Abstract
The double-stranded RNA-binding domain (dsRBD) is a common RNA-binding motif found in many proteins involved in RNA maturation and localization. To determine how this domain recognizes RNA, we have studied the third dsRBD from Drosophila Staufen. The domain binds optimally to RNA stem–loops containing 12 uninterrupted base pairs, and we have identified the amino acids required for this interaction. By mutating these residues in a staufen transgene, we show that the RNA-binding activity of dsRBD3 is required in vivo for Staufen-dependent localization of bicoid and oskar mRNAs. Using high-resolution NMR, we have determined the structure of the complex between dsRBD3 and an RNA stem–loop. The dsRBD recognizes the shape of A-form dsRNA through interactions between conserved residues within loop 2 and the minor groove, and between loop 4 and the phosphodiester backbone across the adjacent major groove. In addition, helix α1 interacts with the single-stranded loop that caps the RNA helix. Interactions between helix α1 and single-stranded RNA may be important determinants of the specificity of dsRBD proteins.
Keywords: dsRBD/NMR/RNA localization/RNA–protein/Staufen
Introduction
The double-stranded RNA-binding domain (dsRBD) is among the most common RNA-binding motifs, and is found in single or multiple copies in many eukaryotic and prokaryotic proteins involved in RNA processing, maturation and localization (Green and Matthews, 1992; St Johnston et al., 1992). Three-dimensional structures of dsRBDs from several proteins have shown that the domain folds into a compact αβββα structure (Bycroft et al., 1995a; Kharrat et al., 1995; Nanduri et al., 1998). As in the other two major eukaryotic RNA-binding protein domains (Varani, 1997), the α–helical surface of the dsRBD structure packs through a conserved hydrophobic core against an antiparallel β–sheet. In vitro studies have shown that dsRBD proteins bind to dsRNA, but not to single-stranded RNA or DNA, nor dsDNA (St Johnston et al., 1992; Bass et al., 1994; Clarke and Matthews, 1995; Bevilacqua and Cech, 1996). These studies have shown that dsRBDs bind any dsRNA of sufficient length, regardless of its base composition, and therefore they represent general dsRNA-binding modules.
The dsRBD was first identified in the Drosophila protein Staufen, which contains five copies of this motif (St Johnston et al., 1992). Staufen plays an essential role in the formation of the anterior–posterior axis in Drosophila and represented the first protein factor to be identified as critical for mRNA localization (St Johnston, 1995). Staufen protein associates with oskar mRNA during oogenesis and is required for its transport to the posterior pole of the oocyte, where it defines where the abdomen and germline will develop (Ephrussi et al., 1991; Kim-Ha et al., 1991; St Johnston et al., 1991). After the egg has been laid, Staufen accumulates at the anterior pole of the egg, and anchors the anterior determinant bicoid mRNA (St Johnston et al., 1989; Ferrandon et al., 1994). Staufen plays a role in RNA localization in somatic cells as well, by associating with prospero mRNA during the asymmetric divisions of the embryonic neuroblasts, and by mediating its segregation to the smaller daughter cell produced by this division (Broadus et al., 1998; Schuldt et al., 1998). In common with most other systems where mRNA localization has been studied, the cis-acting signals required for oskar, bicoid and prospero localization all reside within the 3′–untranslated regions (3′–UTR) of these mRNAs (MacDonald and Struhl, 1988; Kim-Ha et al., 1993). Staufen protein associates in vivo with the 3′–UTRs of bicoid and prospero mRNAs to form ribonucleoprotein particles (Ferrandon et al., 1994; Schuldt et al., 1998). The bicoid RNA sequences required for this interaction have been mapped to three largely double-stranded regions (Schuldt et al., 1998). However, it remains to be proven whether Staufen interacts directly with these RNAs in vivo and, if so, how Staufen recognizes these specific transcripts.
The binding of the dsRBD to dsRNA represents an example of protein–nucleic acid recognition distinct from the other common RNA-binding motifs characterized so far (Varani, 1997). To determine the nature of the dsRBD–dsRNA interaction, we have conducted extensive mutagenesis on the third dsRBD from Staufen (dsRBD3) and have used nuclear magnetic resonance (NMR) to determine the structure of the complex between this domain and an RNA stem–loop containing an optimal Staufen-binding site. We have mutated five critical interfacial residues located at the RNA–protein interface into full-length Staufen protein. These mutations abolish the RNA-binding activity of dsRBD3 in vitro and prevent Staufen-dependent RNA localization in vivo. The present results provide a description at the atomic level of the interactions between the dsRBD and RNA and demonstrate their physiological significance for Staufen-dependent RNA localization.
Results
RNA binding by Staufen dsRBD3
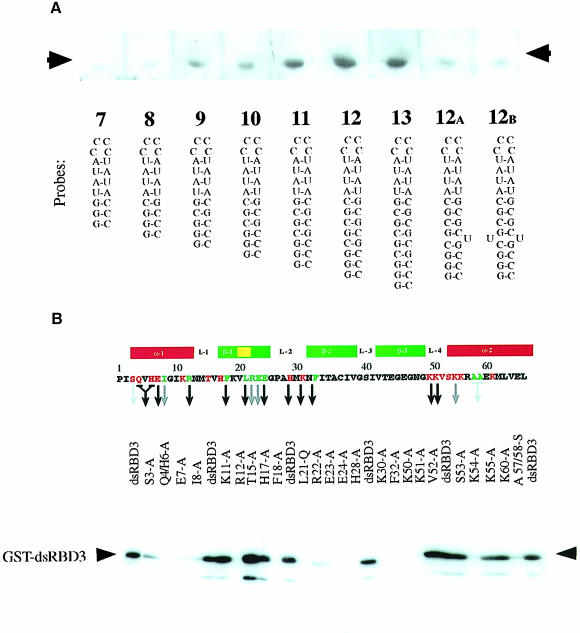
The third dsRBD from Staufen (dsRBD3) binds dsRNA with micromolar affinity, and conforms particularly well to the consensus sequence of the dsRBD motif (Gibson and Thompson, 1994). We therefore chose this domain to analyse the structural basis of dsRBD–RNA interaction. As a first step, we determined the minimal and optimal length of dsRNA required for binding by a dsRBD by performing North-western blots with RNA hairpin substrates containing double–helical stems of increasing length. RNAs containing >8 bp of dsRNA bind to dsRBD3, but optimal binding is observed with RNAs of 12 bp or longer (Figure 1A). Since further increases in the length of the double–helical region do not improve binding, we conclude that dsRBD3 binds optimally to stem–loops containing 12 bp. Disruption of the helical structure of the RNA by the introduction of unpaired bases significantly reduces binding. These results are consistent with studies on polypeptides derived from the two dsRBDs of RNA-activated protein kinase (PKR) (Schmedt et al., 1995; Bevilacqua and Cech, 1996). The full-length polypeptide binds to RNAs that contain at least 16 bp, but each dsRBD was found to cover ∼11 bp of RNA.
Fig. 1. Biochemical characterization of Staufen dsRBD3 interaction with RNA stem–loops. North-western blots showing (A) binding of wild-type dsRBD3 to RNA stem–loops and (B) alanine-scanning mutagenesis of dsRBD3. The top panel shows the positions of the amino acids that were substituted, with a diagram of the secondary structure of the domain underneath. The numbering refers to the general dsRBD alignment scheme; Pro1 corresponds to Pro579 in Drosophila Staufen. Green residues correspond to mutated amino acids required for the correct folding of the domain, while red residues are surface exposed. A black arrow indicates no RNA binding; grey arrow, reduced binding; no arrow indicates that the mutation had no effect. The lower panel shows a representative blot of the alanine substitution mutants probed with double-stranded VA1 RNA. The same amount of protein was present in each lane (data not shown).
The identity of amino acids within dsRBD3 involved in RNA recognition was established by systematic alanine-scanning mutagenesis using the same North-western assay (Figure 1B). Several mutations involved amino acids whose identity is crucial for the structure of the dsRBD. Ile8, Phe18, Ala57 and Ala58 form part of the hydrophobic core of the domain. Mutations in Leu21, Arg22, Glu23 and Glu24 were introduced to disrupt the β–bulge within the first strand of the β–sheet, a highly conserved feature that is also present in ribosomal protein S5, a protein that is very similar in both sequence and structure to the dsRBD (Bycroft et al., 1995). As expected, mutation of each of these amino acids strongly reduces or abolishes RNA binding. Mutations in Arg12 and Phe32 are also likely to fall into this class, even though these amino acids are partially exposed on the surface of the domain. Arg12 caps the N–terminal α–helix, and its replacement with alanine might disrupt RNA binding by extending the helix into the following tight turn. Evidence presented below indicates that Phe32 anchors loop 2 and loop 4.
The most informative mutations were changes in surface residues that affect RNA binding without altering the conformation of the domain, as demonstrated by circular dichroism. Mutations of this type cluster in three regions of the domain: Ser3, Gln4 plus His6, and Glu7 within the N–terminal helix α1; His28 and Lys30 within loop 2; and Lys50, Lys51 and Lys54 within loop 4 and the beginning of helix α2. It is notable that mutations in Lys50, Lys51 or Lys54 abolish RNA binding, whereas two non-basic amino acids in this loop, Val52 and Ser53, can be mutated to alanine without loss of binding. This suggests that electrostatic interactions mediated by basic residues play an important role in dsRBD–RNA recognition.
Staufen dsRBD3 binds RNA using a highly conserved surface and without altering the RNA conformation
Having established the biochemical properties of dsRBD3–RNA recognition, we used high-resolution NMR spectroscopy to characterize this interaction in structural detail. A stem–loop of 12 bp capped by an exceptionally stable C(UUCG)G loop was chosen to represent an optimal substrate, as defined by the experiments reported in Figure 1A. Since the dsRBD–dsRNA interaction is not sequence specific, the double–helical region was made fully symmetrical to simplify the NMR spectral analysis.
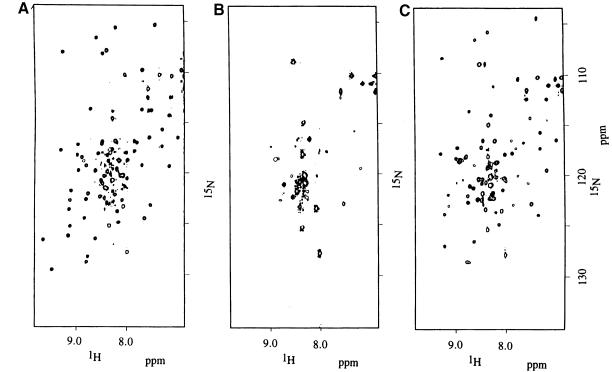
Many protein resonances broadened considerably at subsaturating ratios of RNA when dsRBD3 was titrated with RNA, then sharpened up again when the RNA was added in stoichiometric amounts (Figure 2). This behaviour is found when the interconversion between free and bound forms occurs with intermediate exchange kinetics. This result strongly suggests that the off rate of binding is in the millisecond time scale, consistent with a micromolar dissociation constant and with the on rate being diffusion limited. Essentially complete spectral assignments were obtained for the bound form of dsRBD3 in the presence of RNA by applying standard procedures utilizing 15N- and 13C-15N-labelled dsRBD3 samples mixed with unlabelled RNA. Changes in chemical shift upon RNA binding define the footprint of the RNA on dsRBD3. The regions of the protein where large changes in the NMR spectrum occur upon RNA binding cluster at the N–terminus of the protein, in loop 2 and loop 4 and in the region where α1 packs against α2 and the β–sheet. No significant changes were observed for β2 and β3 or in the C–terminal region of the protein. Two conserved lysine residues within loop 4 (Lys50 and Lys51) are particularly interesting. The backbone amide resonances of Lys50 and Lys51 are invisible in the free protein spectra, presumably due to the accessibility of solvent molecules to this exposed region of the structure. However, the same resonances become visible upon complex formation. These residues are protected from exchange with solvent by the RNA, confirming their role in RNA recognition revealed by the alanine-scanning experiment.
Fig. 2. 1H-15N HSQC spectrum of the free protein (A) and of a sample containing an ∼2–fold protein excess (B). Most protein resonances broaden beyond detection when the protein is in excess, and only resonances from the unfolded regions at either end of the domain remain visible. Under stoichiometric conditions (1:1), resonances from the protein become sharp again (C).
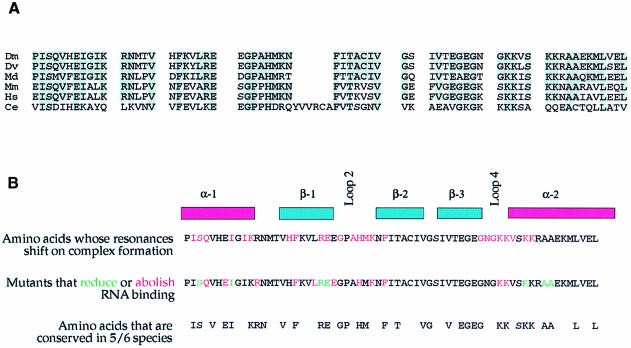
The NMR data demonstrate that the folding of the domain does not change significantly upon RNA binding. Residues that display significant changes in the NMR spectrum of dsRBD3 upon RNA binding were therefore mapped on the structure of the free protein domain (Bycroft et al., 1995). The results unambiguously demonstrate that the face of the dsRBD formed by the N–termini of both helices, and by loops 2 and 4 along the edge of the first strand of the β–sheet, represents the RNA-binding surface of dsRBD3. Thus, the results of the alanine-scanning mutagenesis and the NMR footprint identify the same face of Staufen dsRBD3 as the surface where RNA recognition occurs. This protein surface contains exposed residues that are almost completely conserved among Staufen proteins from Drosophila to humans (Figure 3).
Fig. 3. Evolutionary conservation of the amino acids required for RNA binding in Staufen dsRBD3. (A) Alignment of dsRBD3 sequences from Staufen homologues of Drosophila melanogaster (Dm), Drosophila virilis (Dv), Musca domestica (Md), Mus musculus (Mm), Homo sapiens (Hs) and Caenorhabditis elegans (Ce). Grey boxes highlight positions where two or more other species have the same amino acid as Drosophila melanogaster. (B) Comparison of the results of the NMR analysis of the dsRBD3–RNA complex, the alanine-scanning mutagenesis and the positions of amino acids that have been highly conserved during evolution.
Essentially complete spectral assignments were obtained for both free and bound RNA using isotopically labelled RNA samples. Remarkably, only a few residues displayed significant changes in their NMR properties upon formation of the complex, and the changes were generally of modest magnitude. This result demonstrates that dsRBD3–RNA interaction occurs with only small rearrangements of a preformed RNA structure.
In order to establish the orientation of dsRBD3 with respect to the RNA, we measured residual dipolar couplings in a partially oriented sample. Dipolar interactions assume finite values when the sample is partially oriented, and these values provide absolute information on the orientation of NH and CH bonds (Tjandra and Bax, 1997). Residual NH dipolar couplings for dsRBD3 in complex with RNA show negative values for amino acids within the two α–helices and positive values for the β–strands (data not shown). When residual coupling constants were measured for the RNA in the complex, we found instead positive couplings for base NH and CH bonds. In double-stranded nucleic acids, the bases are approximately perpendicular to the double helix axis. Therefore, positive values of CH and NH couplings within the RNA, compared with the negative couplings for the protein α–helices NHs, show that the protein is bound to the RNA with the α–helices approximately parallel to the RNA double–helical axis.
The dynamic character of the Staufen dsRBD–RNA interaction
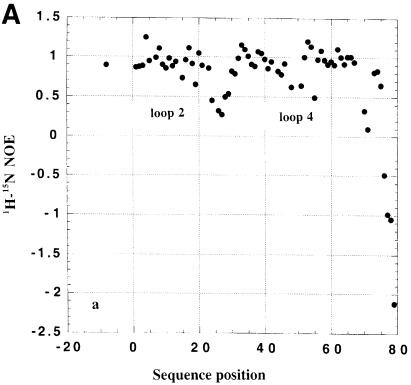
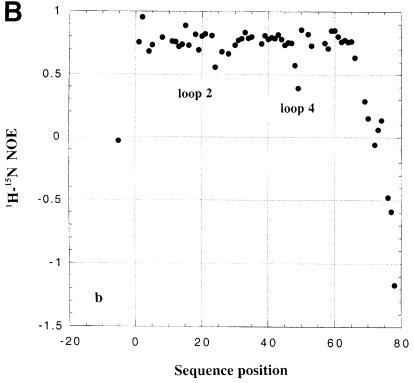
The analysis of 15N NMR relaxation properties for dsRBD3 was used to study the existence of conformational flexibility in the free and RNA-bound protein domain. As shown in Figure 4, 1H-15N heteronuclear NOEs are ∼0.7–0.8 in the well-folded core of the dsRBD. However, low heteronuclear NOE values are observed for the flexible tail at the end of the construct, reflecting complete disorder. Lower than average heteronuclear NOEs are also observed for residues within loop 2 and loop 4 both in the free and RNA-bound dsRBD3, reflecting residual conformational flexibility. Analysis of additional relaxation parameters reveals the existence of conformational exchange within loop 2, loop 4 and the N–terminus of helix α2 (data not shown). Furthermore, some NH resonances within loop 2 and loop 4 could not be analysed in the complex due to exchange broadening. These results demonstrate that loop 2 and loop 4, two of the three regions of the protein that form the RNA interface (see below), are highly mobile in the free protein and retain significant conformational flexibility in the complex.
Fig. 4. Heteronuclear 1H-15N NOE for free (A) and RNA-bound (B) Staufen dsRBD3.
Structure of the Staufen dsRBD3–RNA complex
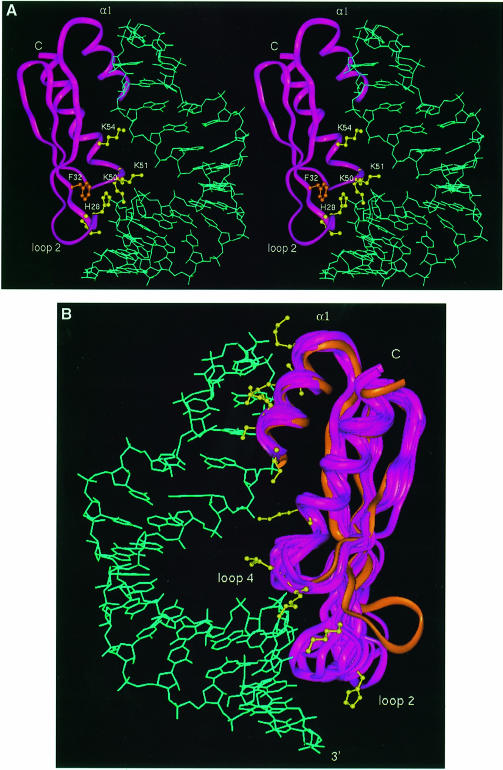
The structure of dsRBD3 in complex with the stem–loop RNA was determined using a protocol very similar to that used in the determination of the structure of the U1A complex (Allain et al., 1996; Howe et al., 1998). No assumption was made at any stage of the data collection or structure calculation about the nature of the interaction or about the protein or RNA structures. The structure was based on the identification of intermolecular NOE interactions and on the definition of the relative orientation of the protein and RNA achieved by measuring residual dipolar couplings in partially oriented samples. The majority of NOE contacts involved sugar resonances in the sugar–phosphate backbone, suggesting that the domain does not contact the RNA bases intimately. However, the observation of NOE contacts from Ade3 H2 demonstrates that the protein binds the minor groove of the double helix, as suggested (Bevilacqua and Cech, 1996). Experimental and structural statistics are summarized in Table I. A stereo view of the structure is shown in Figure 5A and superposition of 10 low energy structures is shown in Figure 5B.
Table I. Experimental constraints and structural statistics.
| Protein | RNA | |
|---|---|---|
| (a) Distance and dihedral angle NMR constraints | ||
| Total | 665 | 588 |
| Intra-residue | 202 | 215 |
| Sequential | 175 | inter residue 206 |
| Medium/long range | 210 | |
| Hydrogen bonds | 24 | 35 |
| Dihedral restraints | 0 | 127 |
| Residual dipolar couplings | 54 | 5 |
| Protein–RNA distance restraints | 10 | |
| Total No. constraints | 1263 | |
| (b) Structure statistics NOE violations | ||
| number >0.2 Å | 3 ± 2.2 | |
| maximum violation | 0.94 Å | |
| Angle violations | ||
| number >5° | 0.1 ± 0.3 | |
| Mean deviation from ideal covalent geometry | ||
| bond length | 0.005 Å | |
| bond angles | 0.79° | |
| impropers | 0.41° | |
| (c) R.m.s. deviations from average structure (Å) | ||
| Protein–regular secondary structure (1–20, 30–47, 51–66) | ||
| backbone | 0.87 Å | |
| heavy atoms | 1.53 Å | |
| RNA (superposed on all heavy atoms for the indicated residues) | ||
| all residues | 2.44 Å | |
| tetraloop | 1.19 Å | |
| double–helical stem | 2.08 Å | |
| Protein–RNA (all heavy atoms for indicated residues) | ||
| all ordered regions | 2.56 Å | |
| complex interface | loop 2 | 2.53 Å |
| loop 4 | 2.14 Å | |
| helix α1 | 1.56 Å | |
Fig. 5. (A) Stereo view of a low energy structure of the dsRBD3–RNA complex; Phe32 and five interfacial residues critical for Staufen function are shown explicitly. (B) Superposition of 10 converged structures and the free protein (orange); this image was prepared by superimposing the protein backbone between residues 1 and 65, but excluding loop 2; residues involved in intermolecular contacts with the RNA are shown explicitly in yellow for one representative structure.
The structure of the dsRBD3–RNA complex is of lower precision than that of the U1A complex (Allain et al., 1996; Howe et al., 1998). This is a consequence of the smaller number of intermolecular distance constraints, which could only be partially compensated by introducing absolute orientational information derived from residual dipolar couplings (Bayer et al., 1999). The small number of intermolecular NOEs is due to three distinctive properties of the dsRBD–RNA interaction, reflecting the weak, non-specific association between dsRBD and RNA. First of all, the majority of intermolecular interactions between Staufen dsRBD3 and RNA involve the RNA backbone, where there are relatively few resonances, and these are difficult to assign to specific nucleotides due to spectral overlap. Secondly, the intermolecular dsRBD3–RNA interface is small and involves relatively few protein residues. The area buried upon complex formation is only ≈1450 Å2, 12% of the total surface area. In contrast, protein side chains and RNA bases form an intricate intermolecular interface in the U1A complex that buries a much larger surface (Allain et al., 1996). Thirdly, the interface retains significant conformational flexibility (Figure 4), and this is likely to quench at least some intermolecular NOE interactions.
Staufen dsRBD3 contacts the RNA stem–loop through the same sites identified by alanine-scanning mutagenesis and NMR chemical shift analysis (Figure 3): helix α1, loop 2 and loop 4 plus the N–terminal part of helix α2. The distance between loop 2 and loop 4 corresponds to the spacing between the minor groove and the phosphate across the intervening major groove in A-form RNA. The distance between the loop 2–RNA interactions and the tetraloop, the site of helix α1–RNA contacts, is 12 bp. This spacing is in perfect agreement with the optimal substrate length (Figure 1A).
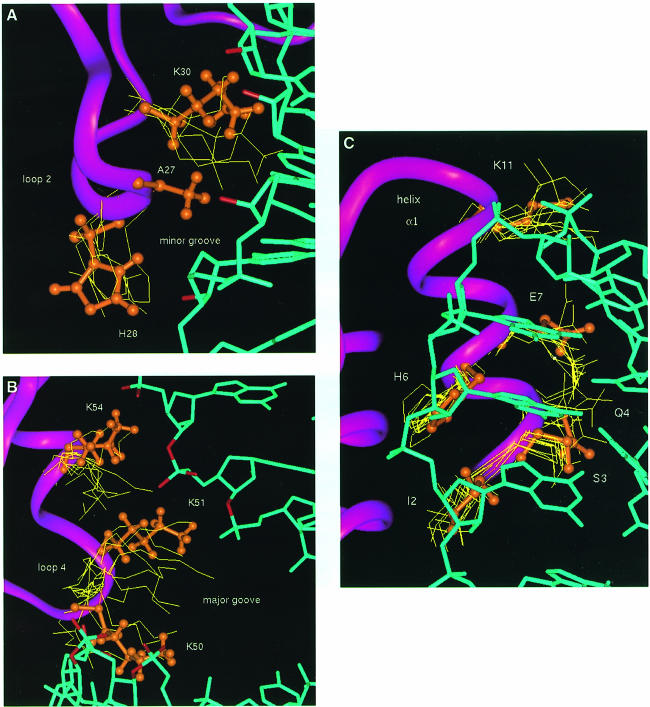
Helix α1 interacts with the C(UUCG)G tetraloop. This interaction is well defined by the experimental data (Figure 6C), and the relaxation data confirm that this region of the protein is rigid. However, only few intermolecular contacts can be interpreted as specific to the UUCG sequence. Ser3 interacts with the 2′–OH and phosphate oxygen of C13, the last nucleotide on the 5′ side of the stem. Glu7 interacts with the 2′–OH of U15 and stacks with the aromatic ring of G17, while Lys11 makes electrostatic interactions with the phosphate of residue 16. Ile10 is in van der Waals contact with G17. Intermolecular interactions involving loop 2 and loop 4 are less precisely defined, due to the residual conformational flexibility in this region of the RNA–protein interface revealed by NMR relaxation measurements. Residues within loop 2 interact with 2′–OH and phosphate oxygens within the minor groove close to the bottom of the double–helical stem (Figure 6A). The side chain amide of Lys30 interacts with 2′–OH groups in the minor groove of the RNA, while the heteroaromatic ring of His28 is positioned almost perpendicularly with respect to a phosphate oxygen. In the majority of converged structures, the phosphate oxygen is directed towards the centre of the ring of His28. The position of loop 4 with respect to the RNA is defined indirectly by the interactions observed between helix α1 and the UUCG loop, and between loop 2 and the RNA minor groove. These intermolecular interactions and the structure of the protein unambiguously position loop 4 near the phosphodiester backbone across the major groove from the site of loop 2 interactions with the minor groove. Three critical lysine residues within loop 4 and the N–terminus of helix α2, Lys50, Lys51 and Lys54, interact with phosphate oxygens and one 2′–OH group (Figure 6B) across the major groove from the sites of loop 2–minor groove interaction. The side chains of Lys50 and Lys51 bridge the major groove by interacting with RNA phosphates across the major groove from each other, while Lys54 reinforces these contacts by interacting with the phosphate immediately following the site of interaction of Lys51.
Fig. 6. Intermolecular interactions between dsRBD and the RNA stem–loop in the superposition of 10 converged structures; one structure is represented in orange for clarity. (A) Interaction between loop 2 and the minor groove of the double–helical stem; Ala27 and His28 from the conserved GPAH sequence and the Lys30 side chains are shown explicitly; 2′–OH groups in close proximity to amino acids side chains are highlighted in red. (B) Interactions between loop 4 and the N–terminus of helix α2 and RNA phosphates (in red). (C) Interaction between helix α1 and the UUCG tetraloop.
Comparison of the structure of dsRBD3 free and in the RNA complex confirms that the structure of the protein does not change significantly on RNA binding, with the exception of loop 2. The rotation of loop 2 (towards the RNA in Figure 5B) is necessary to allow interactions between this region of the protein and the RNA. The RNA double–helical region preserves the A-form structure throughout the double–helical stem, and the UUCG tetraloop is in its well characterized conformation in the presence or absence of the protein. The only significant change in RNA structure upon protein binding is a kink at the stem–loop junction, resulting in the bent appearance of the RNA in the complex (Figure 5A). The presence of this distortion is supported indirectly by the observation of significantly shifted resonances in this region of the RNA. The bend allows the interaction between helix α1 and the tetraloop to occur at the same time as the contacts between loop 2 and the RNA minor groove.
dsRBD mutagenesis in vivo
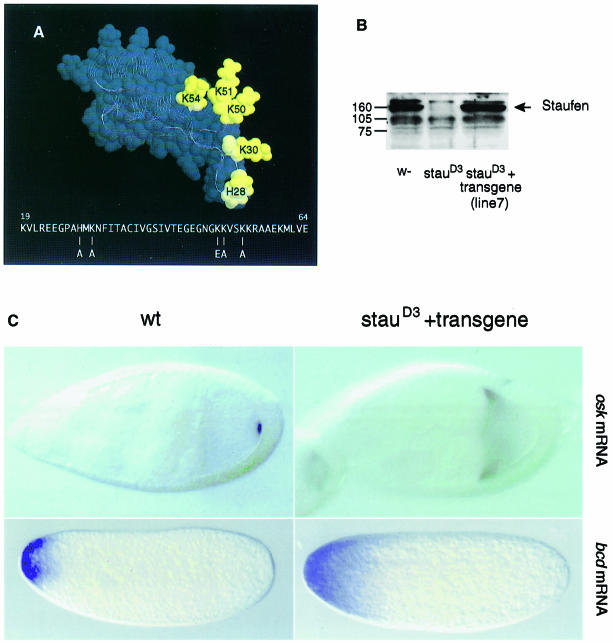
The biochemical and structural data on dsRBD3 described above provide a framework to analyse whether the RNA-binding activity of this domain is required for Staufen function. Five highly conserved basic amino acids within loop 2 and loop 4 (His28, Lys30, Lys50, Lys51 and Lys54) are required for RNA binding in vitro, and lie at the RNA–protein interface where they interact with the RNA (Figure 5A). To generate a form of domain 3 that is completely null for RNA binding, we replaced all five of these amino acids with uncharged or negatively charged residues. 1H-15N HSQC spectra of the bacterially expressed quintuple mutant dsRBD3 are very similar to that of the wild-type protein (data not shown), demonstrating that mutant and wild-type proteins adopt the same conformation. Consistent with this observation, the domain displayed normal solubility and stability when expressed in Escherichia coli, but its in vitro RNA-binding activity was abolished. The DNA encoding this mutant domain was inserted into a staufen cDNA in place of the wild-type domain, and then transformed into the Drosophila germline in a vector that directs expression of the transgene in the female ovary (Micklem et al., 1997). In control flies, a single copy of the wild-type staufen transgene completely rescues the maternal effect of a staufen null mutation, and restores the wild-type localization of both oskar and bicoid mRNAs. In contrast, 10 independent insertions of the dsRBD3 mutant construct give no rescue of the staufen phenotype. In one line that was examined in detail, the mutant Staufen protein is expressed in the female germline at the same level as the wild-type protein (Figure 7B). Nevertheless, the mutant protein does not rescue the localization of oskar mRNA to the posterior of the oocyte, nor the anchoring of bicoid mRNA at the anterior of the egg (Figure 7C), and 100% of embryos die with head defects and no abdomen or pole cells (data not shown). Thus, in the transgenic flies, oskar mRNA is not transported to the posterior of the oocyte, and bicoid mRNA fails to be anchored at the anterior of the egg, showing that the transgene does not rescue the staufen mutant phenotype. Thus, the amino acids in dsRBD3 that interact with dsRNA in vitro are required for the in vivo function of Staufen. These results demonstrate for the first time that the dsRNA-binding activity of dsRBD3 is essential for the interaction of Staufen protein with bicoid and oskar mRNAs, strongly suggesting that Staufen binds directly to these transcripts in vivo.
Fig. 7. The RNA-binding activity of Staufen dsRBD3 is required for localization of oskar and bicoid mRNAs. (A) View of the dsRBD3 structure showing the position of the five amino acids that have been mutated to abolish the RNA-binding activity of the domain. The same amino acids are highlighted in the protein–RNA complex in Figure 5A. (B) Western blot probed with an anti-Staufen antibody of ovary extracts from wild-type, staufen null mutant, and flies carrying one copy of the quintuple mutant dsRBD3 staufen transgene in a staufen null mutant background. (C) The localization of oskar mRNA at stage 9 of oogenesis and bicoid mRNA in the freshly laid egg, in wild-type and staufen mutant flies carrying one copy of the transgene.
Discussion
We have studied how the third dsRBD from Drosophila Staufen protein recognizes RNA and have described features of the dsRBD–RNA interaction that are very likely to be of general relevance to RNA recognition by all dsRBD-containing proteins. We have also shown for the first time that direct interactions between individual dsRBDs and RNA are essential for Staufen function in RNA localization and early development. These results suggest very strongly that Staufen binds directly to the oskar, bicoid and prospero 3′–UTRs to mediate the localization of these mRNAs in vivo, and describe the molecular interactions that are necessary for this to occur.
Molecular basis of the interaction of dsRBDs with dsRNA
The biochemical and structural data presented here identify the three regions of the dsRBD that mediate the binding of the domain to RNA: helix α1, loop 2 and loop 4. Mutations of amino acids in each of these regions abolish or reduce RNA binding significantly, whereas mutations in surface residues in other regions of the protein have no effect. Furthermore, the amino acids in these regions are highly conserved in Staufen homologues from Drosophila to humans, indicating that they play an essential role in the function of the domain. These results are likely to be applicable to other dsRBD-containing proteins, since mutagenesis studies have shown that analogous regions of other dsRBDs are required for interaction with RNA. For example, the RNA-binding activity of PKR is significantly reduced by mutations in the first α–helix or in the lysine-rich loop 4 of the first dsRBD of this protein (Green and Matthews, 1992; Green et al., 1995). Similarly, the RNA-binding activity of dsRBD2 of Xlrbpa is severely compromised by mutation of a histidine in loop 2 that is equivalent to His28 in Staufen dsRBD3 (Krovat and Jantsch, 1996).
The crystallographic structure of the complex between dsRBD2 of Xlrbpa and dsRNA (Ryter and Schultz, 1998) identified the same three regions of the domain that contact RNA as our NMR structure. Both structures show that loop 2 interacts with the minor groove of the RNA (Figure 6A), and loop 4 interacts with the phosphodiester backbone across the major groove from the sites of loop 2 contacts (Figure 6B). However, the two structures provide very different descriptions of the interaction between helix α1 and RNA. In the crystal structure, helix α1 interacts with the minor groove of a second RNA duplex that abuts the first RNA molecule to form a pseudo-continuous double helix. As a consequence, the Xlrbpa dsRBD covers 16 bp across the junction between the two RNA molecules. In contrast, helix α1 of Staufen dsRBD3 binds to a tetraloop that caps a 12 bp stem of perfect A-form RNA, but this interaction requires bending of the RNA at the stem–loop junction. This structural difference is significant. As shown in Figure 1, Staufen dsRBD3 would not bind the 10 bp RNA duplex used in the crystallographic studies, but binds optimally to RNA hairpins with a stem of 12 bp. This length requirement is likely to be important because RNA duplexes of 16 bp do not exist within the bicoid 3′–UTR.
Within loop 2 and loop 4, most differences between the NMR and crystallographic structures are attributable to the lower precision of the NMR structure and the different dynamic behaviour of the protein–RNA complexes in the two systems, as discussed below. However, one important difference concerns His28. In the crystal structure of the Xlrbpa–dsRNA complex, the side chain of His141 stacks on Phe145 (corresponding to His28 and Phe32 in the present numbering system) and interacts with a 2′–OH group (Ryter and Schultz, 1998). However, this interaction requires a backbone conformation inconsistent with the NMR data. In the present structure, the histidine–phenylalanine stacking interaction is not present, and the histidine–phosphate interaction we observe is instead similar to a contact reported between a phenylalanine side chain and a DNA phosphate, as observed in the structure of the P22 Arc repressor–DNA complex (Schildback et al., 1999). The phenylalanine–DNA interaction plays a prominent role in determining the specificity of recognition by modulating the structure of the protein–DNA interface. Mutation of His28 to alanine in Staufen dsRBD3 abolishes RNA binding (Figure 1B), suggesting that interactions between heteroaromatic side chains and the phosphates could play important roles in RNA recognition as well.
Specificity of the dsRBD for dsRNA
Since NMR and X-ray structures reveal different interactions between helix α1 and RNA, it seems very likely that the common interactions involving loop 2 and loop 4 account for the specificity of both domains for dsRNA. Further support for this view comes from the analysis of the N–terminal domain of bacterial ribosomal protein S5, which has a very similar fold to the dsRBD and contains many of the conserved residues that form the hydrophobic core of the domain but lacks the N–terminal α–helix found in the dsRBD (Bycroft et al., 1995). S5 interacts with helix 34 in 16S rRNA (Heilik and Noller, 1996; Davies et al., 1998). The present results suggest that S5 binds to rRNA through loops 2 and 4 alone. The ability of these two loops to discriminate between dsRNA and DNA can be attributed firstly to interactions with 2′–OH groups in the RNA minor groove, as originally described in the Xlrbpa crystal structure (Ryter and Schultz, 1998). In addition, the spacing between loop 2 and loop 4 corresponds well with the spacing and groove distances found in the A-type helix formed by dsRNA, but would not fit the B-type helix of dsDNA. Consistent with this interpretation, mutation of Phe32 abolishes RNA binding completely. Phe32 is buried between loop 2 and loop 4 (Figure 5A); the present structure suggests very strongly that its identity is essential to position these loops with respect to the RNA.
Dynamic nature of the dsRBD–dsRNA interaction
An important aspect of dsRBD interaction revealed by the NMR analysis is the dynamic character of the interface. Broadening of side chain and backbone resonances, the results reported in Figure 4 and other relaxation parameters determined in the course of this study all demonstrate that loop 2, loop 4 and the N–terminus of helix α1 retain significant conformational flexibility in the protein–RNA complex. When the NMR structures are compared, amino acid side chains from loop 2 and loop 4 are found to interact with different acidic groups on the RNA. Electrostatic interactions similar to those observed in the crystal structure can be observed in all structures that satisfy the NMR data (Figure 6A and B), but the same protein side chain sometimes interacts with different 2′–OH or phosphate groups. This description of the intermolecular interface provided by the NMR data is entirely consistent with the observation of a disordered loop 2 interface in the second of the two Xlrbpa dsRBD2 molecules in the crystallographic asymmetric unit. This observation suggests that interactions mediated by loop 2 are dynamic in the crystal as well.
The absence in the dsRBD–RNA structure of significant reorganizations of the RNA or protein structures was a surprise, since induced fit has so far been a nearly universal feature of RNA recognition by proteins and small molecules (Varani, 1997). Furthermore, the dsRBD–RNA complex does not contain a tightly packed intermolecular interface. In both respects, dsRBD–RNA recognition differs substantially from the paradigm for RNA recognition established by human U1A protein (Oubridge et al., 1994; Allain et al., 1996). Formation of the U1A–RNA complex requires significant rearrangements in RNA and protein structures, resulting in a tightly packed intermolecular interface. Staufen dsRBD3 sits instead on the edge of the RNA double helix and interacts with the RNA sugar–phosphate backbone, without making intimate contacts with the bases. The absence of direct contacts with the RNA bases, the lack of a requirement for distortion in RNA structure and the residual conformational flexibility present at the RNA–protein interface all contribute to the lack of sequence specificity in recognition of dsRNA.
The significance of the interaction between helix α1 and the single-stranded loop
The unexpected observation of interactions between helix α1 and the single-stranded loop raises the question of whether these are physiologically significant. The alanine-scanning data demonstrate that surface-exposed residues within helix α1 make essential contributions to RNA binding. Mutation of Gln4, Glu7 and Arg12 abolishes binding, while substitution of Ser3 reduces binding significantly. Furthermore, Ser3, Gln4, Glu7, Lys11 and Arg12 have been conserved during the evolution of Staufen dsRBD3, suggesting that these exposed amino acids play an important role in Staufen function. This hypothesis could be addressed by mutating these residues in full-length Staufen. The observation in our structure of well defined intermolecular interactions mediated by these residues (Figure 6C) raises the possibility that these amino acids play a critical role in RNA recognition.
Intriguing clues as to the diverse functional role of helix α1 compared with loops 2 and 4 are provided by the extension of the phylogenetic comparison with other dsRBDs. The key residues within loops 2 and 4 (His28, Lys/Arg30, Lys50, Lys51 and Lys54) are highly conserved across species in Staufen dsRBD3 (Figure 3), as well as Staufen dsRBD1, a second domain of the protein that binds dsRNA (J.Adams, S.Grünert and D.St Johnston, unpublished results). In contrast, interfacial residues from helix α1 are highly conserved for each domain when different species are examined, but are significantly divergent when the two domains are compared with each other, even within the same species. dsRBD1 contains the conserved Glu7, but has serine–cysteine substitution at position 3, a conserved leucine instead of Glu4, phenylalanine or tyrosine in place of Lys11, and a conserved glutamine in place of Arg12. Similarly, Xlrbpa dsRBD2 contains the same key residues in loops 2 and 4 as found in Staufen dsRBD1 and dsRBD3, but differs from both Staufen domains in four out of five of the exposed positions in helix α1.
The preceding observations indicate that the identity of residues within helix α1 is conserved and domain specific, raising the possibility that different dsRBD domains can contribute to specificity in Staufen–RNA recognition by forming different helix α1–loop interactions. It is very likely that multiple domains from Staufen bind bicoid 3′–UTR, but this does not rule out at all the possibility that helix α1 contributes to specificity. The data presented in Figure 7 demonstrate that dsRBD3–RNA interactions are critical for Staufen function. However, the RNA used in the present study was optimized for affinity in order to facilitate structural studies, and does not correspond to any of the stem–loops within bicoid 3′–UTR. Therefore, we cannot yet identify the binding site for Staufen dsRBD3 in bicoid 3′–UTR.
Support for the importance of helix α1–loop interactions in dsRBD proteins is provided by yeast Rnt1 protein, the eukaryotic RNase III. This dsRBD-containing enzyme cleaves pre-rRNA and a set of snoRNA precursors at sites defined by a conserved hairpin loop (Elela et al., 1996; Chanfreau et al., 1998). Mutation of the AG sequence within the loop reduces Rnt1 binding and severely affects RNA processing. The site of cleavage is always separated from the apical hairpin loop by 14–16 bp, often interrupted by internal loops or bulges (Chanfreau et al., 1998). This separation is just a few base pairs longer than the footprint of Staufen dsRBD3 on our stem–loop structure (12 bp). Although it is not yet possible to exclude a role for the catalytic domain in Rnt1–substrate recognition, it is tempting to suggest that interactions between helix α1 of the Rnt1 dsRBD and the tetraloop, analogous to those described here, could be important in defining the substrate specificity of Rnt1.
The RNA-binding activity of dsRBD proteins has been defined by in vitro studies as non-sequence-specific recognition of perfect dsRNA substrates. However, some of the most extensively investigated dsRBD-containing proteins, such as Staufen, Rnt1 and PKR, regulate the activity of very specific RNAs in vivo. The structural and phylogenetic analysis presented here suggest that domain-specific interactions between helix α1 and single-stranded RNA loops could modulate the specificity of individual dsRBD domains and provide selectivity in the recognition of cellular RNAs.
Materials and methods
Protein and RNA preparation
The dsRBD3 fragment (residues 579–646 of Drosophila Staufen protein) and the various mutant proteins described in the present report were expressed in E.coli using appropriate isotope-labelled nutrients and purified as described (Bycroft et al., 1995). The RNA used in NMR studies was: 5′–GGACAGCUGUCC(CUUCGG)GGACAGCUGUCC–3′ (the tetraloop sequence and flanking base pair is within parentheses). Unlabelled and isotopically labelled RNA oligonucleotides were synthesized in vitro using T7 RNA polymerase and synthetic DNA templates (Price et al., 1998).
Mutagenesis of dsRBD3
Mutagenesis was performed using a two-step PCR mutagenesis protocol. Briefly, we used two primers (sequences available upon request) flanking the dsRBD3 with BamHI and EcoRI restriction sites, which enabled cloning of the dsRBD3-derived PCR fragments in-frame with GST into pGEX2–T (Promega). For each mutation, we synthesized a third mutagenic primer of the desired sequence, including silent mutations to create novel restriction sites in the mutated PCR product. PCRs were performed as described (Bycroft et al., 1995) using Pfu polymerase (Stratagene). The resulting PCR products were subcloned into pGEX2–T and minipreparations were screened for the newly introduced restriction sites. The presence of the desired mutations was verified by sequencing.
RNA-binding assay
North-western binding assays were performed as described (St Johnston et al., 1992). Radiolabelled short hairpins were synthesized by in vitro transcription of synthetic oligonucleotides. Approximately 100 000 c.p.m./ml were used for each probe.
Identification of staufen homologues in other species
staufen homologues from Drosophila virilis and Musca domestica were identified by screening genomic libraries at low stringency with probes derived from the five dsRBDs of the Drosophila melanogaster gene; positive clones were sequenced using the μ transposon strategy (Strathmann et al., 1991). A human staufen homologue was identified from the homology of EST HFBDQ83 (DDBJ/EMBL/GenBank accession Nos T06248 and T06429) to Drosophila staufen; the corresponding cDNA clone was sequenced in its entirety. Four cDNAs encoding a mouse homologue of staufen were identified by screening a 7.7 days post-conception mouse embryonic cDNA library with the human cDNA HFBDQ83. The analysis of the sequence of these homologues will be reported in detail elsewhere (D.R.Micklem, J.Adams, S.Grünert and D.St Johnston, in preparation).
Generation of mutant staufen transgenic lines
Five mutations were introduced into dsRBD3 in a full-length staufen cDNA clone by performing two consecutive rounds of mutagenesis as described in the supplementary material (available in The EMBO Journal Online). An XhoI–MluI fragment containing the mutated dsRBD3 was cloned into the wild-type staufen cDNA, and this was then inserted into transformation vector D277 as described (Schuldt et al., 1998). This produced a construct in which the female germline-specific α4–tubulin promoter drives the expression of a fusion protein that contains amino acids 1–9 of α4–tubulin, a 16 amino acid myc epitope and amino acids 18–1026 of Staufen. This construct was introduced into the germline of w–; cn stauD3 sp/Cy O flies by P element-mediated transformation (Rubin and Spradling, 1982). Multiple independent insertions of the transgene were tested for their ability to rescue the staufen maternal effect phenotype. This was done by performing cuticle preparations on the progeny of stauD3 homozygous females that carry one copy of the transgene, and by examining the localization of bicoid and oskar mRNAs in the ovaries and eggs of these females by in situ hybridization. The expression of the mutant protein was monitored by performing Western blots on ovary extracts of females of the same genotype, and by staining with a rabbit anti-Staufen antibody (St Johnston et al., 1991).
NMR
NMR spectra were recorded on a Bruker DMX-600 spectrometer. Several RNA–protein samples were prepared: [15N]dsRBD3 and [15N-13C]dsRBD3 bound to unlabelled RNA and 15N-13C-labelled RNA with unlabelled dsRBD3. Samples were ∼1 mM in each component; the RNA to protein ratio was adjusted by integrating the intensity of well resolved protein and RNA resonances. An extensive set of two- and three-dimensional NMR spectra was recorded on free protein and RNA components and on the different preparations of RNA–protein complexes. A thorough description of methodological aspects of this work will be presented elsewhere. In total, 98% of protein backbone resonances, 70% of side chain resonances and >95% of all RNA resonances were assigned unambiguously in the complex. Assignments of free and bound RNA spectra were obtained by analysing an extensive set of two- and three-dimensional NMR spectra, as described previously (Varani et al., 1996). Heteronuclear 1H-15N NOE, T1 and T2 relaxation times for dsRBD3 backbone amide resonances were recorded and analysed essentially as reported (Farrow et al., 1994). Partially oriented samples for the extraction of NH and CH bond orientations were prepared by mixing samples of the complex with appropriately labelled phospholipid solutions, as described previously (Tjandra and Bax, 1997; Bayer et al., 1999). Residual dipolar couplings and the orientation tensor were obtained by a variational procedure (Bayer et al., 1999).
Structure determination
Distance constraints for the RNA and protein components of the complex were obtained by analysing three-dimensional 13C- and 15N-edited NOESY spectra recorded at 100 ms mixing time and two-dimensional NOESY spectra recorded at mixing times of 50 and 100 ms. The experimental constraints were generated and interpreted exactly as in the determination of the U1A protein–RNA complex (Howe et al., 1998). Intermolecular NOE interactions were identified in 1/2× filtered NOESY spectra (Otting and Wüthrich, 1990). Ten NOEs could be identified unambiguously and were used in the structure calculation; these are listed in the supplementary material online. Additional NOE cross-peaks involving protein side chains and RNA sugar resonances are observable and could often be assigned to specific residue types (e.g. H2′, H3′ or H4′), but could not be assigned unambiguously to individual RNA nucleotides or protein side chains due to spectral overlap.
The structure of the protein–RNA complex was calculated using an X-PLOR-based simulated annealing protocol optimized for the calculation of the structure of the U1A complex (Howe et al., 1998). The use of residual dipolar couplings in the refinement step necessitated the introduction of a modified refinement protocol (Tjandra et al., 1997; Bayer et al., 1999) (see supplementary material online). Thirty-six converged and were clearly identified from a significant difference in total energy from other structures (Howe et al., 1998), and were energy minimized by introducing the electrostatic component of the potential. Differences between structures calculated before and after minimization were smaller than the uncertainty in the structures themselves. Statistics for the experimental constraints and structural statistics are reported in Table I.
Supplementary data
Supplementary data to this paper are available in The EMBO Journal Online.
Acknowledgments
Acknowledgements
It is a pleasure to thank Dr Peter Bayer for help in measurement of residual dipolar couplings and their incorporation in the structure determination protocol, and Lisa Elphick for help in generating the transgenic Drosophila lines. J.A. was supported by a Boehringer Ingelheim studentship; D.R.M. by a Wellcome Trust Prize studentship; D.StJ. and S.G. by the Wellcome Trust; and A.R. by an EU training fellowship and by the MRC.
References
- Allain F.H.-T., Gubser, C.C., Howe, P.W.A., Nagai, K., Neuhaus, D. and Varani, G. (1996) Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature, 380, 646–650. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Hurst, S.R. and Singer, J.D. (1994) Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol., 4, 301–314. [DOI] [PubMed] [Google Scholar]
- Bayer P., Varani, L. and Varani, G. (1999) Refinement of the structure of protein–RNA complexes by residual dipolar coupling analysis. J. Biomol. NMR, 14, 149–155. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C. and Cech, T.R. (1996) Minor-groove recognition of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry, 35, 9983–9994. [DOI] [PubMed] [Google Scholar]
- Broadus J., Fuestenberg, S. and Doe, C.Q. (1998) Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature, 391, 792–795. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Grünert, S., Murzin, A.G., Proctor, M. and St Johnston, D. (1995) NMR solution structure of a dsRNA binding domain from Drosophila Staufen protein reveals homology to the N–terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Legrain, P. and Jacquier, A. (1998) Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Clarke P.A. and Matthews, M.B. (1995) Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA, 1, 7–20. [PMC free article] [PubMed] [Google Scholar]
- Davies C., Bussiere, D.E., Golden, B.L., Ramakrishnan, V. and White, S.W. (1998) Ribosomal proteins S5 and L6; high-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J. Mol. Biol., 279, 873–888. [DOI] [PubMed] [Google Scholar]
- Elela S.A., Igel, H. and Ares, M.J. (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson, L.K. and Lehmann, R. (1991) oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell, 66, 37–50. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., et al. (1994) Backbone dynamics of a free and a phosphopeptide complexed Src homology 2 domain studied by 15N relaxation. Biochemistry, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick, L., Nüsslein-Volhard, C. and St Johnston, D. (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell, 79, 1221–1232. [DOI] [PubMed] [Google Scholar]
- Gibson T.J. and Thompson, J. (1994) Detection of dsRBA-binding domains in the RNA helicase A and Drosophila Maleless: implications for monomeric RNA helicases. Nucleic Acids Res., 22, 2552–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S.R. and Matthews, M.B. (1992) Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev., 6, 2478–2490. [DOI] [PubMed] [Google Scholar]
- Green S.R., Manche, L. and Mathews, M.B. (1995) Two functionally distinct RNA-binding motifs in the regulatory domain of the protein kinase DAI. Mol. Cell. Biol., 15, 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilik G.M. and Noller, H.F. (1996) Site-directed hydroxyl radical probing of the rRNA neighborhood of ribosomal protein S5. Science, 272, 1659–1662. [DOI] [PubMed] [Google Scholar]
- Howe P.W.A., Allain, F.H.-T., Varani, G. and Neuhaus, D. (1998) Determination of the NMR structure of the complex between U1A protein and its RNA polyadenylation inhibition element. J. Biomol. NMR, 11, 59–84. [DOI] [PubMed] [Google Scholar]
- Kharrat A., Macias, M.J., Gibson, T.J., Nilges, M. and Pastore, A. (1995) Structure of the dsRNA binding domain of E.coli RNase III. EMBO J., 14, 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Smith, J.L. and Macdonald, P.M. (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell, 66, 23–35. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Webster, P., Smith, J. and MacDonald, P. (1993) Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development, 119, 169–178. [DOI] [PubMed] [Google Scholar]
- Krovat B.C. and Jantsch, M.F. (1996) Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J. Biol. Chem., 271, 28112–28119. [DOI] [PubMed] [Google Scholar]
- MacDonald P.M. and Struhl,G. (1988) Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature, 336, 595–598. [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Dasgupta, R., Elliott, H., Gergely, F., Davidson, C., Brand, A., González-Reyes, A. and St Johnston, D. (1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol., 7, 468–478. [DOI] [PubMed] [Google Scholar]
- Nanduri S., Carpick, B.W., Yang, Y., Williams, B.R.G. and Qin, J. (1998) Structure of the double stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J., 17, 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G. and Wüthrich, K. (1990) Heteronuclear filters in two-dimensional [1H-1H]-NMR spectroscopy: combined use with isotope labeling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys., 23, 39–96. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito, N., Evans, P.R., Teo, C.-H. and Nagai, K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Price S.R., Oubridge,C., Varani,G. and Nagai,K. (1998) Preparation of RNA–protein complexes for X-ray crystallography and NMR. In Smith,C. (ed.), RNA–Protein Interactions: A Practical Approach. Oxford University Press, Oxford, UK, pp. 48–74. [Google Scholar]
- Rubin G. and Spradling, A.C. (1982) Genetic transformation of Drosophila with transposable elements vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz, S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildback J.F., Karzai, A.W., Raumann, B.E. and Sauer, R.T. (1999) Origins of DNA-binding specificity: role of protein contacts with the DNA backbone. Proc. Natl Acad. Sci. USA, 96, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmedt C., Green, S.R., Manche, L., Taylor, D.R., Ma, Y. and Mathews, M.B. (1995) Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J. Mol. Biol., 249, 29–44. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams, J.H.J., Davidson, C.M., Micklem, D.R., Haseloff, J., St Johnston, D. and Brand, A.H. (1998) Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. (1995) The intracellular localization of messenger RNAs. Cell, 81, 161–170. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Driever, W., Berleth, T., Richstein, S. and Nüsslein-Volhard, C. (1989) Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development Suppl., 107, 13–19. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Beuchle, D. and Nüsslein-Volhard, C. (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell, 66, 51–63. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown, N.H., Gall, J.G. and Jantsch, M. (1992) A conserved double-stranded RNA binding domain. Proc. Natl Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann M., Hamilton, B.A., Mayeda, C.A., Simon, M.I., Meyerowitz, E.M. and Palazzolo, A. (1991) Transposon-facilitated DNA sequencing. Proc. Natl Acad. Sci. USA, 88, 1247–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N. and Bax, A. (1997) Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science, 278, 1111–1114. [DOI] [PubMed] [Google Scholar]
- Varani G. (1997) RNA–protein intermolecular recognition. Acc. Chem. Res., 30, 189–195. [Google Scholar]
- Varani G., Aboul-ela, F. and Allain, F.H.-T. (1996) NMR investigations of RNA structure. Prog. NMR Spectrosc., 29, 51–127. [Google Scholar]