Abstract
Acetylation is an essential post-translational modification featuring an acetyl group that is covalently conjugated to a protein substrate. Histone acetylation was first proposed nearly half a century ago by Dr. Vincent Allfrey. Subsequent studies have shown that the acetylated core histones are often associated with transcriptionally active chromatin. Acetylation at lysine residues of histone tails neutralizes the positive charge, which decreases their binding ability to DNA and increases the accessibility of transcription factors and coactivators to the chromatin template. In addition to histones, a number of non-histone substrates are acetylated. Acetylation of non-histone proteins governs biological processes, such as cellular proliferation and survival, transcriptional activity, and intracellular trafficking. We demonstrated that acetylation of transcription factors can regulate cellular growth. Furthermore, we showed that nuclear receptors (NRs) are acetylated at a phylogenetically conserved motif. Since our initial observations with the estrogen and androgen receptors, more than a dozen NRs have been shown to function as substrates for acetyltransferases with diverse functional consequences. This review focuses on the acetylation of NRs and the effect of acetylation on NR function. We discuss the potential role of acetylation in disease initiation and progression with an emphasis on tumorigenesis.
1. Nuclear receptors and acetylation
1.1. Nuclear receptors
The nuclear receptors (NRs) are comprised of an activation function-1 (AF-1) domain located in the amino terminus, a DNA binding domain (DBD), a hinge region, and a ligand-binding domain (LBD) located in the carboxyl terminus. The NR superfamily includes ligand-activated transcription factors with a wide array of functions in development, homeostasis, and cellular metabolism. NRs have been classified into four categories (Type I, II, III, and IV), two of which being the main types (Type I and II) and two additional types (Type III and IV) [1,2]. Type I consists of steroid hormone receptors, such as progesterone receptor (PR), glucocorticoid receptor (GR), estrogen receptor (ER), androgen receptor (AR), and mineralcorticoid receptor (MR). Type II receptors include the thyroid hormone receptor (TR), all-trans-retinoic acid receptor (RAR), 9-cis-retinoic acid receptor (RXR), and Vitamin D3 receptor (VDR). These receptors are located in the nucleus regardless of the presence of a ligand. The third class of NRs called orphan receptors recognizes distinct DNA sequences in response ligands, most of which have yet to be identified.
In the absence of a ligand, the Type I NRs, as well as several of the orphan receptors, normally associate with heat shock proteins (HSP) to form an inactive complex in the cytoplasm. Ligand treatment results in the dissociation of HSPs from the receptor/HSPs complex [3]. The receptor rapidly shuttles to the nucleus, dimerizes, and binds to the hormone response element (HRE) to initiate gene transcription through disengagement of the corepressors and subsequent recruitment of transcriptional coregulators. Several coactivators have the ability to bind NRs, including SRC1 (steroid receptor coactivator-1), AIB1 (amplified in breast cancer 1, also known as ACTR or SRC2), GRIP1 (glucocorticoid receptor interacting protein 1, also known as TIF-2 or SRC3), p300/CBP and p/CAF (p300/CBP-associated factor) [4,5]. NRs interact with corepressors in the absence of agonists and/or presence of antagonists. These corepressors are either composed of or recruit histone modifying enzymes, such as HDACs, in order to silence target gene expression through post-translational modification. NR corepressors include NCoR (NR corepressor), SMRT (silencing mediator of retinoid and thyroid hormone receptor), Sin3, HDACs, DACH1 [6], TURP (thyroid hormone receptor uncoupling protein), BRCA1, NuRD, Suv39h1, DNMT1, pRB2/p130, and E2F4/5.
1.2. Acetylation of nuclear receptors
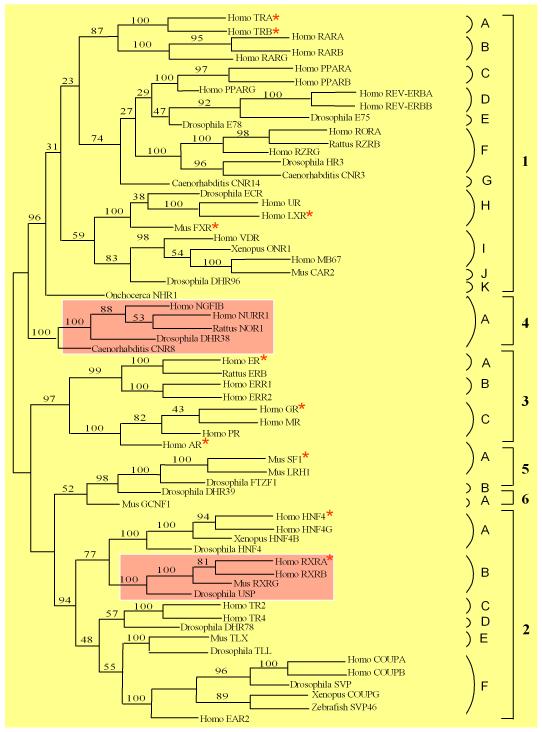
Since our initial identification of NR acetylation with the estrogen receptor alpha (ERα) and androgen receptor (AR) a decade ago [7,8], emerging evidence has demonstrated that other NRs are acetylated as well (Fig. 1). We predicted that the NRs would be acetylated at a conserved acetylation motif that we identified in the ERα and AR.
Figure 1. Phylogenetic tree of nuclear receptor family.
NRs containing the putative acetylation motif are shown in yellow and NRs lacking the motif in the 4A and 2B subgroups are shown in pink. (Adopted from our prior publication [8] with modifications). NRs marked with a star (*) have been shown to be acetylated.
Currently, eleven NRs have been shown to undergo acetylation (Table 1), including the Thyroid Hormone Receptor-like (Thyroid hormone receptor α and β, Liver X receptor α and β, and Farnesoid X receptor), Retinoid X Receptor-like (Hepatocyte nuclear factor 4α and Retinoid X receptor α), Estrogen Receptor-like (Estrogen receptor α, Glucocorticoid receptor, and Androgen receptor), Steroidogenic Factor-like (Steroidogenic factor 1). These studies have established that NR acetylation governs a variety of cellular functions, which include NR activity; DNA binding affinity; ligand sensitivity; receptor stability; and subcellular distribution.
Table 1.
The list of NRs that have been reported to be acetylated
| Nuclear receptor | Acetylase | Deacetylase | Amino acid | Refs |
|---|---|---|---|---|
| Thyroid hormone receptor-α | CBP | K128, K132, K134 | [9] | |
| Thyroid hormone receptor-β | [10] | |||
| Liver X receptor-α | SIRT1 | K432 | [11] | |
| Liver X receptor-β | SIRT1 | K433 | [11] | |
| Farnesoid X receptor | p300/CBP | SIRT1 | K157, K217 | [12,13] |
| Hepatocyte nuclear factor-4-α | CBP | K97, K99, K117, K118 | [14] | |
| Retinoid X receptor-α | p300 | K145 | [15] | |
| Estrogen receptor-α | p300 | SIRT1 | K302, K303, K268, K266 | [8,16] |
| Glucocorticoid receptor | HDAC2 | K494, K495 | [17] | |
| Androgen receptor | p300, P/CAF, Tip60 | SIRT1, HDAC1 | K630, K632, K633 | [7,18] |
| Steroidogenic factor 1 | p300, GCN5 | K102, K105, K106 | [19,20] |
1.2.1. Estrogen receptor
The estrogen receptor α (ERα) is a nuclear receptor involved in the regulation of development and reproduction and plays a role in disease, such as breast cancer, osteoporosis, cardiovascular, and Alzheimer’s diseases. This receptor has two activation function domains, AF-1 and AF-2. The N-terminus, located of AF-1 has a constitutive activity that is independent of ligand and is regulated by p300/CBP, p68 RNA helicase A, and MAPKs [21-23]. The activation of the AF-2 domain is dependent on the presence of a ligand. Upon ligand binding the receptor recruits coactivator proteins, such as the p160 family (SRC1, TIF2/GRIP1, AIB1/ACTR), as well as p300, CBP, and P/CAF [24,25]. Acetylation of the ERα has been shown both in vitro and in vivo. p300-mediated acetylation at lysine residues occurs within the hinge domain of the ERα [8]. Substitution of lysine amino acids with arginine or glutamine in this region leads to enhanced ligand (E2)-dependent function, suggesting that the ERα acetylation represses its transcriptional activity [8].
Since our group’s identification of K302/302R mutation as crucial acetylation sites, the function of ERα and its regulation by acetylation at this site has been investigated by many other groups. Yukun et al. have initially shown the significance of K303 in relation to a known phosphorylation site on S305 by PKAs on ERα. Mutation of the S305 residue, which mimics a constitutively phosphorylated receptor, inhibits acetylation at K303 thereby blocking the inhibition of ERα by acetylation [26]. More recently, a publication by the same group showed that the metastasis associated protein (MTA2) participates in deacetylation of ERα through association with histone deacetylases. This further emphasizes the importance of inhibiting RRα activity via ERα acetylation at K303 [27]. Lysine residue K302 has been shown to function as a site for monoubiquitination by BRCA1 in vitro [28]. ERα with single or double point mutations at K302/303 is resistant to BRCA1-mediated ubiquitination [28]. Furthermore, breast cancer cells expressing K303R mutation convey resistance to an aromatase inhibitor via altered activation of the PI3K/Akt kinase pathway [29].
Additional ERα lysine residues can be acetylated in vitro using p300 and SRC1 as an enzyme source [16]. These lysine residues (K266,268) do not resemble the conserved acetylation motif easily identifiable within distinct NRs studied by others (Table 1). Mutation at these two lysines does not alter neither the ligand, SRC2 coactivator binding, nor subcellular distribution. The ERα acetylation increases DNA binding activity, while the acetylation mimic mutant (K266/268Q) exhibits enhanced transactivation [16]. The lack of a similar lysine motif in other NRs raises the possibility that ERα (K266/268) may be a unique characteristic of ERα.
1.2.2. Androgen receptor
The androgen receptor (AR) plays an important role in the development of secondary sexual characteristics. The androgen receptor is a key regulator of prostate cancer onset and progression. The activity of this receptor can be regulated through both ligand-dependent and ligand-independent mechanisms. Several coactivators are known to enhance AR’s activity including SRC coactivators, p300/CBP, Ubc9, ARA70, ARA55, and TIP60 [30]. Conversely, there are a number of corepressors that down-regulate its activity, such as NAD-dependent HDAC, SIRT1 [31,32], and the cell fate determination factor DACH1 [33]. Acetylation and phosphorylation affect AR activity through modification of local chromatin, as well as through modification of the AR protein [34,35].
The AR is directly acetylated at lysine residues within the hinge domain, located in proximity to the second zinc finger of the DNA binding domain (DBD) [7]. The point mutations of lysine residues on the AR (K630, 632, 633A) abrogate the p300-mediated regulation and reduce ligand-induced activity, revealing receptor acetylation as a key step in ligand-dependent receptor activation [7]. The lysine to glutamine substitution (K630Q), which mimics acetylated lysines, increased ligand-induced transcriptional activity compared to wild-type AR. This mutant also exhibits enhanced activity at lower concentrations of ligand when compared to the wild-type AR and a relative resistance to the antagonist, flutamide [34].
The significance of AR acetylation was investigated in detail initially by our group and later by others. Mutation of the AR acetylation site abrogated MEKK1-induced apoptosis, DHT-response, and regulation by coactivators that include SRC1, p300, Ubc9, and TIP60 [34]. Moreover, AR acetylation mutants K630Q and K630T, which mimic the constitutively acetylated receptor, enhanced p300 binding, reduced association with NCoR/HDAC/Smad3 corepressor complex, and increased AR transcriptional activity. More importantly, the AR acetylation mimics promoted prostate cancer cell growth and survival and increased transcription of AR target genes [36]. All together, these studies constitute the first piece of direct evidence that a single acetylation residue directly promotes contact independent growth in vivo. Mutation of the AR acetylation site abrogated the regulation of the receptor by the HDAC inhibitor TSA (trichostatin A) and by cAMP and AKT signaling [35]. Furthermore, mutation of the AR phosphorylation site showed a direct relationship with HDAC responsiveness, defining acetylation and phosphorylation of the AR as functionally convergent events [35].
1.2.3. Hepatocyte nuclear factor-4 (HNF-4)
HNF-4, originally classified as an orphan nuclear receptor, is an essential transcriptional regulator of hepatocyte differentiation and function. Activation of HNF-4 requires interaction with CREB-binding protein (CBP) through both the AF-1 and AF-2 domains [37]. HNF-4 is acetylated by CBP at conserved lysines 97, 99, 117 and 118 in in vitro as well as in vivo assays. Substitution of these four lysines with alanines abolishes the acetylation and HNF-4-dependent transactivation in the context of local chromatin [14]. Acetylated HNF-4 increases DNA binding ability, enhances CBP interaction, and is required for nuclear retention.
1.2.4. Steroidogenic factor 1 (SF-1)
SF-1 belongs to the orphan NR superfamily. Activation of SF-1 plays a critical role in regulating the expression of genes governing the steroidogenic pathways. Several factors with HAT activity, including p300/CBP, cAMP response element binding protein (CREB), and SRC1, regulate HNF-1 activity. Lysines 34, 38 and 72 that are within the DNA binding domain are acetylated by the acetyltransferase, GCN5 (general control of amino-acid synthesis-5) [20]. Acetylation of SF-1 at these residues increases its transactivation and stability and regulates its cytoplasmic distribution [20]. In addition to these residues located in the DNA binding domain, the lysine-rich motif also serves as an acetylation substrate, preferentially for p300 rather than P/CAF. These findings are in agreement with predictions made in our previous study and show similarities to similar to that of ERα acetylation [8]. In contrast to the previous observation in which acetylation of NRs affects transcriptional activity, acetylation of SF-1 by p300 does not alter transactivation, but rather increases SF-1 DNA binding activity and enhances localization of SF-1 to p300 foci in response to cAMP-dependent signaling [19].
1.2.5. Glucocorticoid receptor (GR)
Acetylation of the GR is dependent on ligand binding [17]. The acetylation sites of the GR are within the lysine-rich motif shared by other acetylated NRs. The mutation of lysine residues 494 and 495 abolishes GR acetylation and reduces activation of the GR-responsive gene SLPI in the presence of glucocorticoids. The anti-inflammatory effects of glucocorticoids are partly due to the inhibition of cytokines, such as GM-CSF. The acetylation mutant does not alter the repression of GR-dependent GM-CSF release upon IL-1β stimulus, indicating that GR acetylation may negatively regulate NFκB signaling. HDAC2 deacetylates the GR, which is a prerequisite for binding to the NFκB subunit, p65.
1.2.6. Thyroid hormone receptor (TR)
The TR regulates the transcription of thyroid hormone-responsive genes when stimulated by thyroid hormone, which regulates physiological and developmental processes. The TR interacts with acetyltranferases and deacetylases and functions as a heterodimer with the 9-cis-retinoic acid X receptors (RXR). Unliganded heterodimers bind to thyroid hormone response elements (TREs) and repress transcription, whereas binding of a ligand activates transcription [38]. The repressive action of unliganded receptors involves interactions with corepressors SMRT and HDACs as well as reduced levels of histone acetylation.
Treatment of cells with both thyroid hormones T3 and T4 leads to the acetylation of the TR, which is dependent on the activation of ERK1/2 MAP kinase [9,10]. The acetylation sites are mapped to the protein translocation domain (D-domain) between amino acid 128 and 142, the lysine-rich motif which is highly conserved among the NRs. Detailed mapping identified lysines 128, 132, and 134 as acetylation sites [9]. Increased acetylation of the TR upon hormone treatment was accompanied by enhanced DNA binding, while mutation of these three lysine residues (triple mutant) abolished the hormone-dependent receptor/DNA binding. These findings suggest that acetylation of TR enhances its DNA binding [9]. Acetylation of TR increases the recruitment of the ACTR coactivator to the TR-RXR hetero-complex. The triple acetylation site mutant is able to bind to the transcriptional repressor SMRT regardless of ligand, suggesting that acetylation is a key step in switching TR from an inactive to an active complex via cofactor associations. The triple acetylation site mutant impairs ligand binding and is incapable of trans-activating gene expression in the presence of a ligand.
1.2.7. Retinoid X receptors (RXRs)
RXRs function through forming homodimer or heterodimers with many NRs including RARs, PPARs, vitamin D receptor, TR, and Nur77. Both RXRα and RXRγ are acetylated by p300. Acetylation of RXRα at lysine 145 increases its DNA binding and transcriptional activity. 9-cis retinoic acid, a ligand for RXRs, enhances RXR binding to the orphan receptor Nur77. Nur77 serves as an inhibitor of RXRα acetylation, which is consistent with the observation that 9-cis retinoic acid reduces RXRα acetylation. Ligand treatment decreases the association of RXRα with p300. Although the RXRα mutant K145R remains in the nucleus, the diffuse pattern distinguishes this mutant from its wild type form, which shows a speckle-like distribution [15]. The K145R mutant, which mimics the unacetylated RXRα, does not increase DNA synthesis by BrdU incorporation in the presence of p300, suggesting that RXRα acetylation by p300 may promote cellular proliferation.
1.2.8. Liver X receptor
LXR, through heterodimer formation with RXRs, regulates cholesterol and fat metabolism in the liver [39-41]. Gene targets of LXR include the ATP-binding cassette transporter A1 (ABCA1) and SREBP-1. 9-cis-RA treatment recruits SIRT1 to the LXR response elements of these genes and enhances their transcription [42]. LXR acetylation increases as a result of SIRT1 deficiency but decreases due to overexpression of SIRT1, suggesting that LXRs are targets for SIRT1. SIRT1 deacetylates and activates LXRs through direct protein-protein interaction. Deacetylation of LXR by SIRT1 promotes LXR ubiquitination and subsequent proteosomal degradation [11].
1.2.9. Farnesoid X receptor (FXR)
Activated by bile acids, FXR regulates cholesterol and bile acid metabolism through direct regulation of gene expression in the liver and intestine [39,43]. FXR acetylated by p300 results in regulation of small heterodimer partner (SHP) gene transcription [12]. The lysine 217, located in the hinge region of FXR, is a major acetylation site targeted by p300 and SIRT1 [13]. As with ERα, mutation of FXR on lysine 217 (K217R) results in increased transactivation. The additional lysine residue (K157) is also acetylated. Mutation of the acetylation sites of FXR increases its DNA binding, while FXR acetylation inhibits it. This could be explained by the inhibitory effect of FXR acetylation on the assembly of FXR/RXRα heterodimers. SIRT1 interacts with and deacetylates FXR. FXR ligand treatment releases SIRT1 from the promoter region of Shp gene, which is the target of FXR [13]. It is worth noting that FXR acetylation is increased in a leptin-deficient ob/ob mice and chronic mouse model induced by feeding mice with western style diet. In both animal models, the percentage of acetylated FXR increased.
2. Acetylases and deacetylases
2.1. Histone Acetylases (acetyltransferases)
Historically classified as type A, histone acetyl transferases (HATs) are now known to belong to two distinct categories. Type A HATs reside in the nucleus and are known to acetylate nucleosomal histones in the context of local chromatin. Type B HATs are located in the cytoplasm with “housekeeping” role that functions to acetylate free histones in the cytoplasm. A list of known HATs and their substrates, divided into six groups, has been previously shown [44]. The GNAT (Gcn5-related N-acetyltansferase) super family includes the best characterized member, yeast Gcn5. Mutagenesis of yeast Gcn5 demonstrated an important role for HAT activity in gene transcription [45]. In mammals, P/CAF was identified through homology to Gcn5 and was found to associate with the p300 coactivator protein [46,47]. Intriguingly, P/CAF inhibited cell cycle progression and acetylated several substrates in addition to histones, including HMG17, p53 and the androgen receptor [34,48]. Tip60, the first member of the human MYST protein family (MOZ, Ybf2/Sas3, Sas2 and Tip60), conveys intrinsic HAT activity [49]. Tip60 interacts with and enhances NR transactivation [18,34,35,50,51]. MOZ (monocytic leukemia zinc finger protein) was initially identified as part of a chromosomal translocation in acute myeloid leukemia. The fusion protein, MOZ-CBP, possesses HAT activity. Nuclear receptor coactivators of the p160 family acetylate either free histones or histones located in mononucleosomes. AIB1/ACTR is amplified in human breast cancers and is induced by mitogenic signaling [52,53].
Acetylation- and deacetylation-dependent regulation of chromatin structure results in indirect regulation of gene transcription. The coactivators, CREB-binding protein (CBP) and the related functional homologue p300, regulate transcription of a number of genes through the actions that have been linked to their histone acetyltransferase activity. Acetylation facilitates the binding of transcription factors to specific DNA sequences by targeting nucleosomes bound to the promoter regions of the gene [54-56]. Acetylation also occurs on non-histone substrates including the Kruppel-like factor (EKLF) [57], the tumor suppressor p53 [58], the erythroid cell differentiation factor GATA-1 [59], NRs (Table 1), and HATs [60]. The identification of non-histone substrates of HATs (also known as FATs, factor acetyl transferase) provided important insight into the mechanism by which acetylation may directly regulate gene expression [44].
The binding of HATs to target substrates assessed by chromatin immunoprecipitation assays is quite transient [61], and coactivator-substrate interactions can lead to transcriptional attenuation [62]. The attenuation of coactivator signaling by acetylation of coactivators themselves illustrates the importance of feedback loops in controlling acetylation signaling pathways. Several studies have identified a mutation of ERα at its acetylated lysine residue (K303R) in breast cancer [63], which escapes normal acetylation and repression by corepressors and functions in a constitutively active manner [26-28].
2.2. Histone deacetylases
2.2.1. HDACs
HDACs are divided into two families: the classical HDAC family and the Sir2 family of NAD+-dependent HDACs. The classical family is further divided into two classes. Class I is comprised of HDACs 1, 2, 3, and 8, all of which exhibit high similarity to the yeast (Saccharomyces cerevisiae) transcriptional regulator RPD3. Class II, made up of HDACs 4, 5, 6, 7, 9, and 10, exhibit homology to the yeast protein HDA1. Whereas Class I HDACs are found in the nucleus and are expressed in most cell types, Class II HDACs are located in both the nucleus and cytoplasm and their expression patterns are more restricted to certain tissue types. For instance, HDAC5, HDAC7, and HDAC9 are expressed in the heart, while HDAC4, HDAC8, and HDAC9 seem to be expressed more in tumor tissues than normal tissue [36]. The recent identification of HDAC11, which more closely resembles Class I HDACs, shows certain deviation from the other member, as it does not reside in any of the known HDAC complexes, such as Sin3, NCoR, or SMRT [64,65].
2.2.2. Sirtuins
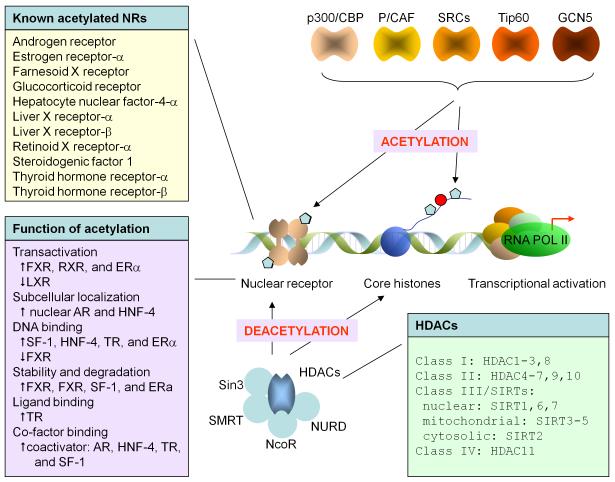
Classified as type III HDACs, the sirtuin family is a group of NAD+-dependent deacetylases conserved from archaeobacteria to eukaryotes [66]. SIRT1 is the human homolog of the yeast silent information regulator 2 (Sir2) gene, which conveys NAD+ dependent ADP-ribosyltransferase activity [67]. Sirtuins couple the removal of the acetyl group from the protein substrate with the cleavage of a high-energy bond in NAD, thus synthesizing a novel protein 2′-O-acetyl-ADP-ribose. The enzymatic activity of sirtuins is regulated by NAD+ and the endogenous nicotinamide inhibits SIRT1 activity. There are seven mammalian homologues of the yeast Sir2 gene identified with distinct functions which include three nuclear sirtuins (SIRT1, SIRT6, and SIRT7), three mitochondrial sirtuins (SIRT3, SIRT4, and SIRT5), and cytosolic SIRT2. SIRT1 deacetylates more than a dozen substrates including the NRs (AR, ERα, RXR and LXR) [11,13,16,32] (Fig. 2), the NR coactivators (PGC1, p300) [60,68], retinoblastoma protein (pRB), and several transcription factors that include p53, NFκB, and Forkhead protein. [67,69,70].
Figure 2. Post-translational modification of nuclear receptor by acetylation.
More than 10 NRs (top left panel) have been shown to undergo acetylation by acetyltransferases, including p300/CBP, P/CAF, SRCs, Tip60, and GCN5. Acetylated NRs are deacetylated by HDACs (bottom right panel). Acetylation-mediated regulation of NRs includes transactivation, subcellular localization, DNA binding, stability and degradation, ligand binding, and cofactor binding (bottom left panel)
SIRT1 inhibits androgen-dependent gene expression in both ligand- and receptor-dependent manners. The SIRT1 inhibitor, nicotinamide, induces androgen-dependent gene expression. SIRT1 inhibits contact-independent growth of cancer cells expressing AR [32,71]. The SIRT1-mediated repression of the AR is abrogated by a point mutation of the conserved acetylation motif, AR-K630T. A point mutation of the core histidine residue of SIRT1, which abolishes its deacetylase activity, abrogates the repression of androgen receptor signaling. SIRT1 interacts with and deacetylates the AR, thereby reducing the recruitment of the AR coactivators P/CAF and p300 [32]. SIRT1-dependent deacetylation of FXR decreases the receptor’s stability and correlates with deleterious metabolic outcomes, such as changes in gene expression (bile acid transporters, BSEP and MRP2), bile acid pool size, and the levels of VLDL, LDL, and HDL levels in serum [13].
3. Acetylation regulates nuclear receptor function
3.1. Activation
Ligand-mediated NR transactivation results in the transcriptional up-regulation of their target genes. Enhanced activity is seen in a point mutant substitution of the ERα acetylation sites (K303R). The lysine to arginine mutant of the FXR acetylation sites (K157,217R) increases FXR activity [13]. The acetylation of lysine residue 145 of RXRα by p300 increases the transcriptional activity in a p300-dependent manner [15]. The substitution of ERα lysine residues K302 and K303 increases the ligand sensitivity and enhances transactivation [8]. The K266/268Q, but not the K266/268R, mutant of ERα exhibits enhanced transactivation, suggesting the ERα acetylation on these residues positively regulates ERα activity [16]. Deacetylation of LXR by SIRT1 activates LXR [11], which further supports the notion that acetylation augments NR activity.
3.2. Localization
The acetylation sequences identified within the ERα, AR, GR, TR, SF-1, and HNF-4 are located at conserved motif in the hinge regions. These lysine-rich motifs are conserved among the majority of NRs (Figure 3), reinforcing the functional importance of NR acetylation. The sites of HNF-4 acetylation are located in the nuclear localization signal (NLS). Wild type HNF-4 in the presence of HAT deficient CBP mutant is located exclusively in the cytoplasm, indicating the importance of HNF-4 acetylation in subcellular localization. Non-acetylated HNF-4 retains in the cytoplasm due to the active export of HNF-4 from the nucleus [14]. Acetylation of the AR at this conserved motif is also critical to its subcellular localization and trafficking [72].
Figure 3. Alignment of NR acetylation motif showing conserved lysine residues.
A star (*) indicates the NR that is acetylated. The NR in bold shows that the acetylation occurs on the conserved lysine residue(s) [8].
3.3. DNA binding
Acetylated lysine residues have been identified in the DNA binding domain of SF-1, ERα, and FXR. Mutation of the KQQKK motif (lysine to alanine) of SF-1 reduces DNA binding activity [19], whereas the ERα acetylation mutant (K266,268Q) and the FXR mutant (K157,217R), show enhanced DNA binding [13,16]. Acetylation of the TR and HNF-4 increases DNA binding although these receptors show no acetylation site in their DNA binding domains [14].
Several studies have been conducted to determine the effect of the NR acetylation on DNA binding in the context of local chromatin using chromatin immunoprecipitation (ChIP) assays. The AR acetylation site regulates the sensitivity and specificity of cofactor recruitment. HDAC inhibition by TSA enhances AR occupancy and acetylation mimics show increased recruitment to canonical AREs site in ChIP assays. The AR acetylation site also regulates specificity of promoter recruitment. The enhanced growth associated with the AR acetylation mimic mutant may be partly due to acetylated receptors being recruited to the promoters of target genes involved in cell growth [73]. In prostate cancer cell lines expressing the activating acetylation site mutation of AR, both cyclin D1 and cyclin E protein levels and activity is increased. In ChIP assays, the acetylation mimic AR mutant shows increased recruitment to the CCND1 gene [74].
3.4. Stability and degradation
The unacetylated FXR with an arginine substitution for the acetylated lysine residues is less stable than wild type form. SIRT1 expression enhances the degradation rate of FXR, suggesting FXR acetylation may stabilize the receptor [13]. Inhibition of SIRT1 with nicotinamide increases LXRα protein levels. The Sirt1 gene deficient mice exhibit a significant increase in LXRα protein expression although the levels of mRNA of LXRα exhibit no change [11]. The SIRT1-dependence of LXRα protein ubiquitination suggests that acetylation prevents LXRα from ubiquitin-mediated degradation. The alanine substitution of ERα acetylation lysines (K320 and 303A), previously identified by our group, displays enhanced basal turnover and rapid poly-ubiquitination [75]. Compared to the wild type, this mutant had increased binding activity to the carboxyl terminus of the HSC70-interacting protein and BAG1 (Bcl-2 associated athanogene 1) [75]. It seems that acetylation of SF-1 also stabilizes the receptor, the HDAC inhibition significantly increases SF-1 protein abundance [20].
3.5. Ligand binding
The acetylation mutant of the TR is defective in both coactivator recruitment and corepressor dissociation. Although the wild type TR binds to T3 ligand efficiently, the acetylation mutant exhibits no detectable binding activity [10]. ERα and AR acetylation does not change the ligand binding affinity [16].
3.6. Interaction with other proteins
The AR acetylation site is a key regulator of coactivator and corepressor recruitment. The acetylation-dead mutants AR K630A and K632/633A are selectively defective in DHT-induced transactivation of androgen-responsive genes and coactivation by SRC1, Ubc9, TIP60, and p300 [18,34]. These mutant receptors exhibit a 10-fold increase in NCoR binding when compared to the wild-type. The AR acetylation gain-of-function mutant (K630Q and K630T) displays enhanced binding to the p300 coactivator and reduced NCoR/HDAC/Smad3 binding. These observations suggest that the acetylated residues provide a “docking” site to the AR for both corepressors and coactivators. Acetylation of HNF-4 increases its affinity for binding CBP, the acetylase for HNF-4 [14]. TRα-associated coactivator ACTR increases upon ligand T3 treatment, while the acetylation mutant of TRα has defective binding to ACTR in the presence of T3 [9]. The T3 receptor ligand releases SMRT from the wild type but not from the TR acetylation mutant, suggesting that acetylation contributes to cofactor binding. GR deacetylation by HDAC2 is a prerequisite for NFκB subunit p65 binding [17]. SF-1 acetylation may contribute to coactivator binding given the factors that the colocalization of SF-1 with p300 in nuclear foci is enhanced by cAMP and cAMP increases SF-1 acetylation.
3.7. Acetylation and other forms of post-translational modification
The lysine residue can be modified by acetylation, methylation, sumoylation, ubiquitination, and neddylation. Acetylation of NRs at conserved lysine residues raises the possibility that acetylation competes with other post-translational modifications to regulate NR action. Phosphorylation on different amino acid residue also affects acetylation. This interaction has been clearly demonstrated for the p300 coactivator, as the p300 acetylation site in the CRD domain is also the site of sumoylation [60]. The serine residue S305 of ERα, a potential protein kinase A (PKA) phosphorylation site, is located adjacent to the acetylation site K303. Mutation of this serine to aspartic acid (S305D) to mimic constitutive phosphorylation blocks acetylation of K303 [26].
4. Deregulation of nuclear receptor acetylation in diseases
4.1. NR acetylation and cancer progression
The AR acetylation site (K630T mutation) and the ERα acetylation site (K303R mutation) have been identified in human prostate and breast cancer, respectively. The ERα K303 is known to be regulated in >30% of atypical breast hyperplasia [63]. The AR acetylation mimic mutant enhances cellular proliferation and evades apoptosis mediated via MEKK1 and JNK. Similarly, the ERα acetylation mutant exhibits stimulation by sub-physiological levels of estradiol [8] and the MCF-7 cells expressing the ERα K303R mutant conveyed a growth advantage at low concentrations of estradiol (1 pM) [63]. This mutation also escapes the repression by metastasis associated protein 2 (MTA-2) and BRCA1 [27,28], suggesting the ERα K303R mutation may play a role in cancer progression. TRα inhibits oncogene Ras-induced cellular transformation; however, the TRα acetylation mutant fails to inhibit Ras-induced NIH-3T3 cell transformation [10].
4.2. NR acetylation and metabolic diseases
Both LXRs and FXR belong to the Liver X receptor-like group in the thyroid hormone receptor-like subfamily of NRs. LXRs are important regulators of cholesterol, fatty acid, and glucose homeostasis [39]. The bile acid receptor FXR plays a critical role in bile acid synthesis, lipid and lipoprotein metabolism, and glucose homeostasis [39,43]. FXR deficiency causes increased serum glucose and impaired glucose and insulin tolerance, leading to severe fatty liver development and an increase in circulating free fatty acids [76]. p300/CBP enhances FXR transactivation partially through direct interaction and acetylation [12,13]. In mouse models of metabolic diseases, increased p300 and reduced SIRT1 binding to FXR were observed, consistent with the elevated FXR acetylation levels in obese and diabetic ob/ob mice and western-style diet fed mice [13]. SIRT1 deacetylation of LXRs activates the receptor by reducing its ubiquitination and degradation [13]. The expression levels of LXR target genes are decreased in SIRT1-deficient cells and tissues. Given the importance of this NR subfamily in regulating metabolism, it will be of interest to determine whether PPARs and RARs are acetylated.
5. Targeting nuclear receptor acetylation in the clinical setting
NRs are critical for normal development and homeostasis. Deregulation of NR-mediated signaling pathways and receptor expression can lead to disease progression. For example, increased ERα or AR expression levels are associated with both the progression and metastasis of breast and prostate cancer. It is conceivable that pharmacological manipulation of NR acetylation could represent an alternative therapeutic approach in abrogating aberrant nuclear NR signaling pathways.
HDAC inhibitors have been shown to decrease cell growth in breast cancer cell lines. This inhibition appears to be related to the ERα status of the cancer [77,78]. Several genes encoding NR coactivators, including SRCs and AIB1, are overexpressed and/or amplified in cancers, which can lead to enhanced cell growth [79-81].
Resveratrol, naturally found in red grapes and pathogen-infected plants, has diverse biological effects, [82] including antioxidant properties; inhibition of mitochondrial ATPases and competition with coenzyme Q. Resveratrol functions as a SIRT1 activator [83] and as an agonist for the ERα [84]. Resveratrol has beneficial effect on metabolic outcome, partly via reduced PGC-1 acetylation [85]. Resveratrol has chemo-preventative properties and inhibits pre-neoplastic mammary gland lesions [86], although the role of resveratrol in breast cancer cellular proliferation is still under debate [87,88]. Further clarification is necessary to determine whether resveratrol functions through SIRT1 in regulating NR activity and whether this agent could have a clinical benefit for patients with cancer and metabolic diseases.
6. Conclusions and perspectives
NR and coactivator acetylation plays a key role in cellular growth control. As a single residue substitution of the AR and ERα acetylation site can promote contact-independent growth, these residues represent practical targets for intervention. SIRT1 transduction of AR expressing prostate cancer cells blocks contact-independent growth almost completely [32]. SIRT1 inhibits the activity of the mutant AR that arises in patients who have failed androgen ablation therapy. Therapies that target SIRT1-dependent interaction with the AR acetylation site may provide novel approaches for patients who exhibit resistance to the therapies that are currently available. In addition, acetylation of NR affects receptor stability, which raises the possibility that acetylation may regulate proteasome degradation of NR. Given the evidence that both acetylation and ubiquitination occur at the same lysine residue in FXR, ERα and SF1 [13,20,75], further investigation in this field should address the functional interaction between acetylation and ubiquitination-mediated NR degradation in normal development and diseases. This is important because proteasome-mediated degradation is coupled to the transactivation of NR [89,90], which is often de-regulated in diseases, such as cancer.
Acknowledgements
This work was supported by grants from National Institute of Health [R01CA70896, R01CA75503, and R01CA86072 to R.G.P.]. Work conducted at the Kimmel Cancer Center was supported by the NIH Cancer Center Core grant [P30CA56036 to R.G.P.]. This project is funded in part by the Dr. Ralph and Marian C. Falk Medical Research Trust and a grant from Pennsylvania Department of Health (to R.G.P. and C.W.). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Novac N, Heinzel T. Nuclear receptors: overview and classification. Curr Drug Targets Inflamm Allergy. 2004;3:335–46. doi: 10.2174/1568010042634541. [DOI] [PubMed] [Google Scholar]
- [3].Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–60. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- [4].Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–51. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor coactivator (SRC) family. Nat Rev Cancer. 2009;9:615–30. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Popov VM, Zhou J, Shirley LA, Quong J, Yeow WS, Wright JA, Wu K, Rui H, Vadlamudi RK, Jiang J, Kumar R, Wang C, Pestell RG. The cell fate determination factor DACH1 is expressed in estrogen receptor-alpha-positive breast cancer and represses estrogen receptor-alpha signaling. Cancer Res. 2009;69:5752–60. doi: 10.1158/0008-5472.CAN-08-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–60. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- [8].Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–83. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- [9].Sanchez-Pacheco A, Martinez-Iglesias O, Mendez-Pertuz M, Aranda A. Residues K128, 132, and 134 in the thyroid hormone receptor-alpha are essential for receptor acetylation and activity. Endocrinology. 2009;150:5143–52. doi: 10.1210/en.2009-0117. [DOI] [PubMed] [Google Scholar]
- [10].Lin HY, Hopkins R, Cao HJ, Tang HY, Alexander C, Davis FB, Davis PJ. Acetylation of nuclear hormone receptor superfamily members: thyroid hormone causes acetylation of its own receptor by a mitogen-activated protein kinase-dependent mechanism. Steroids. 2005;70:444–9. doi: 10.1016/j.steroids.2005.03.001. [DOI] [PubMed] [Google Scholar]
- [11].Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- [12].Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008;283:35086–95. doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–51. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- [15].Zhao WX, Tian M, Zhao BX, Li GD, Liu B, Zhan YY, Chen HZ, Wu Q. Orphan receptor TR3 attenuates the p300-induced acetylation of retinoid X receptor-alpha. Mol Endocrinol. 2007;21:2877–89. doi: 10.1210/me.2007-0107. [DOI] [PubMed] [Google Scholar]
- [16].Kim MY, Woo EM, Chong YTE, Homenko DR, Kraus WL. Acetylation of estrogen receptor a by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activies of the receptor. Molecular Endocrinology. 2006;20:1479–1493. doi: 10.1210/me.2005-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904–13. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- [19].Chen WY, Juan LJ, Chung BC. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol Cell Biol. 2005;25:10442–53. doi: 10.1128/MCB.25.23.10442-10453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jacob AL, Lund J, Martinez P, Hedin L. Acetylation of Steroidogenic Factor 1 Protein Regulates Its transcriptional Activity and Recruits the Coactivator GCN5. J. Biol Chem. 2001;276:37659–37664. doi: 10.1074/jbc.M104427200. [DOI] [PubMed] [Google Scholar]
- [21].Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–72. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [22].Greger JG, Fursov N, Cooch N, McLarney S, Freedman LP, Edwards DP, Cheskis BJ. Phosphorylation of MNAR promotes estrogen activation of phosphatidylinositol 3-kinase. Mol Cell Biol. 2007;27:1904–13. doi: 10.1128/MCB.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [23].Kobayashi Y, Kitamoto T, Masuhiro Y, Watanabe M, Kase T, Metzger D, Yanagisawa J, Kato S. p300 Mediates Functional Synergism between AF-1 and AF-2 of Estrogen Receptor and by Interacting Directly with the N-terminal A/B Domains. J. Biol. Chem. 2000:15645–15651. doi: 10.1074/jbc.M000042200. [DOI] [PubMed] [Google Scholar]
- [24].Edwards DP. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J Mammary Gland Biol Neoplasia. 2000;5:307–24. doi: 10.1023/a:1009503029176. [DOI] [PubMed] [Google Scholar]
- [25].Ratajczak T. Protein coregulators that mediate estrogen receptor function. Reprod Fertil Dev. 2001;13:221–9. doi: 10.1071/rd01023. [DOI] [PubMed] [Google Scholar]
- [26].Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64:9199–208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- [27].Cui Y, Niu A, Pestell R, Kumar R, Curran EM, Liu Y, Fuqua SA. Metastasis-Associated Protein 2 is a Repressor of Estrogen Receptor {alpha} Whose Overexpression Leads to Estrogen-Independent Growth of Human Breast Cancer Cells. Mol Endocrinol. 2006 doi: 10.1210/me.2005-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, Pestell RG, Tomita YA, Rosen EM. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barone I, Cui Y, Herynk MH, Corona-Rodriguez A, Giordano C, Selever J, Beyer A, Ando S, Fuqua SA. Expression of the K303R estrogen receptor-alpha breast cancer mutation induces resistance to an aromatase inhibitor via addiction to the PI3K/Akt kinase pathway. Cancer Res. 2009;69:4724–32. doi: 10.1158/0008-5472.CAN-08-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Culig Z, Comuzzi B, Steiner H, Bartsch G, Hobisch A. Expression and function of androgen receptor coactivators in prostate cancer. J Steroid Biochem Mol Biol. 2004;92:265–71. doi: 10.1016/j.jsbmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [31].Wang L, Hsu CL, Chang C. Androgen receptor corepressors: an overview. Prostate. 2005;63:117–30. doi: 10.1002/pros.20170. [DOI] [PubMed] [Google Scholar]
- [32].Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–35. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu K, Katiyar S, Witkiewicz A, Li A, McCue P, Song LN, Tian L, Jin M, Pestell RG. The cell fate determination factor dachshund inhibits androgen receptor signaling and prostate cancer cellular growth. Cancer Res. 2009;69:3347–55. doi: 10.1158/0008-5472.CAN-08-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fu M, Wang C, Wang J, Zhang X, Sakamaki T, Yeung YG, Chang C, Hopp T, Fuqua SA, Jaffray E, Hay RT, Palvimo JJ, Janne OA, Pestell RG. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol Cell Biol. 2002;22:3373–88. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fu M, Rao M, Wu K, Wang C, Zhang X, Hessien M, Yeung YG, Gioeli D, Weber MJ, Pestell RG. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J Biol Chem. 2004;279:29436–49. doi: 10.1074/jbc.M313466200. [DOI] [PubMed] [Google Scholar]
- [36].Fu M, Rao M, Wang C, Sakamaki T, Wang J, Di Vizio D, Zhang X, Albanese C, Balk S, Chang C, Fan S, Rosen E, Palvimo JJ, Janne OA, Muratoglu S, Avantaggiati ML, Pestell RG. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol. 2003;23:8563–75. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–21. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- [38].Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a corepressor. Nature. 1995;377:451–4. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- [39].Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–91. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- [40].Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–4. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–14. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Feige JN, Auwerx J. DisSIRTing on LXR and cholesterol metabolism. Cell Metab. 2007;6:343–5. doi: 10.1016/j.cmet.2007.10.003. [DOI] [PubMed] [Google Scholar]
- [43].Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020–30. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- [44].Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Histone acetylation and the cell-cycle in cancer. Frontiers in Bioscience. 2001;6:D610–29. doi: 10.2741/1wang1. [DOI] [PubMed] [Google Scholar]
- [45].Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5 functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes and Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Reid JL, Bannister AJ, Zegerman P, Martinez-Balbas MA, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–77. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vassilev A, Yanauchi, Kotani T, Prives C, Avantaggiati ML, Qi J, Nakatani Y. The 400 kDa subunit of the P/CAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- [48].Reutens AT, Fu M, Watanabe G, Albanese C, McPhaul MJ, Balk SP, Janne OA, Palvimo JJ, Pestell RG. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- [49].Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- [50].Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN. Tip60 Is a Nuclear Hormone Receptor Coactivator. J. Biol. Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- [51].Gaughan L, Brady ME, Cook S, Neal DE, Robson CN. Tip60 is a coactivator specific for class I nuclear hormone receptors. J Biol Chem. 2001;276:46841–8. doi: 10.1074/jbc.M103710200. [DOI] [PubMed] [Google Scholar]
- [52].Bautista S, Valles H, Walker RL, Anzick S, Zeillinger R, Meltzer P, Theillet C. In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity. Clin Cancer Res. 1998;4:2925–9. [PubMed] [Google Scholar]
- [53].Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–71. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Herrera JE, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–73. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- [56].Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- [57].Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- [59].Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–8. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- [60].Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of P300 involves lysine residues 1020/1024 within the cell-cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- [61].Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- [62].Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- [63].Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O’Connell P, Allred DC. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60:4026–9. [PubMed] [Google Scholar]
- [64].Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, Pinilla-Ibarz J, Seto E, Sotomayor EM. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–55. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- [66].Kruszewski M, Szumiel I. Sirtuins (histone deacetylases III) in the cellular response to DNA damage--facts and hypotheses. DNA Repair (Amst) 2005;4:1306–13. doi: 10.1016/j.dnarep.2005.06.013. [DOI] [PubMed] [Google Scholar]
- [67].Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- [68].Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- [69].Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Van Der Horst A, Tertoolen LG, De Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2SIRT1. J Biol Chem. 2004 doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- [71].Dai Y, Ngo D, Forman LW, Qin DC, Jacob J, Faller DV. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–21. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Thomas M, Dadgar N, Aphale A, Harrell JM, Kunkel R, Pratt WB, Lieberman AP. Androgen receptor acetylation site mutations cause trafficking defects, misfolding, and aggregation similar to expanded glutamine tracts. J Biol Chem. 2004;279:8389–95. doi: 10.1074/jbc.M311761200. [DOI] [PubMed] [Google Scholar]
- [73].Fu M, Wang C, Zhang X, Pestell RG. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- [74].Fu M, Rao M, Bouras T, Wang C, Wu K, Jiao X, Zhang X, Li Z, Rajnarayanan RV, Pattabiraman N, Yao T-P, Pestell RG. Recruitment of HDACs by Cyclin D1 Inhibits PPAR-gama Mediated Adipogenesis. J. Biol Chem. 2004;279:24745–24756. [Google Scholar]
- [75].Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005;19:2671–84. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- [76].Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–44. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- [77].Duong V, Licznar A, Margueron R, Boulle N, Busson M, Lacroix M, Katzenellenbogen BS, Cavailles V, Lazennec G. ERalpha and ERbeta expression and transcriptional activity are differentially regulated by HDAC inhibitors. Oncogene. 2006;25:1799–806. doi: 10.1038/sj.onc.1209102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Margueron R, Duong V, Bonnet S, Escande A, Vignon F, Balaguer P, Cavailles V. Histone deacetylase inhibition and estrogen receptor alpha levels modulate the transcriptional activity of partial antiestrogens. J Mol Endocrinol. 2004;32:583–94. doi: 10.1677/jme.0.0320583. [DOI] [PubMed] [Google Scholar]
- [79].Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- [80].Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, Ittmann M, Tsai SY, Tsai MJ. SRC3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–83. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- [81].Zhou G, Hashimoto Y, Kwak I, Tsai SY, Tsai MJ. Role of the steroid receptor coactivator SRC3 in cell growth. Mol Cell Biol. 2003;23:7742–55. doi: 10.1128/MCB.23.21.7742-7755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Granados-Soto V. Pleiotropic effects of resveratrol. Drug News Perspect. 2003;16:299–307. doi: 10.1358/dnp.2003.16.5.829318. [DOI] [PubMed] [Google Scholar]
- [83].Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- [84].Lin JN, Lin VC, Rau KM, Shieh PC, Kuo DH, Shieh JC, Chen WJ, Tsai SC, Way TD. Resveratrol modulates tumor cell proliferation and protein translation via SIRT1-dependent AMPK activation. J Agric Food Chem. 2010;58:1584–92. doi: 10.1021/jf9035782. [DOI] [PubMed] [Google Scholar]
- [85].Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- [86].Le Corre L, Chalabi N, Delort L, Bignon YJ, Bernard-Gallon DJ. Resveratrol and breast cancer chemoprevention: molecular mechanisms. Mol Nutr Food Res. 2005;49:462–71. doi: 10.1002/mnfr.200400094. [DOI] [PubMed] [Google Scholar]
- [87].Bowers JL, Tyulmenkov VV, Jernigan SC, Klinge CM. Resveratrol acts as a mixed agonist/antagonist for estrogen receptors alpha and beta. Endocrinology. 2000;141:3657–67. doi: 10.1210/endo.141.10.7721. [DOI] [PubMed] [Google Scholar]
- [88].Bhat KP, Lantvit D, Christov K, Mehta RG, Moon RC, Pezzuto JM. Estrogenic and antiestrogenic properties of resveratrol in mammary tumor models. Cancer Res. 2001;61:7456–63. [PubMed] [Google Scholar]
- [89].Dennis AP, O’Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–51. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- [90].Dennis AP, Lonard DM, Nawaz Z, O’Malley BW. Inhibition of the 26S proteasome blocks progesterone receptor-dependent transcription through failed recruitment of RNA polymerase II. J Steroid Biochem Mol Biol. 2005;94:337–46. doi: 10.1016/j.jsbmb.2004.11.009. [DOI] [PubMed] [Google Scholar]