within short, in vitro-transcribed RNA containing the SL168. Moreover, mutations in Gag that interfere with poly-protein processing and prevent the release of nucleocapsid have a severe impact on viral RNA stability37,98. The form of nucleocapsid (NC) released by the initial cleavage event is NCp15. This protein coats the RNA at about 1 molecule per 7 nt110. Subsequent cleavage, enhanced by the presence of RNA, of NCp15 into NCp9 and finally into NCp7 by the successive removal of p6, then p1, results in further condensation of the genomic RNA, presumably due to a greater chaperone capacity of the NCp7 protein over NCp15, and potentially due to bridging of the RNA through multiple RNA contacts of NCp7 and NCp7-NCp7 protein interactions28,37,63,98,100. The temporal regulation of the cleavages that result in NCp7 was proposed to be the mechanism driving the distinct stages of the RNA maturation process. The evolution of protease inhibitor resistance mutations within the p1-p6 cleavage site clearly reinforces the viral requirement for the processing of NCp15 for viral replication15,72. However, proteolytic processing of Gag and Gag-Pol plays a more complex role in RNA maturation than simply unleashing a chaperone protein as the Gag polyprotein itself has chaperone activity26. Moreover, RNA maturation defects can also be caused by perturbation of the protein composition of the virions, such as altering the Gag/Gag-Pol ratio or by removing reverse transcriptase and integrase products96,97. Additionally, in a late-domain mutant for which few defects in Gag processing can be detected and, therefore, nucleocapsid is released, the dimeric RNA again have lower stability29. Interestingly, the maturation of the RNA dimer appears to be linked to the morphological maturation of the virion itself. In the aforementioned late-domain mutant and in other Gag processing mutants, an inability to mature the virion is correlated to a reduction in the stability of the genomic RNA dimer29,98. Although the detailed molecular mechanisms are unclear, it is likely that the formation of the proper core facilitates the complete maturation of the RNA dimer.
Another viral protein with chaperone activity found within HIV-1 virions is the viral infectivity factor, or Vif34,43. Recent work has highlighted the major function of Vif as a viral defense protein against the host-encoded antiviral APOBEC 3G and 3F proteins95. However, there appear to be Vif-binding sites within the 5′ untranslated region of HIV-146, and Vif has been proposed to aid reverse transcription by reducing reverse transcriptase pausing8,34,111. Using in vitro assays, it has been shown that the presence of Vif also has an impact on the formation of dimers, although the effects of its binding to the viral RNA appear to contrast those of nucleocapsid binding34.
Host factors have also been proposed to play a role in RNA dimerization. The replication defect of SL1 mutant viruses within established T-cell or fibroblastic cell lines has been extensively cataloged7,12,51,53,77. However, in contrast to these studies, SL1 was deemed dispensable for replication in primary peripheral blood mononuclear cells36. Although a direct role has not been ascribed, nor a candidate factor identified, the cell type-dependent effect of the SL1-deletion mutant suggests that a host factor, missing in some cell lines, is capable of substituting for the role of SL1 in the replication of HIV-1.
RNA elements proposed to be involved in regulating dimerization
Full-length genomic RNA can serve as a template for the translation of viral proteins or can be packaged as the genomic material in the virion. The regulation of these two functions is difficult to tease apart, particularly as it has been shown that HIV-1 RNA does not have to be translated to be packaged and that the RNA used for translation and packaging originate from the same pool9,10,20,73. A very appealing model, invoking different conformations of the 5′ untranslated region, has been proposed by which HIV-1 could regulate the fate of its genomic RNA40. In this model, the first conformer, long-distance interaction (LDI), has the DIS occluded within a stable stem and, therefore, not capable of kissing loop interactions. In contrast, the alternative conformer, branched multiple hairpins (BMH), adopts the more widely accepted arrangement of the 5′ untranslated region with separate hairpins (Fig. 1). The BMH conformation has SL1 presenting the DIS for interaction with its dimer partner and SL3 remaining intact to drive the Gag-RNA interaction needed for packaging1. Thus, two interchangeable conformations have been proposed, with the LDI potentially more amenable to translation and the BMH having a propensity to dimerize and be packaged. The model posits that the RNA initially adopts the LDI conformer, allowing Gag to be expressed. Once sufficient levels of Gag have accumulated, they bind the RNA, promoting it to adopt the BMH conformation, thereby promoting dimerization and packaging40,74. Although this model is very attractive and is supported by in vitro data, it lacks support from in vivo results. Recent evidence has shown that both conformations are equally capable of translation2; furthermore, chemical structural probing analyses suggest that both cellular and virion HIV-1 RNA adopt conformations similar to that of BMH78. However, sub-genomic RNA may still adopt the LDI conformation, which could be a distinguishing factor between the genomic and sub-genomic RNA54. All HIV-1 RNA species contain the DIS motif; thus, the use of the LDI conformation would be an ideal mechanism to prevent sub-genomic RNA from becoming dimerized. It has been proposed that RNA dimerization occurs prior to packaging; therefore, preventing sub-genomic RNA from dimerizing would provide a mechanism to exclude these RNA from the virion. Contrary to this argument, a recent study showed that sub-genomic HIV-1 RNA are capable of homodimerization as well as heterodimerization with genomic RNA in vitro101. Thus, the DIS within the spliced RNA must be accessible, reducing the likelihood that the LDI conformation exists in vivo.
Selection of co-packaged RNA: does Gag package two monomers or one dimer?
The molecular mechanisms that govern the encapsidation of two copies of HIV-1 RNA into one virion remain unclear89. It is possible for the viral proteins to package one dimeric RNA or, alternatively, two monomeric RNA molecules. Upon actinomycin D treatment, murine leukemia virus-infected cells were shown to produce particles largely without viral genomes; however, the viral RNA molecules packaged by the remaining viruses were dimeric55. Additionally, experimental evidence using rapid-harvest or PR− murine leukemia virus indicated that these RNA are present as loose dimers, again suggesting that dimeric RNA are packaged31. However, recent evidence from rapid harvest PR− HIV-1 suggests that only monomers are present within nascent viral particles103. Other studies in which viral proteins were altered in different manners have also made the similar observation that genomic RNA can be found within the virions as monomers22,96. Although the observation of monomers in virions appears to favor the hypothesis that HIV-1 packages RNA as two monomers, the possibility exists that during the viral RNA isolation procedure, weak dimeric RNA (such as a kissing loop dimer) became dissociated30. In support of this idea, genetic studies have demonstrated that the rate of co-packaging can be modified, a result that indicates dimer partner selection prior to the commitment of the two RNA to be co-packaged. These studies showed that recombination between subtype B and subtype C HIV-1 is significantly lower than intra-subtype recombination11. Additionally, the lower recombination rate is largely restored by matching the DIS of the two viruses. Recombination occurs between the co-packaged RNA of the two viruses39 and, as such, provides a measure of the rate of dimerization between the two RNA. Therefore, these results suggest that the RNA from these two virus subtypes, which contain two different DIS sequences, were co-packaged inefficiently. Furthermore, the recombination rate between two viruses can be significantly increased by modifying their respective DIS sequences to be non-palindromic and, therefore, non-self-associating, while maintaining their complementarity to each other64. In both situations, the assortment of RNA into the released virions was not random, suggesting that RNA partner selection occurred prior to packaging, and that dimeric RNA were packaged.
RNA dimerization as a target for antiviral therapy
RNA dimerization appears to be a requirement for the production of infectious HIV-1 particles. The well-established role of the DIS in the RNA dimerization process makes it an attractive target for the development of antiviral therapies. There are currently two avenues under investigation for inhibition of HIV-1 replication through disruption of the dimerization process. One involves the use of antisense nucleic acid decoys and the other exploits the sequence and conformational similarity between the kissing loop complex of HIV-1 and the natural target of aminoglycoside antibiotics. Antisense oligonucleotides bind their target through the nature of their complementarity and, in doing so, either prevent translation of the RNA by disrupting ribosomal read-through or induce degradation of the target RNA through an RNaseH-mediated pathway. The success of this strategy relies on the accessibility of the target, the efficiency of the binding, and the abundance of the oligonucleotides. Antisense oligonucleotides have been used to target the DIS and HIV-1 dimerization. Although efficient inhibition of dimerization in vitro and translation of Gag in vivo were both demonstrated using this strategy23,41,42,75, the molecules only had a modest effect on the replication capacity of HIV-123. The alternative strategy relies on the discovery that the crystal structure of the HIV-1 kissing loop complex has some unexpected similarities to the bacterial 16S ribosomal A site24, the target of the aminoglycoside family of antibiotics. This revelation encouraged the development of aminoglycosides as potential antiviral compounds. Neomycin and lividomycin, among others, have been shown to specifically bind a kissing loop complex between two GUGCAC palindrome-containing SL1 hairpins. The binding of the aminoglycosides to the kissing loop complex occurs with high affinity and results in an increase in the stability of the kissing loop domain and inhibition of conversion of the kissing loop complex to the extended dimer in vitro24. As a role for, or presence of, the conversion of kissing loop complex to extended dimer in vivo is still unclear, the success of this strategy has yet to be demonstrated, particularly because no inhibitory effect of aminoglycosides on HIV-1 replication has been demonstrated.
Despite these limited initial successes, the HIV-1 DIS does have the characteristics of a promising drug target: it plays an important role in viral replication, and it is conserved, which is indicative of a major evolutionary constraint. Currently, the development of antiviral strategies targeting the DIS is in its infancy and there are many hurdles to be overcome before these strategies can be successful. One major factor is that some DIS and SL1 mutants can replicate and still package dimeric RNA into their particles, suggesting that viable escape mutants may be easily generated when using antiviral strategies targeting this motif. Understanding the putative redundant dimerization pathways and why HIV-1 needs to have a dimeric RNA genome for replication could pave the way for the successful implementation of such strategies.
Role for RNA dimerization: a question of why?
The Retroviridae is the only virus family that packages two copies of the complete viral genome into one virion. Several selective advantages have been proposed for the packaging of two RNA. It has been postulated that viral RNA is sometimes damaged prior to the initiation of reverse transcription. Packaging two copies of the viral genome would allow the reverse transcriptase complex to switch templates when encountering a break in the viral RNA, and thereby complete DNA synthesis and rescue the genetic information, despite the damage to the RNA genome. This is the basis of the copy-choice model of retroviral recombination14. It has also been proposed that the ability of the virus to package two RNA molecules and to switch RNA templates during reverse transcription allows frequent recombination in order to increase genetic diversity107. When two viruses with different genotypes infect the same cell and produce heterozygous virions, there is the potential for recombination between them to result in a mixing of the genetic materials and the creation of a new hybrid genotype. In essence, this allows the virus to replicate sexually107. The dimeric nature of the RNA genome may also play a structural role during replication. Analyses of the murine leukemia virus RNA-nucleocapsid interactions suggest that a major binding site within the RNA is exposed upon RNA dimerization17. Therefore, RNA dimerization can be used by the virus to generate structural components that regulate specific stages of replication. Finally, it is likely that the dimeric nature of the viral RNA genome provides not just one but all of these selective advantages, and possibly others that are not currently identified.
Conclusions
The dimerization of genomic RNA appears to be essential for successful replication of HIV-1. The two RNA initially make contact at the DIS, a 6 nt palindromic sequence within the 5′ untranslated region. The two RNA then undergo conformational alterations in the virion that result in a condensed stable dimer, a process that is dependent on the proteolytic cleavage of Gag by the viral protease. The condensed RNA dimer is the substrate for reverse transcription, from which the viral DNA is generated prior to integration as a provirus.
Interestingly, despite its central role in RNA dimerization, the DIS is not absolutely required for HIV-1 replication or for the presence of dimeric RNA in the virion. Thus, an as yet unidentified but redundant mechanism must be present to ensure that two genomic RNA are packaged and that dimerization occurs between them. Although it is known that the virion RNA dimer undergoes a maturation process, the stages and molecular mechanisms of RNA maturation are very poorly understood. Identifying the structure of the dimeric RNA is inherently difficult, and establishing the significance of the RNA structures with biological function is an added challenge. Unraveling the mystery of the RNA maturation process may require new RNA analysis techniques and novel approaches to investigate the biological activities of these structures. Additionally, there are many unknown factors to HIV-1 RNA biogenesis and encapsidation that require further investigation. It has been proposed that the interplay between translation of the genomic RNA and its packaging into virions may be regulated by the conformation of the RNA species, particularly its dimerization state. Furthermore, whether viral proteins package two monomeric RNA or a dimeric RNA is still being debated.
In summary, it has been four decades since the first experimental evidence indicated that retroviruses package two RNA genomes in a dimeric form. Although much has been learned about the process of RNA dimerization, there is still more to be elucidated about multiple aspects of this process. Our further understanding of HIV-1 RNA dimerization along with other steps of viral replication may provide insights that can help us curb the HIV-1 epidemic.
Acknowledgments
We thank Anne Arthur for editorial help, Drs. Alan Rein, Rebecca A. Russell and Vinay K. Pathak for input and reading of this manuscript. The authors are supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
References
- 1.Abbink T, Berkhout B. A novel long distance base-pairing interaction in HIV-1 RNA occludes the Gag start codon. J Biol Chem. 2003;278:11601–11. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 2.Abbink T, Ooms M, Haasnoot P, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–66. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 3.Adamson C, Freed E. HIV-1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–87. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 4.Andersen E, Contera S, Knudsen B, Damgaard C, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem. 2004;279:22243–9. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 5.Ashorn P, McQuade T, Thaisrivongs S, Tomasselli A, Tarpley W, Moss B. An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc Natl Acad Sci USA. 1990;87:7472–6. doi: 10.1073/pnas.87.19.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout B, van Wamel J. Role of the DIS hairpin in replication of HIV-1. J Virol. 1996;70:6723–32. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernacchi S, Henriet S, Dumas P, Paillart J, Marquet R. RNA and DNA binding properties of HIV-1 Vif protein: a fluorescence study. J Biol Chem. 2007;282:26361–8. doi: 10.1074/jbc.M703122200. [DOI] [PubMed] [Google Scholar]
- 9.Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol. 2002;76:3089–94. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butsch M, Boris-Lawrie K. Translation is not required to generate virion precursor RNA in HIV-1-infected T cells. J Virol. 2000;74:11531–7. doi: 10.1128/jvi.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin M, Rhodes T, Chen J, Fu W, Hu W. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci USA. 2005;102:9002–7. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J, Parslow T. Mutant HIV-1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–14. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J, Wong M, Parslow T. Requirements for kissing-loop-mediated dimerization of HIV RNA. J Virol. 1996;70:5902–8. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 15.Coren L, Thomas J, Chertova E, et al. Mutational analysis of the C-terminal gag cleavage sites in HIV-1. J Virol. 2007;81:10047–54. doi: 10.1128/JVI.02496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen B. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–24. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 17.D’Souza V, Summers M. Structural basis for packaging the dimeric genome of Moloney murine leukemia virus. Nature. 2004;431:586–90. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 18.Damgaard C, Andersen E, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol. 2004;336:369–79. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Darlix J, Gabus C, Nugeyre M, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the HIV-1. J Mol Biol. 1990;216:689–99. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 20.Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in HIV-1, HIV-2, and Moloney murine leukemia virus. J Virol. 2000;74:11413–7. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duesberg P. Physical properties of Rous sarcoma virus RNA. Proc Natl Acad Sci USA. 1968;60:1511–8. doi: 10.1073/pnas.60.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont S, Sharova N, DeHoratius C, et al. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–5. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 23.Elmen J, Zhang H, Zuber B, et al. Locked nucleic acid containing anti-sense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication. FEBS Lett. 2004;578:285–90. doi: 10.1016/j.febslet.2004.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ennifar E, Paillart J, Bodlenner A, et al. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell. Nucleic Acids Res. 2006;34:2328–39. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8:1064–8. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The HIV-1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–6. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Copeland T, Henderson L, et al. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–81. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher R, Fivash M, Stephen A, et al. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–84. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu W, Dang Q, Nagashima K, Freed E, Pathak V, Hu W. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J Virol. 2006;80:1242–9. doi: 10.1128/JVI.80.3.1242-1249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu W, Gorelick R, Rein A. Characterization of HIV-1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–8. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–9. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard F, Barbault F, Gouyette C, Huynh-Dinh T, Paoletti J, Lancelot G. Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex. J Biomol Struct Dyn. 1999;16:1145–57. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 33.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J Mol Biol. 2002;322:543–57. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 34.Henriet S, Sinck L, Bec G, Gorelick R, Marquet R, Paillart J. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–53. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hibbert C, Rein A. Preliminary physical mapping of RNA-RNA linkages in the genomic RNA of Moloney murine leukemia virus. J Virol. 2005;79:8142–8. doi: 10.1128/JVI.79.13.8142-8148.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill M, Shehu-Xhilaga M, Campbell S, Poumbourios P, Crowe S, Mak J. The dimer initiation sequence stem-loop of HIV-1 is dispensable for viral replication in peripheral blood mononuclear cells. J Virol. 2003;77:8329–35. doi: 10.1128/JVI.77.15.8329-8335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill M, Shehu-Xhilaga M, Crowe S, Mak J. Proline residues within spacer peptide p1 are important for HIV-1 infectivity, protein processing, and genomic RNA dimer stability. J Virol. 2002;76:11245–53. doi: 10.1128/JVI.76.22.11245-11253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoglund S, Ohagen A, Goncalves J, Panganiban A, Gabuzda D. Ultra-structure of HIV-1 genomic RNA. Virology. 1997;233:271–9. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 39.Hu W, Temin H. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–60. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–57. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobsen M, Damgaard C, Andersen E, Podhajska A, Kjems J. A genomic selection strategy to identify accessible and dimerization blocking targets in the 5′-UTR of HIV-1 RNA. Nucleic Acids Res. 2004;32:e67. doi: 10.1093/nar/gnh064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakobsen M, Haasnoot J, Wengel J, Berkhout B, Kjems J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology. 2007;4:29. doi: 10.1186/1742-4690-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao S, Akari H, Khan M, Dettenhofer M, Yu X, Strebel K. HIV-1 Vif is efficiently packaged into virions during productive but not chronic infection. J Virol. 2003;77:1131–40. doi: 10.1128/JVI.77.2.1131-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan A, Zack J, Knigge M, et al. Partial inhibition of HIV-1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67:4050–5. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasprzak W, Bindewald E, Shapiro B. Structural polymorphism of the HIV-1 leader region explored by computational methods. Nucleic Acids Res. 2005;33:7151–63. doi: 10.1093/nar/gki1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan M, Aberham C, Kao S, et al. HIV-1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J Virol. 2001;75:7252–65. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan R, Giedroc D. Recombinant HIV-1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–95. [PubMed] [Google Scholar]
- 48.Khandjian E, Meric C. A procedure for Northern blot analysis of native RNA. Anal Biochem. 1986;159:227–32. doi: 10.1016/0003-2697(86)90332-5. [DOI] [PubMed] [Google Scholar]
- 49.Kieken F, Paquet F, Brule F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–52. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kung H, Bailey J, Davidson N, Nicolson M, McAllister R. Structure, subunit composition, and molecular weight of RD-114 RNA. J Virol. 1975;16:397–411. doi: 10.1128/jvi.16.2.397-411.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laughrea M, Jette L. HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5′ splice junction contribute to loose and tight dimerization of HIV RNA. Biochemistry. 1997;36:9501–8. doi: 10.1021/bi970862l. [DOI] [PubMed] [Google Scholar]
- 52.Laughrea M, Shen N, Jette L, Darlix J, Kleiman L, Wainberg M. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of HIV-1; no role for the palindrome crowning the R-U5 hairpin. Virology. 2001;281:109–16. doi: 10.1006/viro.2000.0778. [DOI] [PubMed] [Google Scholar]
- 53.Laughrea M, Shen N, Jette L, Wainberg M. Variant effects of non-native kissing-loop hairpin palindromes on HIV replication and HIV RNA dimerization: role of stem-loop B in HIV replication and HIV RNA dimerization. Biochemistry. 1999;38:226–34. doi: 10.1021/bi981728j. [DOI] [PubMed] [Google Scholar]
- 54.Lever A. HIV-1 RNA packaging. Adv Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 55.Levin J, Grimley P, Ramseur J, Berezesky I. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–61. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levin J, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 57.Levin J, Rosenak M. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA. 1976;73:1154–8. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang C, Rong L, Cherry E, Kleiman L, Laughrea M, Wainberg M. Deletion mutagenesis within the dimerization initiation site of HIV-1 results in delayed processing of the p2 peptide from precursor proteins. J Virol. 1999;73:6147–51. doi: 10.1128/jvi.73.7.6147-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang C, Rong L, Quan Y, Laughrea M, Kleiman L, Wainberg M. Mutations within four distinct gag proteins are required to restore replication of HIV-1 after deletion mutagenesis within the dimerization initiation site. J Virol. 1999;73:7014–20. doi: 10.1128/jvi.73.8.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorenz C, Piganeau N, Schroeder R. Stabilities of HIV-1 DIS type RNA loop-loop interactions in vitro and in vivo. Nucleic Acids Res. 2006;34:334–42. doi: 10.1093/nar/gkj435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marquet R, Paillart J, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of HIV-1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–51. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McBride M, Panganiban A. Position dependence of functional hairpins important for HIV-1 RNA encapsidation in vivo. J Virol. 1997;71:2050–8. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirambeau G, Lyonnais S, Coulaud D, et al. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 64.Moore M, Fu W, Nikolaitchik O, Chen J, Ptak R, Hu W. Dimer initiation signal of HIV-1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–11. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mujeeb A, Clever J, Billeci T, James T, Parslow T. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5:432–6. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 66.Mujeeb A, Parslow T, Zarrinpar A, Das C, James T. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999;458:387–92. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 67.Mujeeb A, Ulyanov N, Georgantis S, et al. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stemloop of HIV-1: sequence effects and mechanism of RNA refolding. Nucleic Acids Res. 2007;35:2026–34. doi: 10.1093/nar/gkm097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muriaux D, De Rocquigny H, Roques B, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–92. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 69.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–82. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 70.Muriaux D, Girard P, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–16. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 71.Neville M, Stutz F, Lee L, Davis L, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–75. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 72.Nijhuis M, van Maarseveen N, Lastere S, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikolaitchik O, Rhodes T, Ott D, Hu W. Effects of mutations in the HIV-1 Gag gene on RNA packaging and recombination. J Virol. 2006;80:4691–7. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in HIV-1 virions. J Virol. 2004;78:10814–9. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ooms M, Verhoef K, Southern E, Huthoff H, Berkhout B. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res. 2004;32:819–27. doi: 10.1093/nar/gkh206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ortiz-Conde B, Hughes S. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J Virol. 1999;73:7165–74. doi: 10.1128/jvi.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paillart J, Berthoux L, Ottmann M, et al. A dual role of the putative RNA dimerization initiation site of HIV-1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–54. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paillart J, Dettenhofer M, Yu X, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem. 2004;279:48397–403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 79.Paillart J, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J Biol Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 80.Paillart J, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–7. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paillart J, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. The use of chemical modification interference and inverse PCR mutagenesis to identify the dimerization initiation site of HIV-1 genomic RNA. Pharm Acta Helv. 1996;71:21–8. doi: 10.1016/0031-6865(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 82.Paillart J, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 83.Peng C, Ho B, Chang T, Chang N. Role of HIV-1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–6. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pollard V, Malim M. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 85.Rein A, Henderson L, Levin J. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 86.Robinson W, Robinson H, Duesberg P. Tumor virus RNA’s. Proc Natl Acad Sci USA. 1967;58:825–34. doi: 10.1073/pnas.58.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Russell R, Hu J, Beriault V, et al. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of HIV-1 RNA. J Virol. 2003;77:84–96. doi: 10.1128/JVI.77.1.84-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Russell R, Hu J, Laughrea M, Wainberg M, Liang C. Deficient dimerization of HIV-1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303:152–63. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 89.Russell R, Liang C, Wainberg M. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Russell R, Roldan A, Detorio M, Hu J, Wainberg M, Liang C. Effects of a single amino acid substitution within the p2 region of HIV-1 on packaging of spliced viral RNA. J Virol. 2003;77:12986–95. doi: 10.1128/JVI.77.24.12986-12995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakuragi J, Iwamoto A, Shioda T. Dissociation of genome dimerization from packaging functions and virion maturation of HIV-1. J Virol. 2002;76:959–67. doi: 10.1128/JVI.76.3.959-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the HIV-1 virion and its potential roles in the early stages of viral replication. J Virol. 2007;81:7985–92. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sakuragi J, Panganiban A. HIV-1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–4. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B, Pavlakis G. Mutational inactivation of an inhibitory sequence in HIV-1 results in Rev-independent gag expression. J Virol. 1992;66:7176–82. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sheehy A, Gaddis N, Choi J, Malim M. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 96.Shehu-Xhilaga M, Crowe S, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for HIV-1 RNA dimerization and viral infectivity. J Virol. 2001;75:1834–41. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shehu-Xhilaga M, Hill M, Marshall J, Kappes J, Crowe S, Mak J. The conformation of the mature dimeric HIV-1 RNA genome requires packaging of pol protein. J Virol. 2002;76:4331–40. doi: 10.1128/JVI.76.9.4331-4340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shehu-Xhilaga M, Kraeusslich H, Pettit S, et al. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for HIV-1 RNA dimer maturation. J Virol. 2001;75:9156–64. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen N, Jette L, Liang C, Wainberg M, Laughrea M. Impact of HIV-1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–35. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sheng N, Pettit S, Tritch R, et al. Determinants of the HIV-1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J Virol. 1996;71:5723–32. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sinck L, Richer D, Howard J, et al. In vitro dimerization of HIV-1 spliced RNAs. RNA. 2007;13:2141–50. doi: 10.1261/rna.678307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skripkin E, Paillart J, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the HIV-1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–9. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J Mol Biol. 2007;371:1084–98. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 104.St Louis D, Gotte D, Sanders-Buell E, et al. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of HIV-1 can recombine and initiate a spreading infection in vitro. J Virol. 1998;72:3991–8. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swanson C, Malim M. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic. 2006;7:1440–50. doi: 10.1111/j.1600-0854.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 106.Swanson C, Puffer B, Ahmad K, Doms R, Malim M. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. Embo J. 2004;23:2632–40. doi: 10.1038/sj.emboj.7600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Temin H. Sex and recombination in retroviruses. Trends Genet. 1991;7:71–4. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- 108.von Schwedler U, Stemmler T, Klishko V, et al. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. Embo J. 1998;17:1555–68. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–27. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.You J, McHenry C. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–27. [PubMed] [Google Scholar]
- 111.Zhang H, Pomerantz R, Dornadula G, Sun Y. HIV-1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–61. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]