Abstract
The 3-phosphoinositide-dependent protein kinase–1 (PDK1) phosphorylates and activates a number of protein kinases of the AGC subfamily. The kinase domain of PDK1 interacts with a region of protein kinase C–related kinase–2 (PRK2), termed the PDK1-interacting fragment (PIF), through a hydrophobic motif. Here we identify a hydrophobic pocket in the small lobe of the PDK1 kinase domain, separate from the ATP- and substrate-binding sites, that interacts with PIF. Mutation of residues predicted to form part of this hydrophobic pocket either abolished or significantly diminished the affinity of PDK1 for PIF. PIF increased the rate at which PDK1 phosphorylated a synthetic dodecapeptide (T308tide), corresponding to the sequences surrounding the PDK1 phosphorylation site of PKB. This peptide is a poor substrate for PDK1, but a peptide comprising T308tide fused to the PDK1-binding motif of PIF was a vastly superior substrate for PDK1. Our results suggest that the PIF-binding pocket on the kinase domain of PDK1 acts as a ‘docking site’, enabling it to interact with and enhance the phosphorylation of its substrates.
Keywords: AGC kinase/AKT/PDK1/PIF/protein kinase A
Introduction
Stimulation of cells with insulin and growth factors generates the second messengers phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] and phosphatidyl-3,4-bisphosphate [PtdIns(3,4)P2] (Leevers et al., 1999), which induce the activation of certain members of the AGC subfamily of protein kinases that include protein kinase B (PKB) (Alessi and Downes, 1998; Shepherd et al., 1998), p70 ribosomal S6 kinase (p70 S6K) (Proud, 1996; Pullen and Thomas, 1997), serum and glucocorticoid-induced kinase (SGK) (Kobayashi and Cohen, 1999; Park et al., 1999) and protein kinase C (PKC) isoforms (Mellor and Parker, 1998). These kinases can then mediate many of the effects of insulin and growth factors by phosphorylating key regulatory proteins (reviewed in Alessi and Downes, 1998; Coffer et al., 1998; Shepherd et al., 1998).
The interaction of PtdIns(3,4,5)P3 with the plekstrin homology (PH) domain of PKB causes PKB to translocate to the plasma membrane where it is activated by phosphorylation of two residues, namely Thr308 and Ser473. Both of these residues need to be phosphorylated for maximal activation, and their phosphorylation in vivo is prevented by inhibitors of phosphoinositide (PI) 3–kinase (Alessi and Downes, 1998; Shepherd et al., 1998). Thr308 lies in the activation loop of the kinase domain, while Ser473 is located C–terminal to the catalytic domain, in a region that displays high homology between different AGC family members. Importantly, p70 S6K (Pearson et al., 1995), PKC isoforms (Mellor and Parker, 1998), SGK (Kobayashi and Cohen, 1999; Park et al., 1999) and p90 ribosomal S6 kinase (Dalby et al., 1998; Frodin and Gammeltoft, 1999) also possess residues lying in sequences equivalent to Thr308 and Ser473 of PKB, whose phosphorylation is necessary for activation of these kinases in vivo. Ser473 of PKB and the equivalent residues of p70 S6K, PKC and SGK lie in a hydrophobic motif Phe-Xaa-Xaa-Phe-Ser/Thr-Phe/Tyr distinct from the sequences surrounding Thr308.
The protein kinase termed 3-phosphoinositidedependent protein kinase-1 (PDK1) plays a central role in activating AGC subfamily members (reviewed in Belham et al., 1999; Peterson and Schreiber, 1999). PDK1 phosphorylates PKB at Thr308 (Alessi et al., 1997a,b; Stokoe et al., 1997; Stephens et al., 1998) and the equivalent residues on PKC isoforms (Chou et al., 1998; Dutil et al., 1998; Le Good et al., 1998), p70 S6K (Alessi et al., 1998; Pullen et al., 1998), SGK (Kobayashi and Cohen, 1999; Park et al., 1999) and the p90 ribosomal S6 kinase (Jensen et al., 1999; Richards et al., 1999). cAMP-dependent protein kinase (PKA) is also phosphorylated by PDK1 at the equivalent residue (Thr197) and this is required for PKA activity (Cheng et al., 1998). However, unlike the other members of the AGC subfamily of protein kinases discussed above, PKA does not possess a residue equivalent to Ser473 of PKB. Instead, its amino acid sequence terminates with the sequence Phe-Xaa-Xaa-PheCOOH corresponding to the first part of the hydrophobic motif Phe-Xaa-Xaa-Phe-Ser/Thr-Phe/Tyr that surrounds Ser473. Nevertheless, this C–terminal region of PKA plays an important role as its mutation or deletion greatly diminishes activity (Etchebehere et al., 1997).
Recently, we discovered that the kinase domain of PDK1 interacts with a region of protein kinase C–related kinase-2 (PRK2) termed the PDK1-interacting fragment (PIF). In the presence of PIF, PDK1 phosphorylates not only Thr308 but also Ser473 (Balendran et al., 1999a). PIF contains a hydrophobic sequence motif (Phe-Xaa-Xaa-Phe-Asp-Tyr) similar to that found in PKB, except that the residue equivalent to Ser473 is an aspartic acid. Mutation of any of the conserved aromatic residues in this motif or mutation of the aspartic acid residue to either alanine or serine greatly weakens the interaction of PIF with PDK1, indicating that PIF binds to PDK1 via these residues (Balendran et al., 1999a).
In contrast to PKB, whose activation by PDK1 is enhanced by PIF, we found that PIF prevents PDK1 from phosphorylating p70 S6K. This suggests that in order for p70 S6K to become phosphorylated by PDK1, it might need to bind to a region of PDK1 that overlaps with the PIF-interacting site (Balendran et al., 1999b). In this study, we identify and partially characterize a hydrophobic pocket on the small lobe of the kinase domain of PDK1 that interacts with PIF and the C–terminal fragment of PKA. We also develop novel assays for PDK1 that can be exploited to identify drugs that activate or inhibit PDK1 by interacting with the binding pocket for PIF.
Results
PDK1 interacts with the C–terminal fragment of PKA
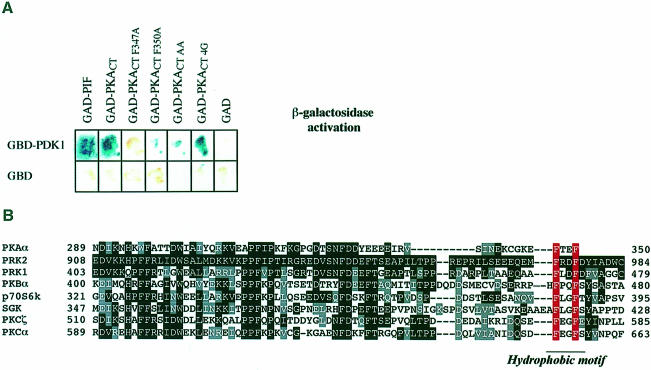
A yeast two-hybrid screen was carried out to identify proteins expressed in human brain that interact with PDK1. We identified a clone corresponding to the C–terminal 223 amino acids of PKA (termed PKACT) that yielded a positive interaction with full-length PDK1 (Figure 1A), but not with the PH domain of PDK1 (data not shown). PKACT includes part of the kinase domain as well as amino acids in the C–terminal non-catalytic region of PKA that show high sequence homology between AGC subfamily kinases (Figure 1B). The C–terminal 62 amino acids of PKA possess significant homology with PIF and terminates in the sequence motif (347-Phe-Xaa-Xaa-PheCOOH). This sequence is similar to the PDK1-interacting motif in PIF [974-Phe-Xaa-Xaa-Phe-Asp-Tyr979, numbering based on the human PRK2 sequence (Balendran et al., 1999a)], except that the aspartic acid residue is replaced by the C–terminal carboxylate group of PKA and the C–terminal tyrosine is missing. This suggested that the interaction of PKACT with PDK1 might be mediated by the C–terminal sequence 347-Phe-Xaa-Xaa-PheCOOH. The mutation of either or both of the C–terminal Phe347 and Phe350 to alanine of PKACT significantly reduced its interaction with PDK1 (Figure 1A), but the addition of four glycine residues to the C–terminus of PKACT to move the free carboxylate group to another position had no effect on the ability of PKACT to interact with PDK1 (Figure 1A). These findings indicate that both phenyl– alanine residues, but not the carboxylate group in this motif, are required for the interaction of PKACT with PDK1.
Fig. 1. Two-hybrid interaction of PDK1 and wild-type and mutant C–terminal fragment of PKA. (A) The Y190 yeast strain was transformed with vectors expressing PDK1 fused to the Gal4 DNA-binding domain (GBD), together with vectors encoding either the C–terminal 26 residues of PIF or the wild-type, or the indicated mutants of a C–terminal fragment of PKA (PKACT residues 129–350) fused to a Gal4 activation domain (GAD). As a control, yeast were also co-transformed with the GBD domain alone and the GAD domain alone. The yeast were grown overnight at 30°C and β–galactosidase filter lift assays performed at 30°C for 4 h. An interaction between GBD–PDK1 and GAD–PKACT induces the expression of β–galactosidase, which is detected as a blue colour in the filter lift assay. (B) Alignment of the amino acid sequence of the C–terminal 77 amino acids of PKA with the equivalent region of the AGC subfamily kinases indicated. Identical residues are denoted by white letters on a black background, and similar residues by grey boxes. The aromatic residues in the hydrophobic motif are indicated in red.
Identification of a putative hydrophobic pocket in the kinase domain of PDK1 that interacts with PIF
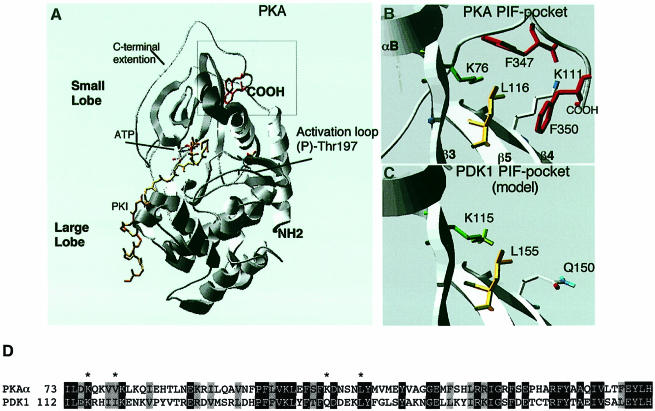
PKA was the first protein kinase whose three-dimensional structure was solved at high resolution (Knighton et al., 1991), and has established a structural framework for the catalytic domain of most protein kinases (reviewed in Taylor and Radzio-Andzelm, 1994). Analysis of the structure of PKA revealed that the non-catalytic C–terminus forms a loop that interacts with the kinase domain (Figure 2A). Most interestingly, the C–terminal residues of PKA implicated above in binding to the kinase domain of PDK1 interact with a deep hydrophobic pocket in the small lobe of the PKA catalytic domain (Figure 2B). This site does not overlap with the ATP- or peptide substrate-binding sites on PKA. The residues that make obvious hydrophobic interactions with the two phenylalanine residues in the terminal 347-Phe-Xaa-Xaa-Phe motif of PKA are Lys76, Val80, Lys111 and Leu116 of PKA (Figure 2B).
Fig. 2. C–terminal Phe-Xaa-Xaa-Phe residues of PKA interact with a hydrophobic pocket on the PKA kinase domain, predicted to be conserved in PDK1. (A) Ribbon structure of the PKA–PKI–ATP ternary complex (Zheng et al., 1993); PKI is shown in yellow, and the ATP molecule is highlighted. The C–terminal Phe347 and Phe350 are shown in red. The position of phospho-Thr197 (the PDK1 phosphorylation site) in the T-loop is indicated. (B) Detailed structure of the hydrophobic pocket on the kinase domain of PKA that interacts with the C–terminal Phe-Xaa-Xaa-Phe residues of PKA. Lys76 (equivalent of Lys115 in PDK1) is shown in green, Leu116 (equivalent of Leu155 in PDK1) is shown in yellow, Phe347 and Phe350 in red, and certain amine residues are in blue. (C) The structure of the PDK1 kinase domain was modelled as described in Materials and methods. The region of PDK1 equivalent to the hydrophobic pocket of PKA termed the PIF-binding pocket is shown. The residues predicted to be involved in binding to PIF are highlighted, including the amide residue of Q150. (D) Alignment of the amino acid residues of PDK1 around the PIF-binding pocket and the equivalent region of PKA. Identical residues are denoted by white letters on a black background, and similar residues by grey boxes. Residues on PKA that interact with the C–terminal Phe-Xaa-Xaa-Phe motif are marked with an asterisk.
A sequence alignment of the kinase domains of PDK1 and PKA indicated that the residues equivalent to Lys76 (Lys115 on PDK1) and Leu116 (Leu155 on PDK1) of PKA are conserved in PDK1 (Figure 2D). Molecular modelling of the structure of the kinase domain of PDK1 based on that of PKA confirmed that PDK1 is likely to possess a hydrophobic pocket in the equivalent region of its kinase domain, and that Lys115 and Leu155 in PDK1 are likely to lie in positions equivalent to Lys76 and Leu116 in PKA. The residues on PKA equivalent to Val80 and Lys111, which form part of the hydrophobic pocket, lie in the same position as Ile119 and Gln150, respectively, of the PDK1 kinase domain. The model of the PDK1 kinase domain indicates that these residues, as well as Lys115 and Leu155, may form part of a hydrophobic binding site (Figure 2D).
Effect of mutation of Lys115 and Leu155 on PIF binding to PDK1
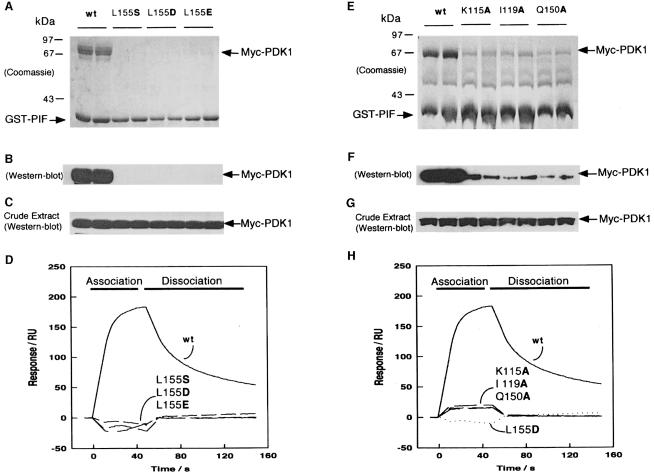
The model for the hydrophobic pocket in PDK1 predicts that Lys115 and Leu155 should participate in a hydrophobic interaction with the residues equivalent to Phe974 and Phe977 of PIF. We therefore mutated Lys115 to alanine and Leu155 to serine, aspartic acid or glutamic acid and compared the ability of these PDK1 mutants and wild-type PDK1 to interact with GST–PIF (Figure 3). As reported previously, a complex was readily observed between GST–PIF and wild-type PDK1. In contrast, the K115A interacted very poorly with PIF, whilst none of the L155 mutants interacted significantly with PIF, although these PDK1 mutants were expressed to the same level as wild-type PDK1 (Figure 3C and G).
Fig. 3. Effect of mutation of conserved residues in the PIF-binding pocket of PDK1 on the ability of PDK1 to interact with PIF. 293 cells were transiently transfected with DNA constructs expressing GST–PIF and either wild-type Myc-PDK1 or the indicated mutants of PDK1. At 36 h post-transfection, the cells were lysed and GST–PIF purified by affinity chromatography on glutathione–Sepharose beads. A 2 μg aliquot of each protein was electrophoresed on a 10% SDS–polyacrylamide gel and either stained with Coomassie Blue (A and E) or immunoblotted using an anti-Myc antibody to detect Myc-PDK1 (B and F). To establish that the wild-type PDK1 and mutant proteins were expressed at a similar level, 10 μg of total cell lysate was electrophoresed on a 10% SDS–polyacrylamide gel and immunoblotted using anti-Myc antibodies (C and G). Duplicates of each condition are shown. Similar results were obtained in three to five separate experiments. (D and H) Surface plasmon resonance measurements were carried out on a BiaCore instrument as described in Materials and methods to measure the interaction of wild-type and mutant GST–PDK1 preparations with the 24 residue synthetic peptide whose sequence encompasses the PDK1-binding site on PIF termed PIFtide (Balendran et al., 1999a). PIFtide was immobilized on an SA SensorChip, and wild-type (wt) or the indicated mutants of PDK1 were injected at a concentration of 40 nM. All data are single determinations from a representative experiment that was repeated at least three times with similar results. For clarity, the bulk refractive index changes associated with the first and last 10 s of the injection have been removed.
Surface plasmon resonance (SPR) measurements confirmed a high affinity interaction between wild-type GST–PDK1 and immobilized biotinylated synthetic peptide termed PIFtide, whose sequence encompasses the PDK1-binding site, as reported previously (Balendran et al., 1999a). However, the L155S, L155D and L155E mutants of PDK1 had no detectable affinity for PIFtide (Figure 3D), whilst the K115A mutant interacted weakly with PIFtide (Figure 3D).
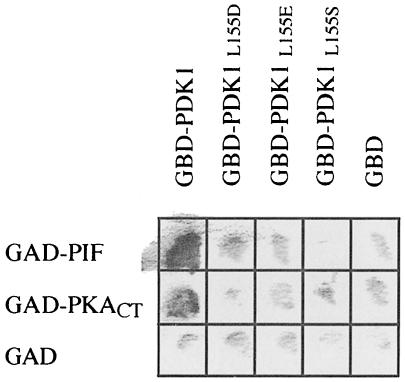
Yeast two-hybrid analysis also confirmed that the L155S, L155D and L155E mutants of PDK1 failed to interact with PIF (Figure 4). Furthermore, the interaction of PKACT with the L155S, L155D or L155E mutants of PDK1 was greatly reduced in a yeast two-hybrid binding assay, further suggesting that the C–terminus of PKA interacts with the PDK1 catalytic domain at the same site as PIF (Figure 4).
Fig. 4. Leu155 mutants of PDK1 do not interact with either PIF or the C–terminal fragment of PKA in the two-hybrid system. The Y190 yeast strain was transformed with vectors expressing the wild-type PDK1 or the indicated mutants of PDK1 fused to the Gal4 DNA-binding domain (GBD) together with vectors encoding the expression of either the 26 C–terminal residues of PRK2 (PIF) or the C–terminal fragment of PKA (PKACT residues 129–350) fused to a Gal4 activation domain (GAD). As a control, yeast were also co-transformed with vectors expressing the GAD and GBD domains only. The yeast were grown overnight at 30°C and β–galactosidase filter lift assays performed at 30°C for 4 h. An interaction between GBD–PDK1 and either GAD–PIF or GAD–PKACT induces the expression of β–galactosidase, which is detected as a blue colour (shown as black in the figure) in the filter lift assay.
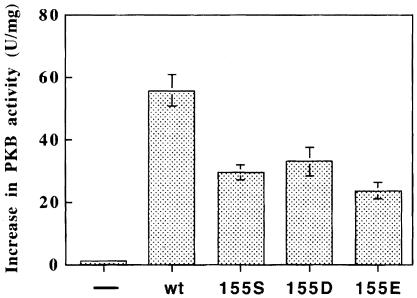
The K115A, L155S, L155D and L155E mutants of PDK1 were 50–60% as efficient as wild-type PDK1 in activating GST–473D-PKBα in the presence of MgATP and PtdIns(3,4,5)P3 (Figure 5). This indicated that the conformation of the active site of PDK1 was not significantly impaired by these mutations.
Fig. 5. Phosphorylation of Thr308 of PKB by wild-type and PIF-binding pocket mutants of PDK1. Wild-type or mutant forms of GST–PDK1 were expressed in 293 cells and purified by affinity chromatography on glutathione–Sepharose beads. Each GST fusion protein (0.2 ng) was incubated for 30 min at 30°C with GST–S473D-PKBα and MgATP in the presence or absence of phospholipid vesicles containing 100 μM phosphatidycholine, 100 μM phosphatidylserine and 10 μM sn-1-stearoyl-2-arachidonoyl-d-PtdIns(3,4,5)P3, and the increase in specific activity of GST–S473D-PKBα was determined relative to a control incubation in which the PDK1 was omitted (average of six determinations, three independent experiments). The basal activity of GST–S473D-PKBα was 1.5 U/mg. Under the conditions used, it was verified that the activation of GST–473D-PKBα was proportional to the amount of PDK1 added to the assay (data not shown). ‘–’ indicates that PDK1 was omitted.
Effect of mutation of Ile119 and Gln150 on PIF binding to PDK1
Ile119 and Gln150, which are also predicted to form part of the PIF-binding pocket on the small lobe of the PDK1 kinase domain, were mutated to alanine. In both pull-down (Figure 3E and F) and SPR experiments (Figure 3H), the I119A and Q150A mutants of PDK1 interacted very weakly with PIF compared with wild-type PDK1. These mutants also activated a GST–473D-PKBα at 60–70% of the rate of wild-type PDK1 (data not shown).
Effect of PIF on the catalytic activity of PDK1 towards a peptide substrate
A recent study by Dong et al. (1999) demonstrated that PDK1 can phosphorylate a synthetic peptide KT*FCG– TPEYLAPEV-RR, here termed T308tide, whose sequence encompasses residues 307–320 of PKBα with two arginine residues added to the C–terminus to make the peptide bind to P81 paper. As it is unlikely that T308tide would interact with the PIF-binding pocket of PDK1, we decided to use this substrate to investigate the effect of PIF binding on the catalytic activity of PDK1. We confirmed that T308tide was phosphorylated in vitro by PDK1 although the Km was very high (>10 mM). We also established that T308tide was phosphorylated at the residue equivalent to Thr308 of PKBα (indicated by an asterisk), by solid phase sequencing of 32P-labelled T308tide phosphorylated by PDK1 (data not shown).
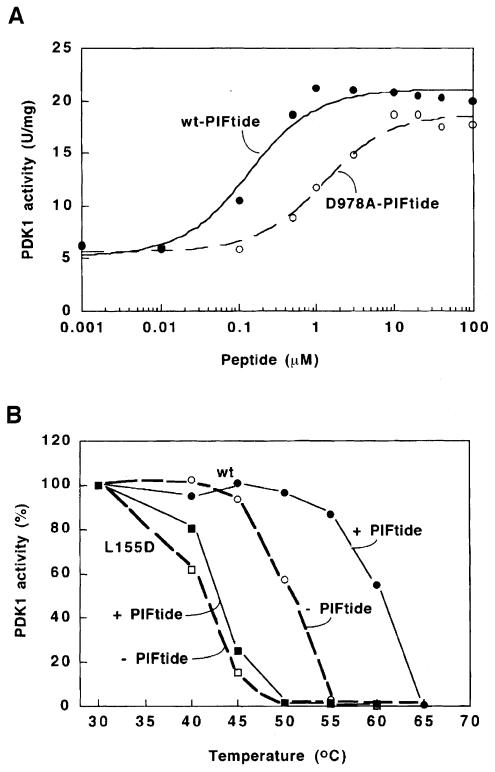
PDK1 activity towards T308tide was increased up to 4–fold in the presence of PIFtide. The concentration required for half-maximal activation was 0.14 μM (Figure 5A) which correlates with the affinity of PDK1 for PIFtide (Kd of ∼0.3 μM; Balendran et al., 1999a). This increase in PDK1 activity was observed with either full-length PDK1 or forms lacking the N– or C–terminal non-catalytic regions (data not shown). The effects of PIFtide on PDK1 activity were unaffected by pre-incubating these components for up to 30 min on ice prior to initiating the assay. Similarly, a mutant D978A-PIFtide, which exhibits a 10–fold reduced affinity for PDK1 (Balendran et al., 1999a), was 8–fold less effective at inducing PDK1 activation (Figure 6A). Several unrelated peptides of similar size were unable to induce any activation of PDK1 (data not shown). This strongly indicates that PDK1 is activated directly by PIF. Furthermore, PIF did not alter the Km of PDK1 for ATP (data not shown).
Fig. 6. PDK1 is activated and stabilized through its interaction with PIFtide. (A) GST–PDK1 activity was measured in the presence of increasing concentrations of wild-type (wt) PIFtide (•) or a mutant D978A-PIFtide (○), using the synthetic peptide substrate termed T308tide, as described in Materials and methods. The data were fitted to a hyperbola using the Kaleidagraph software. The concentration needed to obtain 50% activation of PDK1 was 0.14 μM for wt-PIFtide and 1.1 μM for D978A-PIFtide. The assay shown was performed in triplicate and there was <5% difference between each assay. Similar results were obtained in two further experiments. (B) The wild-type GST–PDK1 (circles) or the L155D mutant of GST–PDK1 (squares) was incubated in the presence (closed symbols) or absence (open symbols) of 100 μM PIFtide and then heated for 2 min at the indicated temperatures, rapidly brought to 0°C (see Materials and methods), and 2 min later assayed at 30°C for 10 min using T308tide as substrate. The activity of PDK1 obtained by incubation at 30°C was taken as 100%. The assay shown was performed in duplicate with similar results obtained in two separate experiments.
GST–PDK1 activity was reduced by 50% if the enzyme was heated for 2 min at 50°C (TM50 value, Figure 6B). However, PDK1 was stabilized in the presence of wild-type PIFtide, the TM50 being increased by 8–10°C. PIF also caused a 6–10°C increase in the TM50 value for all GST–PDK1 mutants tested that lack either the PH domain, the N–terminal 51 residues or both non-catalytic domains (data not shown). The L155D mutant of GST–PDK1 was more heat labile than wild-type PDK1, with a TM50 value of 42°C. As expected, PIF did not stabilize this mutant significantly (Figure 6B).
Activity of PDK1 mutants towards T308tide
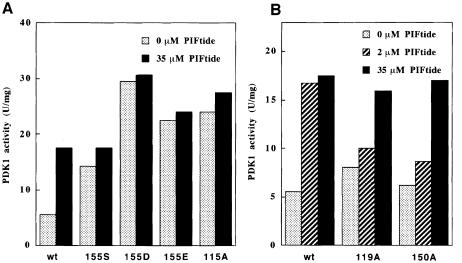
We next tested the specific activities of the PIF-binding pocket mutants towards T308tide. The K115A, L155S, L155D and L155E mutants of PDK1 phosphorylated T308tide 3- to 5–fold more rapidly than wild-type PDK1, i.e. at a rate similar to that of wild-type PDK1 in the presence of PIF (Figure 7). PIFtide did not activate these mutants further, consistent with their inability to bind PIF. In contrast, the I119A and Q150A mutants of PDK1 possessed a specific activity similar to wild-type PDK1 and were stimulated ∼2–fold in the presence of PIF. However, 10–fold more PIF was required for maximal activation compared with wild-type PDK1, consistent with the reduced affinity of these mutants for PIF (Figure 3).
Fig. 7. Effect of PIFtide on PDK1 PIF pocket mutants. Wild-type and the indicated mutants of GST–PDK1 were assayed with T308tide either in the absence (dotted bars) or the presence of 2 mM PIFtide (dashed bars) or 35 μM PIFtide (filled bars). Under the conditions used, the phosphorylation of T308tide by PDK1was linear with time (data not shown). (A) The PDK1 mutants that are activated in the absence of PIFtide and (B) those mutants that are activated by high concentrations of PIFtide. The assay was performed in triplicate with <10% difference between triplicate samples. Similar results were obtained in three separate experiments.
PDKtide is a vastly superior peptide substrate for PDK1
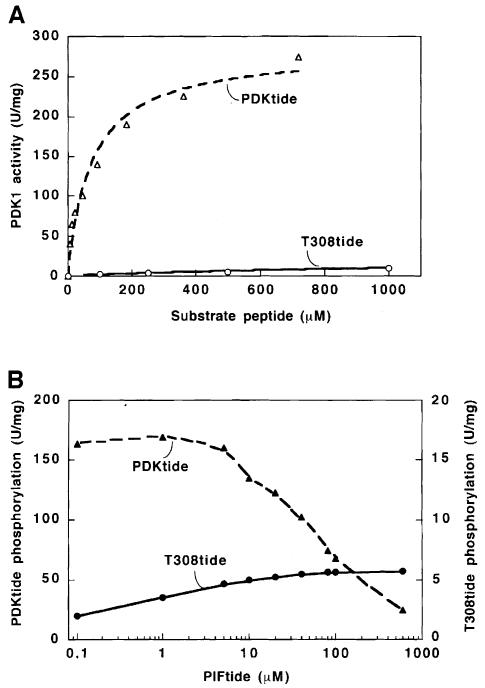
The results presented above suggested that a peptide substrate for PDK1 might be phosphorylated with a much lower Km value if it also contained the PDK1-interacting sequence of PIF. We therefore synthesized a 39 amino acid polypeptide composed of T308tide fused to PIFtide, and termed it PDKtide. This peptide was vastly superior to T308tide as a substrate for PDK1; its Km was ∼80 μM (compared with >10 mM for T308tide) and, when assayed at 100 μM, PDKtide was phosphorylated at a rate >100–fold greater than that using T308tide (Figure 8A). The activity of PDK1 towards PDKtide was inhibited by inclusion of PIFtide in the assay, in contrast to T308tide phosphorylation, which was stimulated by PIFtide (Figure 8B).
Fig. 8. PDKtide is vastly superior to T308tide as a substrate for PDK1 because it interacts with the PIF-binding pocket of PDK1. (A) His-PDK1 was assayed for activity using as substrate the indicated concentration of either PDKtide (▵) or T308tide (○). (B) His-PDK1 was assayed for activity in the presence of PDKtide (25 μM, ▴) or T308tide (100 μM, •) in the presence of the indicated concentrations of PIFtide. The assay was performed in triplicate with <5% difference between the triplicate samples. Similar results were obtained in three separate experiments
Discussion
The C–terminal residues of PKA, Phe-Xaa-Xaa-PheCOOH, correspond to part of the PDK1-binding motif of PIF. These residues are known to interact with the small lobe of the kinase domain of PKA at a location distinct from the ATP- or peptide substrate-binding sites (Figure 2). In this study, we demonstrate that PKACT also interacts with the kinase domain of PDK1 by yeast two-hybrid analysis, and that mutation to alanine of the residues in PKACT equivalent to Phe347 or Phe350 significantly reduces its interaction with PDK1 (Figure 1). As the mutation of the equivalent phenylalanine residues to alanine on PIF also abolishes its interaction with PDK1 (Balendran et al., 1999a), these findings suggested that the PKACT and PIF might interact at the same site in the PDK1 kinase domain.
The residues in the kinase domain of PKA known to interact with the C–terminus of this protein are present in PDK1, and their mutation either abolished or significantly diminished the interaction of PDK1 with both PIF and PKACT. These observations strongly suggest that PDK1 possesses an equivalent hydrophobic pocket in its kinase domain that interacts with PIF and PKACT. PDK1 is itself a member of the AGC subfamily of protein kinases but, in contrast to PKA, it does not possess a hydrophobic Phe-Xaa-Xaa-Phe motif at the equivalent position. PDK1 is therefore likely to possess an unoccupied PIF-binding pocket in its kinase domain, which is available to interact with the C–terminal hydrophobic motifs of PKA and other AGC subfamily members.
PIF enables PDK1 to phosphorylate PKBα at both Thr308 and Ser473 in a PtdIns(3,4,5)P3- or PtdIns(3,4,) P2-dependent manner (Balendran et al., 1999a). The PDK1-binding motif in PIF (Phe-Xaa-Xaa-Phe-Asp-Tyr) could therefore be required as a pseudosubstrate sequence, raising the possibility that PIF interacts with the substrate-binding site of PDK1. However, if this were the case, PIF would be expected to prevent PDK1 from phosphorylating PKBα at Ser473 rather than promoting this reaction. The finding that PIF interacts with a site on the kinase domain of PDK1 that is distinct from the substrate-binding site explains why this is not the case, and suggests that PIF may be capable of inducing conformational changes in the PDK1 catalytic core that alter its substrate specificity.
In order to assess the effect of PIF on the intrinsic catalytic activity of PDK1, we used the peptide substrate T308tide rather than a protein substrate of PDK1 such as p70 S6K, which may interact with PDK1 at a site that overlaps the PIF-binding pocket (Balendran et al., 1999b; see Introduction). Using this assay, we demonstrated that the PIF-binding pocket was likely to be important in regulating the activity of PDK1. When unoccupied, the PIF-binding pocket appears to suppress the activity of PDK1, because the mutation of key residues that form it, Lys115 and Leu155, enhanced PDK1 activity towards T308tide to the level equivalent to that of wild-type PDK1 in the presence of PIF (Figure 6). It is therefore likely that the binding of PIF transduces an allosteric transition, which stabilizes a functionally active conformation of PDK1. Evidence that the C–terminal Phe-Xaa-Xaa-Phe motif of PKA plays an analogous role has been obtained by mutation of the phenylalanine residues in this region, which greatly reduced PKA activity and stability (Etchebehere et al., 1997).
The interaction of PIF with PDK1 requires an aspartic acid residue (Asp978) at the position equivalent to Ser473 of PKBα. An interesting possibility was that the C–terminal carboxylate group of the Phe-Xaa-Xaa-PheCOOH motif of PKACT may have played an analogous function to Asp978 of PIF to enable binding to PDK1. However, this does not seem to be the case as the addition of four glycines to the C–terminus of PKACT did not affect its interaction with PDK1. The C–terminal carboxylate group of Phe350 of PKA does not form any interaction with the hydrophobic pocket on the kinase domain of PKA but instead faces outwards from this site and forms a hydrogen bond with Gln35 in the N–terminal non-catalytic region of PKA (Zheng et al., 1993). The importance of this interaction has not yet been investigated by mutating Gln35 of PKA, but may not be critical, since the addition of six histidine residues to the C–terminus of PKA did not affect PKA activity significantly (R.Sumathipala and R.Clegg, personal communication). Similarly, it is possible that Asp978 of PIF may not interact with the PIF-binding pocket, but with a distinct region of PDK1.
Sequence alignment of PKB, SGK and p70 S6K indicates that these members of the AGC subfamily of kinases are also likely to possess a PIF-binding pocket in their kinase domains. These kinases are all activated by phosphorylation of a Ser/Thr residue at the position equivalent to Asp978 of PIF. It is therefore possible that the introduction of a negative charge at this site by phosphorylation causes the residues of this motif to interact with their own PIF-like binding pockets, thereby leading to increased activity and stability, in a manner similar to the way in which PIF activates and stabilizes PDK1. The observation that phosphorylation of the same site increases the stability of conventional PKC isoforms is consistent with this consensus (Bornancin and Parker, 1997).
The PIF-binding pocket may be the site that enables PDK1 to interact with its substrates. This interaction may also induce a conformational change that enhances the rate at which these substrates are phosphorylated by PDK1. For example, the interaction of PKA with PDK1 via the C–terminal Phe-Xaa-Xaa-PheCOOH motif of PKA may facilitate the phosphorylation of PKA at Thr197. However, we have recently shown that PDK1 is unable to interact with or phosphorylate p70 S6K in the presence of PIF (Balendran et al., 1999b) and this is also true for SGK (T.Kobayashi and P.Cohen, unpublished work) and for PKCζ and PRK2 (see below). These observations suggest that PDK1 substrates such as p70 S6K, PKCζ and PRK2 need to interact with PDK1, at a site that overlaps with the PIF-binding site, before they can become phosphorylated by PDK1. In contrast, this is not the case for PKB as the phosphorylation of PKB by PDK1 is not inhibited by the presence of PIF in vitro or in transfected 293 cells that have been stimulated with insulin-like growth factor–1 (IGF1) (Balendran et al., 1999b). As PKB and PDK1 both interact with 3–phosphoinositides through their PH domains, it is possible that this lipid second messenger is an important determinant for co-localizing these molecules at the plasma membrane, hence allowing PDK1 to phosphorylate PKB (Currie et al., 1999). It is also possible that PKB may also interact with another site on PDK1 besides the PIF-binding pocket, and that the binding of PKB and PDK1 to 3–phosphoinositides may induce a conformational change in the structures of these kinases enabling them to interact with one another.
PKCζ and PRK2 are other protein kinases that interact with PDK1 (Chou et al., 1998; Le Good et al., 1998; Flynn et al., 2000) and possess an acidic residue rather than a Ser/Thr in the C–terminal hydrophobic motif. A recent study in our laboratory has shown that this region of PKCζ and PRK2 interacts with the PIF-binding pocket of PDK1, and this interaction enables PDK1 to phosphorylate and hence to activate PKCζ and PRK2 at their T–loop phosphorylation site (A.Balendran, R.M.Biondi, P.C.F.Cheung, M.Deak and D.R.Alessi, submitted). Thus the C–termini of these kinases are likely to be acting as PDK1 ‘docking sites’ analogous to docking sites that are present in other kinases that are components of kinase cascades such as MAP kinases, CDK2 and JNK (reviewed in Holland and Cooper, 1999). For example, MAP kinase interacts with Phe-Xaa-Phe-Pro motifs present in some of its substrates, and addition of these residues to a short peptide substrate decreased the Km for phosphorylation of this peptide by 85–fold (Jacobs et al., 1999). Consistent with the Phe-Xaa-Xaa-Phe-Asp-Tyr motif of PIF being a docking site for PDK1, the addition of this motif to T308tide greatly increases the rate at which it is phosphorylated by PDK1 (Figure 8).
A major outstanding question is the identity of the kinase that phosphorylates p70 S6K and PKB at their hydrophobic motif. There is controversy as to whether this phosphorylation is mediated by PDK1 (perhaps in complex with a regulatory protein) or by a distinct protein kinase, or whether this reaction could even occur by autophosphorylation as has been proposed for conventional PKC isoforms (BehnKrappa and Newton, 1999). As PDK1 complexed to PIF in the presence of PtdIns(3,4,5)P3 phosphorylated both Thr308 and Ser473 (Balendran et al., 1999b), this originally was interpreted to mean that PDK1 was phosphorylating both Thr308 and Ser473 of PKB. However, recent experiments demonstrate that catalytically inactive mutants of PKB are only phosphorylated at Thr308 and not at Ser473 by PDK1 when complexed to PIF (D.R.Alessi, unpublished). It is therefore possible that the PIF peptide might actually be playing a dual role: by binding to PDK1 it would increase basal activity and stability of PDK1, and by binding to the analogous PIF pocket on PKB it could displace the Ser473 residue from this site enabling it to become phosphorylated by autophosphorylation. It should be noted that this conclusion is not definitive evidence that the phosphorylation of PKB at Ser473 in these experiments is due to autophosphorylation, as the catalytically inactive PKB could be misfolded and therefore not recognized by PDK1 complexed to PIF. Furthermore, we have also not been able to detect a significant interaction between PKB and PIF. As a catalytically inactive mutant of PKB is phosphorylated at Ser473 in response to IGF1 stimulation when transfected into 293 cells (Alessi et al., 1996), this has been taken to rule out the possibility that PKB autophosphorylates at this residue. However, it could be argued that either the kinase-dead PKB transfected into cells is not 100% ‘dead’ and could still possess the ability to autophosphorylate at Ser473 or that this phosphorylation could be mediated by the wild-type endogenous PKB present in these cells.
We have also obtained evidence that overexpression of PIF in cells prevented the p70 S6K from being phosphorylated at its hydrophobic motif (Balendran et al., 1999b), which indicated that PDK1 played a role in preventing this phosphorylation from occurring. However, if PDK1 bound the hydrophobic motif of p70 S6K through its PIF-binding pocket, which is located at >20 Å from the catalytic pocket, it would not be possible for the same molecule of PDK1 to phosphorylate p70 S6K at its hydrophobic motif. It is therefore possible that the key role that PDK1 plays in enabling p70 S6K to be phosphorylated at its hydrophobic motif is to bind to the hydrophobic motif of the p70 S6K and displace it from its hydrophobic pocket on the p70 S6K domain. PDK1 might then dissociate from this site, thus exposing the p70 S6K hydrophobic motif for either autophosphorylation or for phosphorylation by PDK1 itself or by a distinct protein kinase. This might explain why when PDK1 is prevented from interacting with p70 S6K in cells as a consequence of expression of PIF, it can no longer be phosphorylated at its hydrophobic motif.
In summary, PDK1 appears to possess a hydrophobic binding site in the small lobe of the kinase catalytic domain that regulates its activity as well as its interaction with substrates. These findings raise the possibility of developing novel drugs that interact with the PIF-binding pocket on PDK1. Such drugs could either activate or inhibit PDK1, by modulating its interaction with particular substrates, and thus could switch on or switch off signal transduction pathways that are regulated by PDK1. Thus T308tide could be used as a substrate to identify compounds that activate PDK1 by mimicking the effect of PIF, while PDKtide may be the peptide of choice to identify compounds that disrupt the interaction of PDK1 with PIF.
Materials and methods
Materials
Complete protease inhibitor cocktail tablets and anti-Myc monoclonal antibodies were from Roche; tissue culture reagents were from Life Technologies; SensorChips SA were from BiaCore AB; biotinylated reagent and secondary anti-mouse IgG antibodies coupled to horseradish peroxidase were from Pierce; and glutathione–Sepharose and ECL reagent were from Amersham Pharmacia Biotech.
Peptides
The 24 residue synthetic peptide whose sequence encompasses the PDK1-binding site termed PIFtide (REPRILSEEEQEMFRDFDYIADWC), the mutant D978A-PIFtide (numbering based on the human PRK2 sequence REPRILSEEEQEMFRDFAYIADWC), unrelated peptides (YRRAAV– PPSPSLSRHSSPHQAEDEEE and KKVKPPFIPTIRGREDVSNFDD– EFT used in control experiments for Figure 6), the PKB-specific peptide substrate (RPRAATF), and the PDK1 peptide substrates T308tide (KTFCGTPEYLAPEVRR) and PDKtide (KTFCGTPEYLAPEVRREPRILSEEEQEMFRDFDYIADWC) were synthesized by Dr G.Blomberg (University of Bristol, UK).
General methods
Molecular biology techniques were performed using standard protocols. Site-directed mutagenesis was performed using a QuikChange kit (Stratagene) following the instructions provided by the manufacturer. DNA constructs used for transfection were purified from bacteria using a Qiagen plasmid Mega kit according to the manufacturer's protocol, and their sequence verified using an automated DNA sequencer (Model 373, Applied Biosystems). Human kidney embryonic kidney 293 cells were cultured on 10 cm dishes in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Phospholipid vesicles containing phosphatidylcholine, phosphatidylserine and sn-1-stearoyl-2-arachidonoyl-d-PtdIns(3,4,5)P3 (Gaffney and Reese, 1997) were prepared as described previously (Alessi et al., 1997b).
PDK1 constructs
Full-length PDK1 (residues 1–556), PDK1 constructs (residues 52–556), (residues 52–404), (residues 1–360) and (residues 1–426) were expressed in 293 cells with an N–terminal GST tag from the pEBG2T vector (Sanchez et al., 1994) and affinity purified on glutathione–Sepharose (Alessi et al., 1997a). The indicated Lys115, Ile119, Gln150 and Leu155 mutants of PDK1 used in this study were expressed and purified in a similar fashion. Between 0.5 and 1.0 mg of each GST fusion protein was obtained by transfection of twenty 10 cm diameter dishes of 293 cells and each protein was >90% homogeneous as judged by SDS–PAGE (data not shown). PDK1 (residues 52–556) was also expressed in Sf9 cells with a His6 tag at the N–terminus and purified as described previously (Balendran et al., 1999a).
Yeast two-hybrid screen
Myc-tagged human PDKI was subcloned into the EcoRI–SalI site of pAS2-1 (Clontech) as a Gal4 DNA-binding domain (GBD) fusion. A yeast two-hybrid screen was carried out by co-transforming pAS2-1 PDK1 and a pACT2 human brain cDNA library fused to the Gal4 activation domain (GAD) into the yeast strain Y190. The brain library was purchased from Clontech. Transformed yeast cells were incubated for 10 days at 30°C on SD media supplemented with 25 mM 3-aminotriazole and lacking histidine, leucine and tryptophan. Approximately 5 × 106 colonies were screened.
Yeast two-hybrid analysis
The indicated mutants of pAS2-1 PDK1 and the pACT2-PKACT were constructed. The pACT2-PIF vector used in this study encodes the C–terminal 26 amino acids of PRK2, which encompasses the PDK1-binding motif (Balendran et al., 1999a). Y190 yeast strains were co-transformed with the indicated combinations of vectors and grown on SD media lacking histidine, uracil, tryptophan and leucine at 30°C until the appearance of colonies. Yeast colonies were patched onto fresh agar, incubated overnight at 30°C and filter lifts taken. Reporter β–galactosidase activity of the transformants was tested by incubating filters in X-Gal at 30°C for 4 h.
Structural modelling
The structure of the kinase domain of PDK1 (residues 92–341) was modelled using the programme Swiss-PdbViewer [http://www.expasy.ch/spdbv/mainpage.htm (Guex and Peitsch, 1997)] connecting to Swiss Model Automated Protein Modeling Server. Modelling was based on several structures of the PKA catalytic subunit available in the database (Protein Data Bank Identification: 1YDR, 1CTP, 1STC, 1ATP and 1CDK). Sequence identity to PDK1 within the catalytic region (residues 55–297 of mouse PKA) was 40%, with a similarity of 68%.
Binding of PIF to Myc-PDK1
A pEBG2T plasmid encoding GST fused to the last 77 residues of PRK2 termed GST–PIF (10 μg) (Balendran et al., 1999a) and pCMV5 plasmid expressing Myc-PDK1 wild-type or the indicated mutants of PDK1 (10 μg) were co-transfected into a 10 cm diameter dish of 293 cells using a modified calcium phosphate method (Alessi et al., 1996). At 48 h post-transfection, the cells were lysed in 0.6 ml of lysis buffer [50 mM Tris–HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (by mass) Triton X–100, 1 mM sodium orthovanadate, 50 μM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 μM microcystin-LR, 0.1% (by volume) β–mercaptoethanol and one tablet of protease inhibitor cocktail per 50 ml of buffer], cleared by centrifugation, and 0.5 ml of supernatant was incubated for 2 h at 4°C with 30 μl of glutathione–Sepharose. The beads were washed twice in lysis buffer containing 0.5 M NaCl, followed by two further washes in lysis buffer. The beads were resuspended in 1 vol of buffer containing 100 mM Tris–HCl pH 6.8, 4% (by mass) SDS, 20% (by volume) glycerol and 200 mM dithiothreitol (DTT) and subjected to SDS–PAGE. The gels were either stained with Coomassie Blue or analysed by immunoblotting with anti-Myc antibodies.
BiaCore analysis of PIF binding to GST–PDK1
Binding was analysed directly by SPR in an upgraded Bialite system. PIFtide (comprising the last 24 residues of PRK2) was biotinylated though its C–terminal cysteine and bound to an streptavidin-coated Sensor chip SA (Biacore AB, Stevenage, UK) as described previously (Balendran et al., 1999a). Wild-type or the indicated mutants of GST–PDK1 (10–400 nM) were injected in an intracellular type buffer, over the immobilized biotinylated PIFtide at a flow rate of 30 μl per min as previously described (James et al., 1996). The sensor chip surface was regenerated by pulses of 10 mM NaOH.
Measurement of PDK1 catalytic activity
PDK1's ability to phosphorylate Thr308 of PKBα was measured using a mutant of GST–PKBα in which Ser473 was mutated to aspartic acid (GST–473D-PKBα) in the presence of phospholipid vesicles containing sn-1-stearoyl-2-arachidonoyl-d-PtdIns(3,4,5)P3 (Alessi et al., 1997b). The ability of wild-type and mutant PDK1 to phosphorylate the synthetic peptides T308tide or PDKtide was determined in 20 μl assays containing 50 mM Tris–HCl pH 7.5, 0.1% 2-mercaptoethanol, 10 mM MgCl2, 100 μM [∣γ–32P]ATP (∼500 c.p.m./pmol), 0.5 μM microcystin-LR, PDK1 and the peptide concentrations indicated in Results. After incubation for 10 min at 30°C, the reaction was stopped by addition of 20 μl of 150 mM phosphoric acid. Then 35 μl of the resultant mixture was spotted into P81 phosphocellulose paper (2 × 2 cm) and the paper was washed and analysed as described previously for assays of MAP kinase (Alessi et al., 1994). Wild-type PIFtide or the mutant D978A-PIFtide peptides were included in the reactions as indicated. Control assays were carried out in parallel in which either PDK1 or peptide substrate were omitted; these values were always <5% of the activity measured in the presence of these reagents. One unit of PDK1 activity was defined as that amount required to catalyse the phosphorylation of 1 nmol of the T308tide in 1 min. The assays were linear with time up to a final PDK1 concentration of 5 U/ml.
Thermal denaturation
Heat denaturation was carried out by incubating the indicated forms of PDK1 (0.4 mg/ml) for 2 min at temperatures ranging from 30 to 65°C. The heat treatment was terminated by the addition of a 10–fold volume excess of ice-cold buffer (50 mM Tris–HCl pH 7.5, 1 mM DTT and 0.1 mg/ml bovine serum albumin), and the samples were incubated for 2 min in an ice–water bath before a 4 μl aliquot was assayed for activity towards T308tide.
Acknowledgments
Acknowledgements
We would like to thank Philip Cohen and Peter Downes for helpful discussions and comments on the manuscript, Nick Morrice for protein sequencing, Dr Andrew Paterson for preparation of His-PDK1 baculovirus, and Agnieszka Kieloch for culture of 293 cells. This work was supported by the British Diabetic Association (D.R.A.), the UK Medical Research Council (D.R.A.), AstraZeneca, Novo-Nordisk, Pfizer and SmithKline Beecham.
References
- Alessi D.R. and Downes, C.P. (1998) The role of PI 3-kinase in insulin action. Biochim. Biophys. Acta, 1436, 151–164. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Cohen, P., Ashworth, A., Cowley, S., Leevers, S.L. and Marshall, C.J. (1994) Assay and expression of mitogen activated protein kinase, MAP kinase and Raf. Methods Enzymol., 255, 279–290. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Andjelkovic, M., Caudwell, B., Cron, P., Morrice, N., Cohen, P. and Hemmings, B.A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J., 15, 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Alessi D.R., et al. (1997a) 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., James, S.R., Downes, C.P., Holmes, A.B., Gaffney, P.R., Reese, C.B. and Cohen, P. (1997b) Characterization of a 3-phospho– inositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol., 7, 261–269. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Kozlowski, M.T., Weng, Q.P., Morrice, N. and Avruch, J. (1998) 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6K in vivo and in vitro. Curr. Biol., 8, 69–81. [DOI] [PubMed] [Google Scholar]
- Balendran A., Casamayor, A., Deak, M., Paterson, A., Gaffney, P., Currie, R., Downes, C.P. and Alessi, D.R. (1999a) PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol., 9, 393–404. [DOI] [PubMed] [Google Scholar]
- Balendran A., Currie, R.A., Armstrong, C.G., Avruch, J. and Alessi, D.R. (1999b) Evidence that PDK1 phosphorylates p70 S6K in vivo at Thr412 as well as Ser252. J. Biol. Chem., 274, 37400–37406. [DOI] [PubMed] [Google Scholar]
- Belham C., Wu, S. and Avruch, J. (1999) Intracellular signalling: PDK1–a kinase at the hub of things. Curr. Biol., 9, R93–R96. [DOI] [PubMed] [Google Scholar]
- BehnKrappa A. and Newton, A.C. (1999) The hydrophobic phosphorylation motif of conventional protein C is regulated by autophosphorylation. Curr. Biol., 9, 728–737. [DOI] [PubMed] [Google Scholar]
- Bornancin F. and Parker, P.J. (1997) Phosphorylation of protein kinase C–α on serine 657 controls the accumulation of active enzyme and contributes to its phosphatase-resistant state [published erratum appears in J. Biol. Chem., 1997, 272, 13458]. J. Biol. Chem., 272, 3544–3549. [DOI] [PubMed] [Google Scholar]
- Cheng X., Ma, Y., Moore, M., Hemmings, B.A. and Taylor, S.S. (1998) Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl Acad. Sci. USA, 95, 9849–9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.M., Hou, W., Johnson, J., Graham, L.K., Lee, M.H., Chen, C.S., Newton, A.C., Schaffhausen, B.S. and Toker, A. (1998) Regulation of protein kinase Cζ by PI 3-kinase and PDK-1. Curr. Biol., 8, 1069–1077. [DOI] [PubMed] [Google Scholar]
- Coffer P.J., Jin, J. and Woodgett, J.R. (1998) Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J., 335, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie R.A., Walker, K.S., Gray, A., Deak, M., Casamayor, A., Downes, C.P., Cohen, P., Alessi, D.R. and Lucocq, J. (1999) Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem. J., 337, 575–583. [PMC free article] [PubMed] [Google Scholar]
- Dalby K.N., Morrice, N., Caudwell, F.B., Avruch, J. and Cohen, P. (1998) Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J. Biol. Chem., 273, 1496–1505. [DOI] [PubMed] [Google Scholar]
- Dong L.Q., Zhang, R.B., Langlais, P., He, H., Clark, M., Zhu, L. and Liu, F. (1999) Primary structure, tissue distribution and expression of mouse phosphoinositide-dependent protein kinase-1, a protein kinase that phosphorylates and activates protein kinase Cζ. J. Biol. Chem., 274, 8117–8122. [DOI] [PubMed] [Google Scholar]
- Dutil E.M., Toker, A. and Newton, A.C. (1998) Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr. Biol., 8, 1366–1375. [DOI] [PubMed] [Google Scholar]
- Etchebehere L.C., Van Bemmelen, M.X., Anjard, C., Traincard, F., Assemat, K., Reymond, C. and Veron, M. (1997) The catalytic subunit of Dictyostelium cAMP-dependent protein kinase–role of the N–terminal domain and of the C–terminal residues in catalytic activity and stability. Eur. J. Biochem., 248, 820–826. [DOI] [PubMed] [Google Scholar]
- Flynn P., Mellor,H. and Parker,P.J. (2000) Rho-GTPase control of PRK activation by PDK1. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Frodin M. and Gammeltoft, S. (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol., 151, 65–77. [DOI] [PubMed] [Google Scholar]
- Gaffney P.R.J. and Reese, C.B. (1997) Synthesis of 1-[(1-O-stearoyl-2-O-arachidonoyl-sn-glycer-3-yl)-phosphoryl]-d-myo-inositol 3,4,5-trisphosphate [PtdIns (3,4,5)P3] and its stereoisomers. Bioorg. Med. Chem. Lett., 7, 3171–3176. [Google Scholar]
- Guex N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Holland P.M. and Cooper, J.A. (1999) Protein modification: docking sites for kinases. Curr. Biol., 9, R329–R331. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Glossip, D., Xing, H., Muslin, A.J. and Kornfeld, K. (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev., 13, 163–175. [PMC free article] [PubMed] [Google Scholar]
- James S.R., Downes, C.P., Gigg, R., Grove, S.J., Holmes, A.B. and Alessi, D.R. (1996) Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem. J., 315, 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen C.J., Buch, M.B., Krag, T.O., Hemmings, B.A., Gammeltoft, S. and Frodin, M. (1999) 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem., 274, 27168–27176. [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng, J.H., Ten Eyck, L.F., Ashford, V.A., Xuong, N.H., Taylor, S.S. and Sowadski, J.M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science, 253, 407–414. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Cohen, P. (1999) Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J., 339, 319–328. [PMC free article] [PubMed] [Google Scholar]
- Le Good J.A., Ziegler, W.H., Parekh, D.B., Alessi, D.R., Cohen, P. and Parker, P.J. (1998) Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science, 281, 2042–2045. [DOI] [PubMed] [Google Scholar]
- Leevers S.J., Vanhaesebroeck, B. and Waterfield, M.D. (1999) Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr. Opin. Cell Biol., 11, 219–225. [DOI] [PubMed] [Google Scholar]
- Mellor H. and Parker, P.J. (1998) The extended protein kinase C superfamily. Biochem. J., 332, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Leong, M.L., Buse, P., Maiyar, A.C., Firestone, G.L. and Hemmings, B.A. (1999) Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J., 18, 3024–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.B., Dennis, P.B., Han, J.W., Williamson, N.A., Kozma, S.C., Wettenhall, R.E. and Thomas, G. (1995) The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J., 14, 5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.T. and Schreiber, S.L. (1999) Keeping it all in the family. Curr. Biol., 9, R521–R524. [DOI] [PubMed] [Google Scholar]
- Proud C.G. (1996) p70 S6 kinase: an enigma with variations. Trends Biochem. Sci., 21, 181–185. [PubMed] [Google Scholar]
- Pullen N. and Thomas, G. (1997) The modular phosphorylation and activation of p70s6k. FEBS Lett., 410, 78–82. [DOI] [PubMed] [Google Scholar]
- Pullen N., Dennis, P.B., Andjelkovic, M., Dufner, A., Kozma, S.C., Hemmings, B.A. and Thomas, G. (1998) Phosphorylation and activation of p70s6k by PDK1. Science, 279, 707–710. [DOI] [PubMed] [Google Scholar]
- Richards S.A., Fu, J., Romanelli, A., Shimamura, A. and Blenis, J. (1999) Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol., 12, 810–820. [DOI] [PubMed] [Google Scholar]
- Sanchez I., Hughes, R.T., Mayer, B.J., Yee, K., Woodgett, J.R., Avruch, J., Kyriakis, J.M. and Zon, L.I. (1994) Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature, 372, 794–798. [DOI] [PubMed] [Google Scholar]
- Shepherd P.R., Withers,D.J. and Siddle,K. (1998) Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling [published erratum appears in Biochem. J., 1998, 335, 711]. Biochem. J., 333, 471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L., et al. (1998) Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science, 279, 710–714. [DOI] [PubMed] [Google Scholar]
- Stokoe D., Stephens, L.R., Copeland, T., Gaffney, P.R., Reese, C.B., Painter, G.F., Holmes, A.B., McCormick, F. and Hawkins, P.T. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science, 277, 567–570. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and Radzio-Andzelm, E. (1994) Three protein kinase structures define a common motif. Structure, 2, 345–355. [DOI] [PubMed] [Google Scholar]
- Zheng J., Knighton, D.R., ten Eyck, L.F., Karlsson, R., Xuong, N., Taylor, S.S. and Sowadski, J.M. (1993) Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry, 32, 2154–2161. [DOI] [PubMed] [Google Scholar]