Abstract
The γ-aminobutyric acid (GABA) type A receptor (GABAAR) is one of the most important targets for insecticide action. The human recombinant β3 homomer is the best available model for this binding site and 4-n-[3H]propyl-4’-ethynylbicycloorthobenzoate ([3H]EBOB) is the preferred non-competitive antagonist (NCA) radioligand. The uniquely high sensitivity of the β3 homomer relative to the much-less-active but structurally-very-similar β1 homomer provides an ideal comparison to elucidate structural and functional features important for NCA binding. The β1 and β3 subunits were compared using chimeragenesis and mutagenesis and various combinations with the α1 subunit and modulators. Chimera β3/β1 with the β3 subunit extracellular domain and the β1 subunit transmembrane helices retained the high [3H]EBOB binding level of the β3 homomer while chimera β1/β3 with the β1 subunit extracellular domain and the β3 subunit transmembrane helices had low binding activity similar to the β1 homomer. GABA at 3 µM stimulated heteromers α1β1 and α1β3 binding levels more than 2-fold by increasing the open probability of the channel. Addition of the α1 subunit rescued the inactive β1/β3 chimera close to wildtype α1β1 activity. EBOB binding was significantly altered by mutations β1S15’N and β3N15’S compared with wildtype β1 and β3, respectively. However, the binding activity of α1β1S15’N was insensitive to GABA and α1β3N15’S was stimulated much less than wildtype α1β3 by GABA. The inhibitory effect of etomidate on NCA binding was reduced more than 5-fold by the mutation β3N15’S. Therefore, the NCA binding site is tightly regulated by the open-state conformation that largely determines GABAA receptor sensitivity.
Keywords: chimeragenesis, GABAA receptor, insecticide, mutagenesis, non-competitive antagonist, [3H]EBOB
Introduction
The γ-aminobutyric acid (GABA) type A receptor (GABAAR) is a major insecticide target along with the voltage-dependent sodium channel, the nicotinic receptor and acetylcholinesterase (Bloomquist, 1996; Casida and Quistad, 1998). Important insecticides acting at the GABAAR are lindane, α-endosulfan and fipronil. They bind at the picrotoxinin or non-competitive antagonist (NCA) site to block GABA-induced chloride flux. The safe and effective use of GABAergic insecticides requires detailed knowledge about the structural and functional basis of GABAAR-NCA interactions. The NCA site is readily assayed with 4-n-[3H]propyl-4’-ethynylbicycloorthobenzoate ([3H]EBOB) as the radioligand (Casida, 1993; Ratra et al., 2001). Ten years ago in this journal we reported a significant step in establishing the toxicity mechanisms of these insecticides and EBOB by defining that they all bind with very high affinity to the NCA site of human recombinant GABAAR β3 homomer with a specificity approximating that of the similarly-sensitive insect receptor (Ratra et al., 2001). These studies then localized the binding site of the insecticides and EBOB to A2’, T6’ and L9’ of the chloride channel (Chen et al., 2006a). The present investigation uses the human GABAAR β3 subunit as a homomer, heteromers and chimeras to define the unique structural and functional features of NCA action and interactions.
The GABAAR consists of 5 subunits arranged around a central ion-conducting pore and each of the subunits has a long extracellular domain and four transmembrane (TM) helices. There are 19 known human GABAAR subunits (α1-6, β1-4, γ1-3, δ, ε, π, ρ1-3) with sequence identity of about 30% between subunits and 70% between subunit subtypes (Olsen and Sieghart, 2008). Although the GABAAR is expressed in neurons as a heteromeric pentamer containing two or more different subunits, studies of homomeric receptors can reveal important structural determinants for assembly and ligand selectivity. When somatic cells are transfected with α1, β1, β2, β3, or γ2 subunits, only β3 and occasionally β1 subunits are detected on the cell surface with high spontaneous holding current (Bracamontes and Steinbach, 2008; Connolly et al., 1996a, b; Serafini et al., 2000; Taylor et al., 1999). HEK cells transfected with β3 subunits not only form dimers or tetramers but also significant amounts of homopentamers (Barnard et al., 1998). Critical residues in the extracellular domain of β3 are important for surface expression of homomers (Bracamontes and Steinbach, 2008; Sarto-Jackson and Sieghart, 2008; Taylor et al., 1999). However, the results often vary with the expression systems and species from which the subunit is obtained. The present study uses insect Sf9 cells as the expression system for human recombinant receptors (Chen et al., 2006a; Ratra et al., 2001).
The human GABAAR subunits have very different sensitivities to NCA binding. The β subunit is essential for NCA sensitivity in the recombinant multiple-subunit receptors (Ratra et al., 2001). The β3 is the only single subunit highly sensitive to NCA binding at current knowledge (Chen et al., 2006a, b; Ratra et al., 2001). It is surprising that β1 and β2 are insensitive to NCAs (Ratra and Casida, 2001; Ratra et al., 2001). The β1 homomer has little or no binding activity unless expressed with other subunits (Ratra and Casida 2001; Ratra et al., 2001). The β3 subunit assembles to form homomeric surface receptors in somatic cells, but human β1 subunits do not (Taylor et al., 1999). NCA binding studies have focused on TM2 of the GABAAR (Buhr et al., 2001; Chen et al., 2006a, b; Dibas et al., 2002; Jursky et al., 2000; Perret et al., 1999). There are high homologies between β subunits, particularly in TM2. The β3 and β2 subunits contain N15', while the β1 contains S15'. This residue faces away from the ion channel pore and into a water-filled cavity that appears capable of accommodating drugs (Chen et al., 2006b, Miller and Smart 2010). Mutation at 15’ of the β subunit is known to affect anesthetics and alcohol action to potentiate GABAA receptor-mediated electrical responses (Belelli et al., 1997; Hemmings et al., 2005; Jurd et al., 2002).
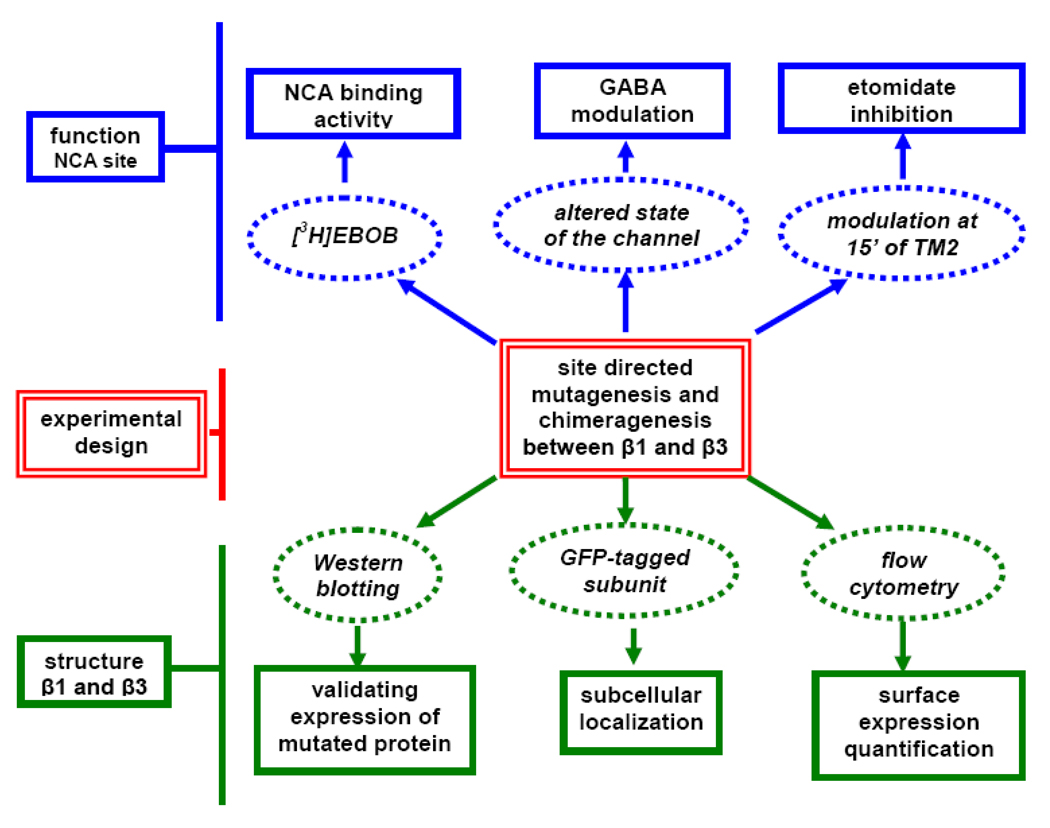
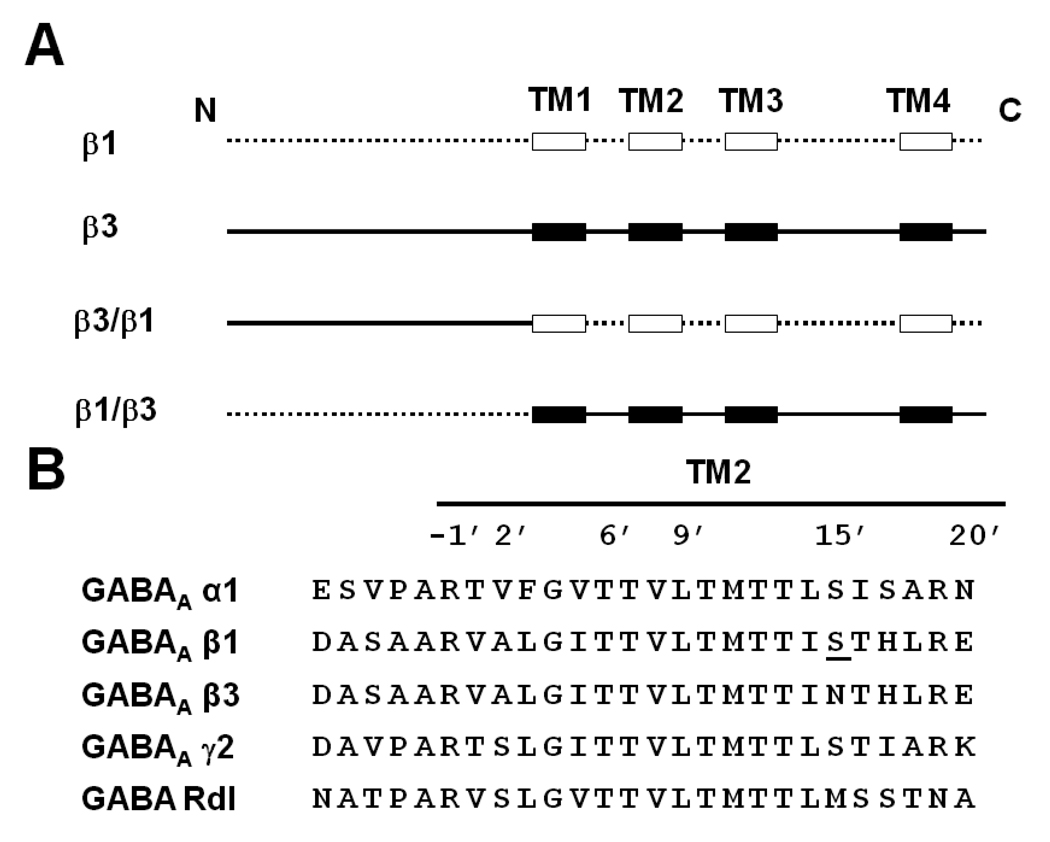
In this investigation, we employ the β3 homomer as a model to study the differential sensitivity conferred by β3 and β1 subunits using chimeragenesis and site-directed mutagenesis (Fig.1). Two chimeras (β3/β1 and β1/β3) (Fig. 2A) were constructed to localize the important domains in the β1 and β3 subunits for surface expression of the GABAA receptor and formation of the NCA site. These chimeras allowed us to study whether lack of surface expression plays a role in the low binding activity of β1. Sequence alignment of the important NCA binding domain of TM2 revealed that the GABAAR across the subunits is highly conserved in this region (Fig. 2B). The only difference between β3 and β1 in TM2 is at position N15’ and S15’, respectively. We therefore chose this position to mutate for exploring the binding activity and anesthetic modulation difference between β3 and β1. The agonist GABA regulates the GABAAR activity or the state of the ion channel which in turn potentially affects the receptor’s NCA binding sensitivity. Finally, by co-expression with the α subunit in order to introduce the agonist binding site, we also consider the effect of GABA (modulating channel state) and general anesthetics (15’ position) on NCA binding sensitivity (Belelli et al., 1997; Hemmings et al., 2005).
Fig. 1.
Strategy and experimental design to define the mechanisms for differential surface expression ability and NCA binding activity between β1 and β3 subunits.
Fig. 2.
(A) Design of chimera β3/β1 and chimera β1/β3 based on β1 and β3 predicted extracellular domain and TM helices. The extracellular domain and the loops between the TMs of the β1 subunit are drawn as a dotted line ( ) and the TM helices as open boxes (
) and the TM helices as open boxes ( ). The β3 subunit’s extracellular domain and the loops between the TMs are drawn as a continuous black line (
). The β3 subunit’s extracellular domain and the loops between the TMs are drawn as a continuous black line ( ) and the TM helices as black boxes (
) and the TM helices as black boxes ( ). (B) TM2 alignment of human GABA receptor subunits α1, β1, β3 and γ2 and Drosophila GABA receptor Rdl mutant (ffrench-Constant et al., 1993). The amino acid numbering system is based on Horenstein et al. (2001). The single amino acid difference in β1 from β3 at position 15’ is underlined and was the site for mutagenesis.
). (B) TM2 alignment of human GABA receptor subunits α1, β1, β3 and γ2 and Drosophila GABA receptor Rdl mutant (ffrench-Constant et al., 1993). The amino acid numbering system is based on Horenstein et al. (2001). The single amino acid difference in β1 from β3 at position 15’ is underlined and was the site for mutagenesis.
Materials and Methods
Chimeragenesis and site-directed mutagenensis
cDNAs encoding the human GABAA receptor α1, β1 and β3 subunits were inserted in the pVL1392 baculovirus transfer vector (Chen et al., 2006a, b). Point mutations were introduced with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Mutagenic oligonucleotides were prepared by Operon (Huntsville, AL). All mutations were confirmed by double-strand DNA sequencing (DNA Sequencing Facility, University of California, Berkeley). A modified overlapping PCR method was used to generate the chimera proteins (Wurch et al., 1998). The primer sequences are listed in Table 1.
Table 1.
Primers used for chimeragenesis and site-directed mutagenesis
| Name | Sequences |
|---|---|
| β3/β1-Aa | 5’-CAAAACTGCAGATGTGGGGCCTTGCGGGAG-3’ |
| β3/β1-Ba | 5' -GAAGTAACCAATGTTCCTCTTCAACCGAAAGCTCAG - 3'b |
| β3/β1-Ca | 5’-CTGAGCTTTCGGTTGAAGAGGAACATTGGTTACTTCATTTTG C-3’ |
| β3/β1-Da | 5’ -ACATGTCTAGATCAGTGTACATAGTAAAGCC-3' |
| β1/β3-Ac | 5’-CAAAACTGCAGATG TGG ACAGTACAAAATC-3’ |
| β1/β3-Bc | 5'-GAAGTATCCAATGTTTCTCTTTAGACGAAAACTTA -3' |
| β1/β3-Cc | 5’-GTCACTAAGTTTTCGTCTAAAGAGAAACATTGGATACTTCATTC-3’ |
| β1/β3-Dc | 5'-ATCAGCATCTAGATCAGTTAACATAGTACAGCC-3' |
| β1S15’Nd | 5’-CAATGACAACCATCAACACCCACCTCAGGGAG ACC-3’ |
| β3N15’Sd | 5’-GACAATGACAACCATCTCAACCCACCTTCGGGAGACC-3’ |
β3/β1-A, -B,-C and -D for construction of β3/β1 chimera containing the β3 extracellular domain and β1 TM helices; the β3/β1 chimera consisted of the complete extracellular domain of the β3 subunit (amino acids 1–241) and the C-terminal part (amino acids 242–474) of the β1 subunit.
The nucleotide in bold is the fusion part to connect two subunits.
β1/β3-A, -B, -C and-D for construction of β1/β3 chimera containing the β1 extracellular domain and β3 TM helices; the β1/β3 chimera had the opposite relationship with the complete N-terminal part of the β1 subunit (amino acids 1–241) and the C-terminal part (amino acids 242–473) of the β3 subunit.
β1S15’N and β3N15’S are primers to introduce 15’N and15’S, respectively. The mutated genetic codons are underlined.
Cell culture, protein expression and membrane preparation
Insect Sf9 cells (serum-free adapted, derived from ovaries of Spodoptera frugiperda) were maintained as described (Chen et al., 2006a, b). Log phase Sf9 cells were infected with recombinant baculovirus at a multiplicity of infection of 5–8. Cells were harvested at 65 h after infection. They were pelleted at 1,500 × g for 5 min and washed once with phosphate-buffered saline (PBS) (155 mM NaCl/3.0 mM NaH2PO4/1.0 mM K2HPO4, pH 7.4). Cell pellets were stored at −80°C until ready to use, then resuspended in PBS and homogenized in a glass tube with a motor-driven Teflon pestle. Cellular debris was removed by centrifugation at 500 × g for 10 min at 4°C. The supernatant was centrifuged at 100,000 × g for 40 min at 4°C, and the resulting pellet was resuspended in PBS and stored at −80°C. Protein concentration was determined with the detergent-compatible Lowry assay (Bio-Rad, Hercules, CA).
Western blotting
Membrane preparations were mixed with Laemmli sample buffer (Bio-Rad). After standard SDS-PAGE and transfer procedures, the membranes were blocked in Tris-buffered saline (Bio-Rad) containing 2% nonfat dry milk with 0.5% Tween 20 for 1 h at room temperature and incubated with the anti-GABAAR, α/β-chain antibodies (Chemicon International, Temecula, CA), at a dilution of 1:1,000, also for 1 h at room temperature. After three 5-min washings in Tris-buffered saline with 0.5% Tween 20, the blots were incubated with anti-mouse horseradish peroxidase-linked secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:2,000 for 1 h at room temperature. Following extensive washing, immunoreactivity was detected by chemiluminescence kit (PerkinElmer, Waltham, MA).
Green fluorescent protein (GFP) fusion constructs
Various constructs of empty vector (EV) and GABAAR subunits were subcloned in frame with GFP at the C terminus of the pcDNA5/FRT expression vector. The GFP-fusion constructs were used to generate stable HEK 293 cell lines with the Flp–In system (Invitrogen, Carlsbad, CA). To localize the subunit, cells were plated on poly-D-lysine-coated glass coverslips in 24-well plates at a density of 1.5×105 cells per well. Twenty–four hours after seeding, cells were fixed with 1% paraformaldehyde and washed three times in PBS. Coverslips, removed from the 24-well plate, were mounted in Vectashield antifade solution (Vector Technologies, Inc., Burlingame, CA) on glass microscope slides. A Retiga CCD-cooled camera and associated QCapture Pro software (QImaging, Surrey, BC Canada) were used to visualize the cells.
Flow cytometry analysis
Flow cytometric analysis was used to determine surface expression measured as membrane immunofluorescence (Xue et al., 2008). Sf9 cells were transfected with recombinant baculovirus for 48 h. Cells were harvested, washed with PBS, and then 106 cells in 100 µl PBS were incubated for 30 min on ice with anti-GABAAR α/β-chain antibody (1:200) or, as a negative control, with PBS only. After washing twice with PBS, Alexa Fluor 488 goat anti-mouse IgG (1:200) (Invitrogen) was added to each sample on ice for 30 min. Following three washes with PBS, the fluorescence density was measured using a Coulter Elite instrument and analyzed with WinMDI 2.8 software provided by Duke University (Durham, NC). The expression was evaluated as the mean fluorescence value.
[3H]EBOB binding
Assay mixtures contained 1 nM [3H]EBOB (48 Ci/mmol) (PerkinElmer) and the recombinant expressed receptor (100 µg protein) in PBS (500 µl final volume) (Chen et al. 2006a, b). After incubation for 90 min at 25 °C, the samples were filtered through GF/B filters (presoaked in 0.2% polyethylenemine for 3 h) and rinsed three times with ice-cold saline (0.9% NaCl). Specific binding was determined as total binding minus non-specific binding in the presence of 1 µM α-endosulfan. Etomidate [(R)-1-(1-phenylethyl)-1H-imidazole-5-carboxylic acid ethyl ester] (Sigma, St. Louis, MO) in inhibition studies was added simultaneously with [3H]EBOB. Each experiment was repeated three or more times with duplicate samples. The binding activity of mutants was expressed as percent [mean ± standard deviation (SD)] of that for the wildtype (WT).
Statistical analysis
Data are expressed as mean ± SD from 3 independent experiments. Multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett’s 1 or 2 two-tailed test. The basis for comparison was WT receptor, unless stated otherwise. Quantitative data were analyzed using GraphPad Prism 4.0 (GraphPad Software Inc.). A P value less than 0.05 was considered statistically significant.
Results
Expression of chimeras and mutants
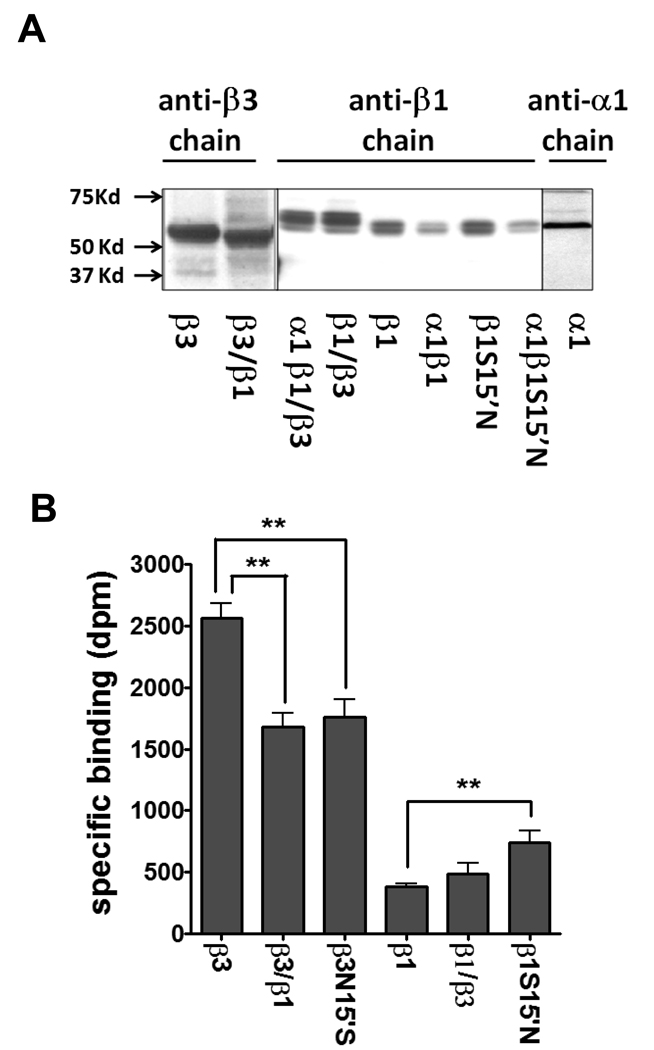
To identify and validate the constructs, a monoclonal antibody against the extracellular half of β3 was used to detect chimera β3/β1 (Fig. 3A) in the total membrane fraction. The chimeric protein β3/β1 showed a smaller size than β3 which was about 55 kD, indicating β3/β1 was successfully expressed without shifting or altering of the reading frame. The expression level was similar between β3 and β3/β1. However, chimera β1/β3 protein had a slightly larger size than the β1 subunit (Fig. 3A). Two nearby or dual bands detected by the polyclonal antibodies in β1 and its chimeric β1/β3 subunit might be due to different extents of glycosylation which is common in the ligand-gated ion channel subunit. The mutant S15’N in the β1 subunit did not affect the expression level when compared to the WT β1 subunit as shown, and also to β3N15’S (data not shown). The β1 subunit was also detected in other forms when combined with the α1 subunit. By adding α1, the β1 and β1S15’N subunit membrane expression was reduced relative to β1 itself. The β1 antibody detected less β1 subunit in the α1β1 heteromer than in the β1 homomer, consistent with the lower β subunit composition in the heteromers (Olsen and Sieghart, 2008). The α1 subunit was clearly detected in the total membrane fraction and had a similar size to the β subunits. The anti-α1 and -β3 subunit antibodies were monoclonal which specifically detect one form of the subunits.
Fig. 3.
Western blotting and [3H]EBOB binding activity of expressed receptors. (A) Individual subunits (α1, β1, β3, β1S15’N, β3/β1 and β1/β3) and co-expressed with α1 subunit (β1, β1/β3 and β1S15’N subunits). Both β3/β1 and β1/β3 are recognized by the anti-β3 chain and anti-β1 chain antibodies, respectively. β3/β1 shows a lower size than β3 WT and β1/β3 a larger size than β1 WT, indicating new proteins are generated. (B) Effect of site-specific mutations and chimeras on specific binding of [3H]EBOB. The data for plotting are given in Table S1 part A. ** P<0.01.
[3H]EBOB binding activity
Consistent with our earlier studies (Ratra and Casida, 2001; Ratra et al., 2001), the specific binding activity of β3 was about 7-fold higher than β1 (Fig. 3B). Interestingly, chimera β3/β1 showed a much increased binding activity relative to β1 itself and had a 66% binding activity relative to that of β3. In contrast, chimera β1/β3 showed similar binding to β1 (Fig. 3B). β3N15’S showed about 31% reduced activity compared with β3. Mutant β1S15’N almost doubled the EBOB binding of the β1 homomer (**P<0.01), which partially rescued the NCA binding to about 30% of that of the β3 homomer. This implies that the single amino acid difference in TM2 between β1 and β3 influenced the NCA binding level in β subunits. The binding parameters of β3/β1 showed similar nH (Hill coefficient) and Kd (apparent dissociation constant) to those of the β3 homomer but the Bmax of β3/β1 was reduced about one-third from that of β3 (*P<0.05) (Table 2). β1/β3 was similar to the β1 homomer, neither one of which had high enough binding activity for kinetic study.
Table 2.
[3H]EBOB binding parameters for β3, β1, β3/β1 and β1/β3 GABAA receptors
| Parametera | Subunit or Chimera | |||
|---|---|---|---|---|
| β3 | β1 | β3/β1 | β1/β3 | |
| specific binding (%) | 86 ± 5 | 21 ± 1 | 72 ± 4 | 18 ± 5 |
| nH | 0.94 ± 0.07 | N.D.b | 0.86 ± 0.09 | N.D. |
| Kd (nM) | 3.1 ± 0.2 | N.D. | 3.6 ± 0.4 | N.D. |
| Bmax (pmol/mg protein) | 1.8 ± 0.15 | N.D. | 1.3 ± 0.07* | N.D. |
nH, Hill coefficient; Kd, apparent dissociation constant; Bmax, maximal binding capacity;
N.D, not determined.
P<0.05 compared with β3 homomer
Subcellular localization of GFP-tagged subunits
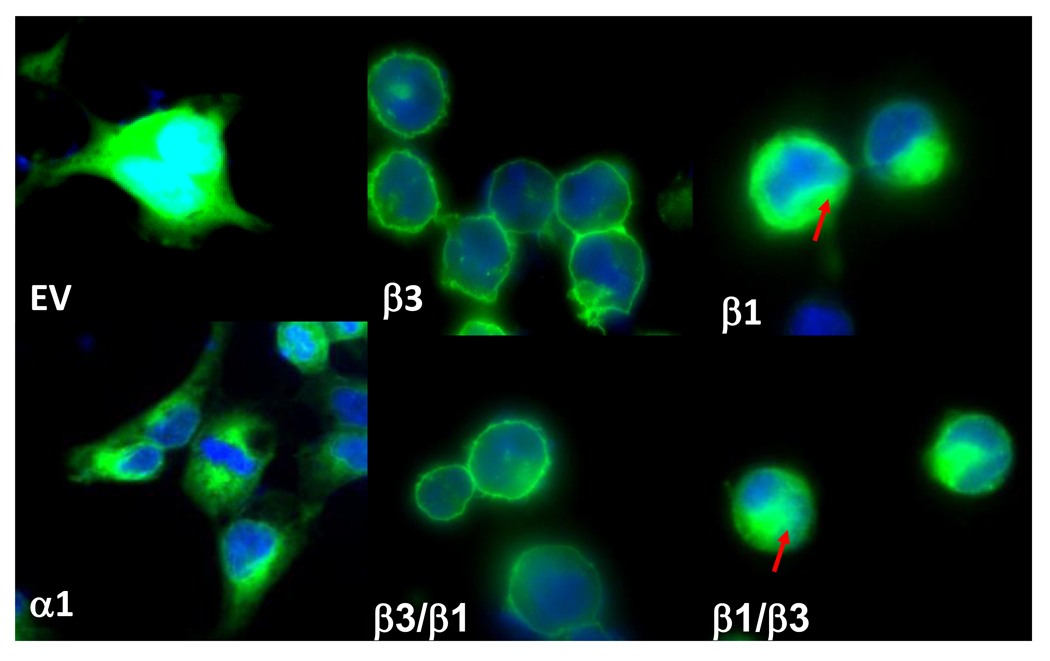
Although Western blotting showed clear expression of various WT or engineered proteins in total membrane preparations (Fig.3A), the surface expression or sub-cellular localization of subunits remained to be determined. We tagged GFP in the C-terminus of α1, β1 and β3, as well as two chimeras. As shown in Fig. 4, GFP empty vector (EV) was expressed in the cytosol and nucleus (cyan color due to overlapping of DAPI (blue) and GFP (green)). β3 was localized in the plasma membrane whereas β1 showed more cytoplasmic than surface expression. α1 was not expressed in the cell surface but instead mainly appeared in cytoplasmic membranes. Chimera β3/β1 with the β3 extracellular domain showed strong cell surface expression similar to the parent β3 subunit. Chimera β1/β3 containing the extracellular domain of subunit β1 did not have the same ability as β3/β1 to express in the cell surface.
Fig. 4.
Subcellular localization of GFP-tagged subunits in HEK293 cells. Stably-expressed GFP EV, α1, β1, β3 and the chimeras. Cell nucleus staining by DAPI (4',6-diamidino-2-phenylindole) is shown in blue. The red arrow indicates that GFP signaling is retained in the cytoplasm of the cell.
Quantification of surface expression of various receptors
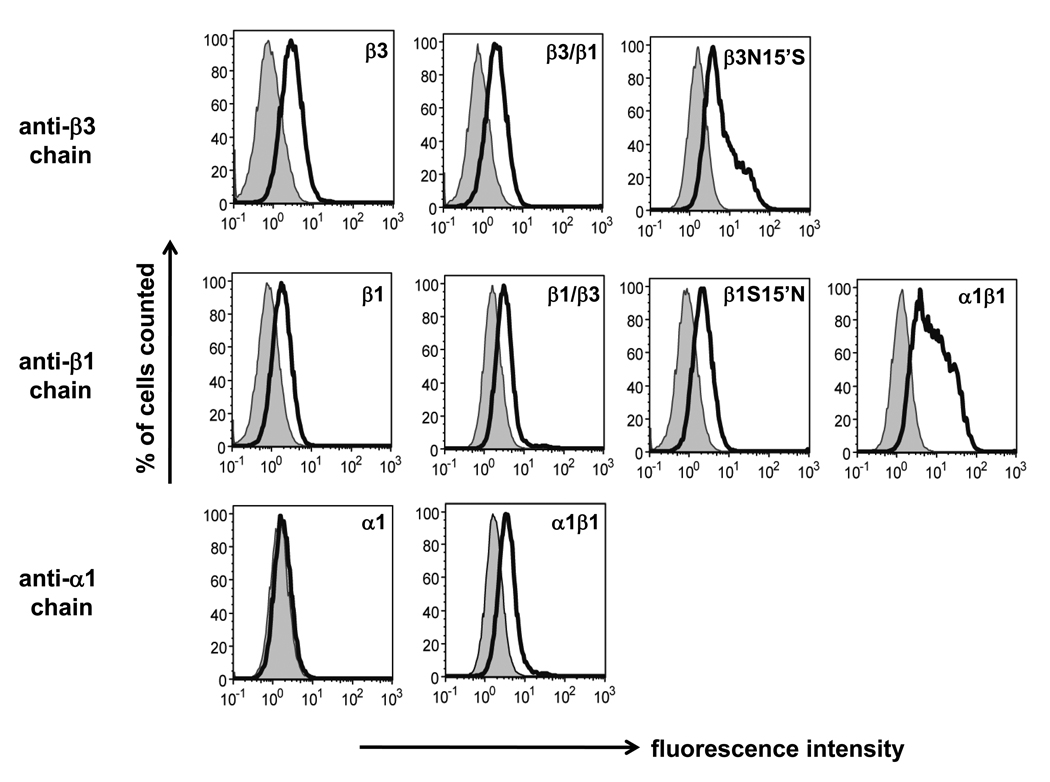
Expression of the subunits and combinations was examined by flow-cytometry with subunit-specific antibodies using live cells without adding any detergent or fixation reagent (Fig. 5). Consistent with the findings on GFP-tagged subunits, the β3 and β3/β1 subunits were well detected in the cell plasma membrane and also showed similar expression levels (mean fluorescent levels: 16.3 and 17.3, respectively). Although the β1 and β1/β3 subunits showed similar receptor expression on the cell surface (mean fluorescence levels 5.71 and 5.46, respectively), they were only about one-third of those for the β3 homomers (mean fluorescence level 16.3). The α1 subunit alone was not detected in the cell surface but showed significant surface expression in combination with the β1 subunit (mean fluorescence level 14.3).
Fig. 5.
Surface expression of various receptors quantified by flow cytometry in Sf9 cells. Anti-β3 chain was used for detection of β3, β3/β1 and β3N15’S, anti-β1 chain for β1, β1/β3, β1S15’N and α1β1 and anti-α1 chain for α1 and α1β1. Plasma membrane expression was measured as fluorescence intensity. Shaded areas were negative controls for proteins labeled with secondary antibody only. Cells were labeled alive so that the antibodies only bound to the specific protein in the cell surface.
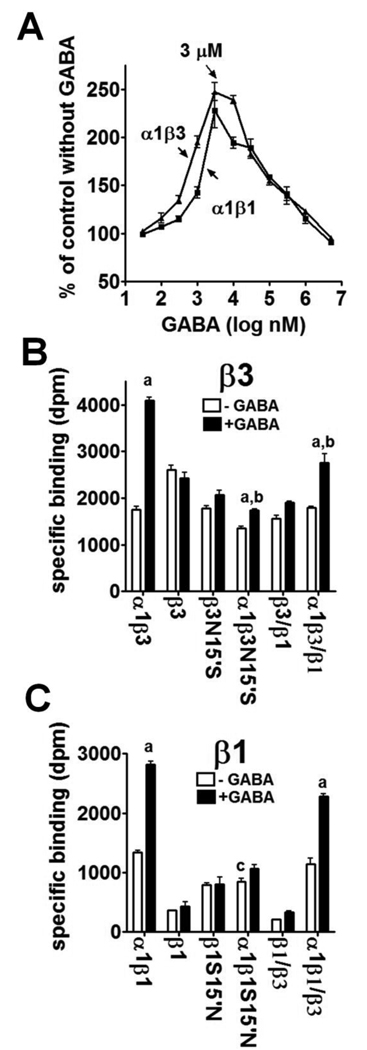
Influence of agonist binding on NCA site
High NCA binding affinity of the β3 homomer is proposed to be due to both the number of binding sites and the spontaneous open state of the channel (Chen et al. 2006a, b). To explore the influence of agonist binding on the NCA site, we applied GABA to co-expressed αβ receptors, which form the agonist binding site from both subunits. With GABA at 3 µM, both α1β1 and α1β3 showed more that 2-fold increase for [3H]EBOB binding activity (Fig. 6A). This concentration of GABA was then examined for effect on the single mutants, chimeras and their αβ combinations. For the β3 related receptors (Fig. 6B), GABA did not change the [3H]EBOB binding activity of the single β3, β3N15’S and β3/β1 chimera but significantly increased those of the corresponding α1β3N15’S (29%), α1β3/β1 (49%) and particularly α1β3 (141%) receptors (a and b, **p<0.01). Apparently, the N15’S mutation impaired the GABA-stimulated NCA binding in β3-related receptors. For the β1-related receptors (Fig. 6C), GABA did not appreciably improve the low [3H]EBOB binding of the single β1, β1S15’N and β1/β3 receptors but significantly increased the binding by α1β1 (111%) and α1β1/β3 (99%) compared with each corresponding receptor without GABA (a, **P<0.01). However, the NCA binding of α1β1S15’N showed little increase with 3 µM GABA (Fig. 6C). It is worth noting that, without GABA, α1β1S15’N significantly reduced the NCA binding activity of α1β1 by 36% (c, **P<0.01) (Fig. 6C).
Fig. 6.
Effect of GABA on [3H]EBOB binding by various receptors. A. Effect of GABA concentration on α1β3 and α1β1 binding. B. Effect of 3 µM GABA on various β3 type receptors. The data for plotting are given in Table 1S part B; C. Effect of 3 µM GABA on various β1 type receptors. The data for plotting are given in Table 1S part C. Statistical comparisons (**P<0.01) as follows: Panel B; a, α1β3, α1β3N15’S and α1β3/β1 with GABA compared to their corresponding receptors without GABA; b, α1β3N15’S and α1β3/β1 with GABA compared to α1β3 with GABA. Panel C; a, α1β1 and α1β1/β3 with GABA compared to the corresponding receptors without GABA; c, α1β1S15’N without GABA compared to α1β1 without GABA.
Interaction of NCA site with etomidate site
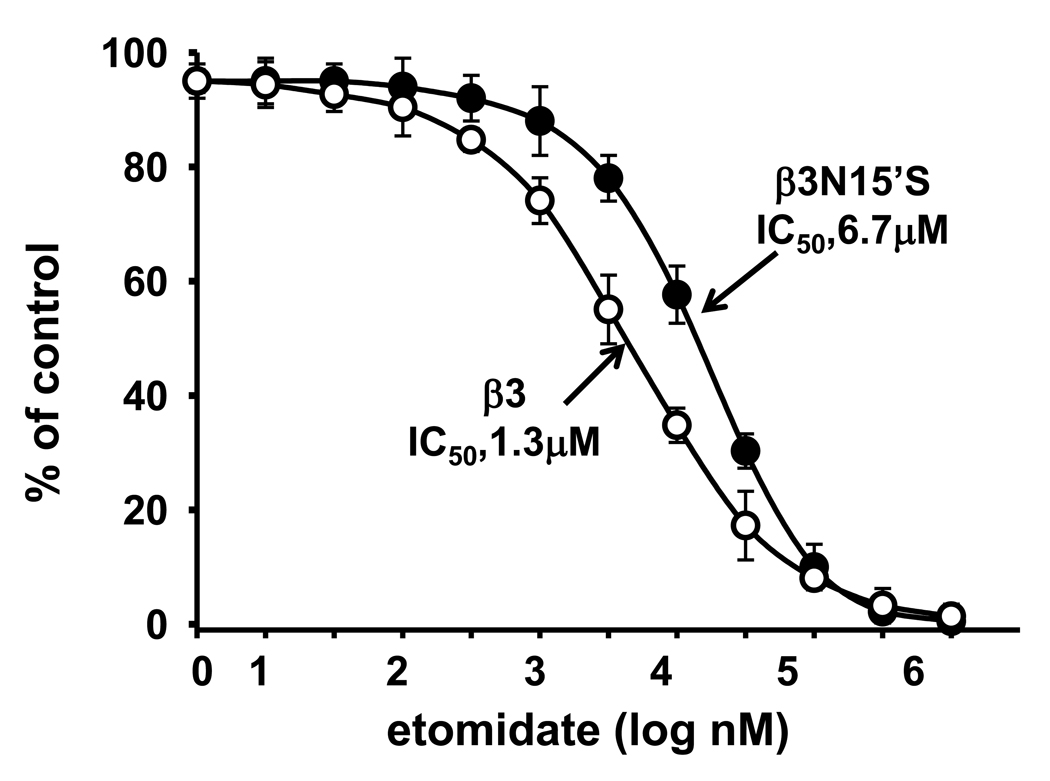
The only different amino acid residue in TM2 between β3 and β1 is 15’, which is an important site in general anesthetics etomidate action (Belelli et al., 1997). Etomidate is a positive allosteric modulator of GABAAR activity. The ability of intravenous etomidate to modulate and activate GABAARs is uniquely dependent upon the β subunit subtype present within the receptor. Receptors containing β2 or β3 but not β1 subunits are highly sensitive to this agent (Belelli et al., 1997). To study the possible interaction between the NCA and anesthetics sites, we determined the inhibition of [3H]EBOB binding by etomidate with β3 and its mutant β3N15’S. The IC50 of β3N15’S (6.7 µM) was increased 5–fold compared to the β3 WT (1.3 µM) (Fig. 7).
Fig. 7.
Effect of β3N15’S mutation on etomidate inhibition of [3H]EBOB binding. The nH values were 1.75 for β3 and 1.36 for β3N15’S.
Discussion
β3 homomer confers favorable expression for NCA binding
The β3 homomer has higher NCA sensitivity than any other single vertebrate GABAAR subunit and any heteromeric combinations that have been tested (Chen et al., 2006b; Ratra and Casida 2001; Ratra et al., 2001). NCAs with excellent fit for the β3 homomer may show less favorable docking in the heteromeric native receptors due to subunit variation at the 2′ position (Fig. 2) (Chen et al., 2006a; Law and Lightstone, 2008). The poorly active β1 and highly active β3 subunits, expressed individually as homomers, 15’ mutations and β3/β1 and β1/β3 chimeras, help define the structural and functional requirements for uniquely high NCA sensitivity. These findings confirm that the extracellular half of the β3 subunit contains the cell surface expression residues essential to form the high affinity NCA binding site. Correspondingly, the low surface expression level of the β1 subunit may partially contribute to its low NCA sensitivity.
β3 homomer confers favorable channel conformation for NCA binding
The extracellular part of the β3 subunit not only controls the receptor assembly and the signal of membrane protein trafficking but also the channel’s open state (Serafini et al., 2000). The β3 homomer can form a spontaneously open ion channel (Serafini et al., 2000; Wooltorton et al., 1997), potentially facilitating ligand fit at its binding site so that chimera β3/β1 has the features of the β3 homomer with a similar binding pocket at TM2 delivering high sensitivity to NCAs. We believe that both high surface expression and open state of the channel account for the high sensitivity of the β3 homomer to NCAs. GABA increases NCA binding more than 2-fold with the α1 subunit-containing α1β1 and α1β3 receptors but not the β3 homomer. This dramatic stimulation indicates that the extracellular domains binding with GABA probably alter the access of NCAs to their binding site. Substantial evidence shows that GABA activates its receptor by interacting with an extracellular ligand-binding site, triggering a rapid conformational change in the channel pore (TM2) that results in opening of the ion channel (Kash et al., 2003; Olsen and Sieghart, 2008) by retraction of TM2 helices (towards TM1, TM3 and TM4) to conduct the chloride ions (Miller and Smart, 2010). The linker between TM2 and TM3 is considered crucial for signal transduction through the extracellular domain to the channel pore and for opening the channel. The coupling of the agonist (GABA) site to the NCA site likely results from the structural change in TM2 induced by GABA, which increases the accessibility of the NCA to its site(s). Notably, mutations in the pore-facing residues (9’, 13’ and 14’) that presumably are the gate of the receptor/channel (Akabas 2004; Miller and Smart, 2010) also eliminate [3H]EBOB binding (Chen et al., 2006a, b). Collectively, NCA binding accessibility and sensitivity may be tightly correlated to the conformational status of TM2. This activation also indicates that GABA may significantly affect NCA binding in the native receptors which are generally heteromers.
The 15’ position modulates GABA and NCA interactions
Mutation at the 15’ position showed opposite NCA binding responses for β1S15’N (increase) and β3N15’S (decrease). GABA does not stimulate [3H]EBOB binding by either α1β3N15’S or α1β1S15’N mutants to the same extent as their WT counterparts. These findings indicate that the 15’ position in TM2 is associated with the sensitivity of the GABAA receptor to GABA or the GABA-mediated channel state. N15’ is a key determinant to transduce the modulator binding signal by facilitating or hindering the tilting of M2 domains during gating (Miller and Smart, 2010). The reduction of GABA-induced NCA binding by the 15’ mutation may be due to damage in channel gating which would dramatically affect NCA binding (Chen et al., 2006b). The magnitude of spontaneous activity of these receptors was correlated with the molecular volume of the residue at 15’ for both homomeric and heteromeric GABAA receptors (Miko et al. 2004). The reduced response to GABA of NCA binding with α1β3N15’S and α1β1S15’N might result from an alteration of the channel gating which impairs GABA-induced opening. The action of general anesthetics and alcohol is modulated by mutations at the 15’ position to alter the GABAAR mediated chloride current (Belelli et al., 1997; Hemmings et al., 2005; Jurd et al., 2002). The IC50 of etomidate for [3H]EBOB binding increased about 5-fold with mutant N15’S, strengthening the role of this position in binding of general anesthetics.
NCA action on GABAA receptors
The β3 homomer is ideal for studying NCA action with its simple subunit composition and high sensitivity. In this study, we clearly show that NCAs can bind at α1β1 and α1β3 heteromers without GABA and GABA greatly enhances NCA binding, suggesting that NCAs have multiple pathways or the receptors have multiple conformations in accessing this site. One pathway is directly through the pore and another one is possibly through water cavities between adjacent subunits (Chen et al., 2006b), similar to the suggested general anesthetics binding pocket in the water crevice behind M2 (Hemmings et al., 2005). In an α1β2γ2 computational modeling study, many of the binding modes are suggestive of a non-competitive allosteric mechanism based on interruption of channel gating rather than directly blocking of the channel and possibly multiple sites for NCA binding (Law and Lightstone, 2008). This computational model is consistent with mutations in residues in the purported channel gate (Chen et al., 2006b) and at 15’ altering NCA binding sensitivity (present study). Direct labeling and/or co-crystallization of a NCA with the GABAAR (especially with the high sensitivity β3 homomer) may ultimately contribute to understanding the complex relationships of NCA and agonist interactions.
Supplementary Material
Acknowledgements
This work was supported by the University of California at Berkeley William Muriece Hoskins Chair in Chemical and Molecular Entomology (to J. E. C). Ann Fischer of the Molecular and Cell Biology Cell Culture Facility at Berkeley provided the Sf9 cells. Ligong Chen is currently a postdoctoral fellow at the Department of Bioengineering and Therapeutic Sciences at the University of California at San Francisco.
Abbreviations
- EV
empty vector
- GABAAR
γ-aminobutyric acid (GABA) type A receptor
- GFP
green fluorescent protein
- [3H]EBOB
4-n-[3H]propyl-4’-ethynylbicycloorthobenzoate
- NCA
non-competitive antagonist
- TM
transmembrane
- WT
wildtype
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest in this study
References
- Akabas MH. GABAA receptor structure-function studies: a reexamination in light of new acetylcholine receptor structures. Int. Rev. Neurobiol. 2004;62:1–43. doi: 10.1016/S0074-7742(04)62001-0. [DOI] [PubMed] [Google Scholar]
- Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson AN, Langer SZ. International Union of Pharmacology. XV. Subtypes of γ-aminobutyric acidA receptors: Classification on the basis of subunit structure and receptor function. Pharmacol. Rev. 1998;50:291–313. [PubMed] [Google Scholar]
- Belelli D, Lambert JJ, Peters JA, Wafford K, Whiting PJ. The interaction of the general anesthetic etomidate with the γ-aminobutyric acid type A receptor is influenced by a single amino acid. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11031–11036. doi: 10.1073/pnas.94.20.11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist JR. Ion channels as targets for insecticides. Annu. Rev. Entomol. 1996;41:163–190. doi: 10.1146/annurev.en.41.010196.001115. [DOI] [PubMed] [Google Scholar]
- Bracamontes JR, Steinbach JH. Multiple modes for conferring surface expression of homomeric β1 GABAA receptors. J. Biol. Chem. 2008;283:26128–26136. doi: 10.1074/jbc.M801292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr A, Wagner C, Fuchs K, Sieghart W, Sigel E. Two novel residues in M2 of the γ-aminobutyric acid type A receptor affecting gating by GABA and picrotoxin affinity. J. Biol. Chem. 2001;276:7775–7781. doi: 10.1074/jbc.M008907200. [DOI] [PubMed] [Google Scholar]
- Casida JE. Insecticide action at the GABA-gated chloride channel: recognition, progress, and prospects. Arch. Insect Biochem. Physiol. 1993;22:13–23. doi: 10.1002/arch.940220104. [DOI] [PubMed] [Google Scholar]
- Casida JE, Quistad GB. Golden age of insecticide research: past, present, or future? Annu. Rev. Entomol. 1998;43:1–16. doi: 10.1146/annurev.ento.43.1.1. [DOI] [PubMed] [Google Scholar]
- Chen L, Durkin KA, Casida JE. Structural model for γ-aminobutyric acid receptor noncompetitive antagonist binding: widely diverse structures fit the same site. Proc. Natl. Acad. Sci. U.S.A. 2006a;103:5185–5190. doi: 10.1073/pnas.0600370103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Durkin KA, Casida JE. Spontaneous mobility of GABAA receptor M2 extracellular half relative to noncompetitive antagonist action. J. Biol. Chem. 2006b;281:38871–38878. doi: 10.1074/jbc.M608301200. [DOI] [PubMed] [Google Scholar]
- Connolly CN, Krishek BJ, McDonald BJ, Smart TG, Moss SJ. Assembly and cell surface expression of heteromeric and homomeric γ-aminobutyric acid type A receptors. J. Biol. Chem. 1996a;271:89–96. doi: 10.1074/jbc.271.1.89. [DOI] [PubMed] [Google Scholar]
- Connolly CN, Wooltorton JRA, Smart TG, Moss SJ. Subcellular localization of γ-aminobutyric acid type A receptors is determined by receptor β subunits. Proc. Natl. Acad. Sci.U.S.A. 1996b;93:9899–9904. doi: 10.1073/pnas.93.18.9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibas MI, Gonzales EB, Das P, Bell-Horner CL, Dillon GH. Identification of a novel residue within the second transmembrane domain that confers use-facilitated block by picrotoxin in glycine α1 receptors. J. Biol. Chem. 2002;277:9112–9117. doi: 10.1074/jbc.M111356200. [DOI] [PubMed] [Google Scholar]
- ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993;363:449–451. doi: 10.1038/363449a0. [DOI] [PubMed] [Google Scholar]
- Hemmings HC, Jr, Akabas MH, Goldstein PA, Trudell JR, Orser BA, Harrison NL. Emerging molecular mechanisms of general anesthetic action. Trends Pharmacol. Sci. 2005;26:503–510. doi: 10.1016/j.tips.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Wagner DA, Czajkowski C, Akabas MH. Protein mobility and GABA-induced conformational changes in GABAA receptor pore- lining M2 segment. Nat. Neurosci. 2001;4:477–485. doi: 10.1038/87425. [DOI] [PubMed] [Google Scholar]
- Jurd R, Arras M, Lambert S, Drexler B, Siegwart R, Crestani F, Zaugg M, Vogt KE, Ledermann B, Antkowiak B, Rudolph U. General anesthetic actions in vivo strongly attenuated by a point mutation in the GABAA receptor β3 subunit. FASEB J. 2002;16:250–271. doi: 10.1096/fj.02-0611fje. [DOI] [PubMed] [Google Scholar]
- Jursky F, Fuchs K, Buhr A, Tretter V, Sigel E, Sieghart W. Identification of amino acid residues of GABAA receptor subunits contributing to the formation and affinity of the tert-butylbicyclophosphorothionate binding site. J. Neurochem. 2000;74:1310–1316. doi: 10.1046/j.1471-4159.2000.741310.x. [DOI] [PubMed] [Google Scholar]
- Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Coupling of agonist binding to channel gating in the GABA(A) receptor. Nature. 2003;421:272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- Law RJ, Lightstone FC. GABA receptor insecticide non-competitive antagonists may bind at allosteric modulator sites. Intern. J. Neurosci. 2008;118:705–734. doi: 10.1080/00207450701750216. [DOI] [PubMed] [Google Scholar]
- Miko A, Werby E, Sun H, Healey J, Zhang L. A TM2 residue in the β1 subunit determines spontaneous opening of homomeric and heteromeric γ-aminobutyric acid-gated ion channels. J. Biol. Chem. 2004;279:22833–22840. doi: 10.1074/jbc.M402577200. [DOI] [PubMed] [Google Scholar]
- Miller PS, Smart TG. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010;31:161–174. doi: 10.1016/j.tips.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International Union of Pharmacology. LXX. Subtypes of γ-aminobutyric acidA receptors: Classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol. Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret P, Sarda X, Wolff M, Wu T-T, Bushey D, Goeldner M. Interaction of non-competitive blockers within the γ-aminobutyric acid type A chloride channel using chemically reactive probes as chemical sensors for cysteine mutants. J. Biol. Chem. 1999;274:25350–25354. doi: 10.1074/jbc.274.36.25350. [DOI] [PubMed] [Google Scholar]
- Ratra GS, Casida JE. GABA receptor subunit composition relative to insecticide potency and selectivity. Toxicol. Lett. 2001;122:215–222. doi: 10.1016/s0378-4274(01)00366-6. [DOI] [PubMed] [Google Scholar]
- Ratra GS, Kamita SG, Casida JE. Role of human GABAA receptor β3 subunit in insecticide toxicity. Toxicol. Appl. Pharmacol. 2001;172:233–240. doi: 10.1006/taap.2001.9154. [DOI] [PubMed] [Google Scholar]
- Sarto-Jackson I, Sieghart W. Assembly of GABAA receptors. Mol. Membr. Biol. 2008;25:302–310. doi: 10.1080/09687680801914516. [DOI] [PubMed] [Google Scholar]
- Serafini R, Bracamontes J, Steinbach JH. Structural domains of the human GABAA receptor β3 subunit involved in the actions of pentobarbital. J. Physiol.-London. 2000;524:649–676. doi: 10.1111/j.1469-7793.2000.00649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PM, Thomas P, Gorrie GH, Connolly CN, Smart TG, Moss SJ. Identification of amino acid residues within GABAA receptor β subunits that mediate both homomeric and heteromeric receptor expression. J. Neurosci. 1999;19:6360–6371. doi: 10.1523/JNEUROSCI.19-15-06360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooltorton JRA, Moss SJ, Smart TG. Pharmacological and physiological characterization of murine homomeric β3 GABAA receptors. Eur. J. Neurosci. 1997;9:2225–2235. doi: 10.1111/j.1460-9568.1997.tb01641.x. [DOI] [PubMed] [Google Scholar]
- Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol. Tech. 1998;12:653–657. [Google Scholar]
- Xue L, Nolla H, Suzuki A, Mak TW, Winoto A. Normal development is an integral part of tumorigenesis in T cell-specific PTEN-deficient mice. Proc. Natl. Acad. Sci.U.S.A. 2008;105:2022–2027. doi: 10.1073/pnas.0712059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.